
园艺学报 ›› 2021, Vol. 48 ›› Issue (5): 849-859.doi: 10.16420/j.issn.0513-353x.2020-0895
• 研究论文 • 下一篇
姚富文1,*, 王枚阁1,*, 宋春晖1, 宋尚伟1, 焦健1, 王苗苗1, 王昆2, 白团辉1,**( ), 郑先波1,**(
), 郑先波1,**( )
)
收稿日期:2020-12-29
修回日期:2021-04-08
出版日期:2021-05-25
发布日期:2021-06-07
通讯作者:
白团辉,郑先波
E-mail:tuanhuibai88@163.com;xianboz@163.com
作者简介:*共同第一作者
基金资助:
YAO Fuwen1,*, WANG Meige1,*, SONG Chunhui1, SONG Shangwei1, JIAO Jian1, WANG Miaomiao1, WANG Kun2, BAI Tuanhui1,**( ), ZHENG Xianbo1,**(
), ZHENG Xianbo1,**( )
)
Received:2020-12-29
Revised:2021-04-08
Online:2021-05-25
Published:2021-06-07
Contact:
BAI Tuanhui,ZHENG Xianbo
E-mail:tuanhuibai88@163.com;xianboz@163.com
摘要:
为了探究热激蛋白(HSP90)基因家族成员在苹果(Malus × domesticaBorkh.)耐热中的作用,利用生物信息学方法对苹果HSP90基因家族成员进行鉴定,并分析其在高温胁迫下和不同组织中的表达规律。结果表明,苹果HSP90基因家族有11个成员,分布于10条染色体上,其氨基酸序列大小介于104 ~ 818 aa,蛋白质分子质量介于11.81 ~ 93.58 kD。转录组分析显示,11个MdHSP90家族成员中,有4个基因,MdHSP90-1、MdHSP90-3、MdHSP90-5和MdHSP90-11在高温胁迫下显著上调;亚细胞定位显示这4个基因中前三者定位在细胞膜与细胞核上,而后者定位在细胞核上;进一步通过qRT-PCR分析发现这4个基因在两种苹果砧木平邑甜茶(Malus hupehensis)和变叶海棠(Malus toringoides)中的表达趋势基本一致,其表达量在高温胁迫早期大幅升高,在中后期有所下降;不同苹果组织中均能检测到这4个基因的表达,高温处理可不同程度地激活这些基因的表达,表明它们可能在苹果耐热胁迫中具有重要作用。
中图分类号:
姚富文, 王枚阁, 宋春晖, 宋尚伟, 焦健, 王苗苗, 王昆, 白团辉, 郑先波. 苹果HSP90家族基因鉴定及高温胁迫下的表达分析[J]. 园艺学报, 2021, 48(5): 849-859.
YAO Fuwen, WANG Meige, SONG Chunhui, SONG Shangwei, JIAO Jian, WANG Miaomiao, WANG Kun, BAI Tuanhui, ZHENG Xianbo. Identification and Expression Analysis of HSP90 Gene Family Under High Temperature Stress in Apple[J]. Acta Horticulturae Sinica, 2021, 48(5): 849-859.
| 用途Useage | 基因 Gene | 引物(5′-3′)Primer |
|---|---|---|
| 实时荧光定量qRT-PCR | MdHSP90-1 | F:CGTCGATTCGGATGATTTGC;R:TGGCTCTGTTCTGGCTGTCC |
| MdHSP90-3 | F:GGAAATCAGCGACGACGAAG;R:CAAGGTGCTCCTCCCAGTCA | |
| MdHSP90-5 | F:ATGTTGCTTCTGACAGCGTGAC;R:CCATATTCCTGCTTAATCTCCTTTT | |
| MdHSP90-11 | F:CGTATTGCTCCGTTGCTTAG;R:AGTTTCTCCAGGAATGGTGTATT | |
| Actin | F:GGATTTGCTGGTGATGATGCT;R:AGTTGCTCACTATGCCGTGC | |
| 亚细胞定位 Subcellular location | GFPHSP90-1 | F:GTGGATCCAAAGAATTCATGGCCGACGTTCAGATGCAC |
| R:CTCCTTTACCCATGAATTCTCAAAATTATATTAACATTATCGAG | ||
| GFPHSP90-3 | F:GTGGATCCAAAGAATTCCCGATGTCAGCATGATTGG | |
| R:CTCCTTTACCCATGAATTCAAAAACCCGAAACATCCACAAACTC | ||
| GFPHSP90-9 | F:GTGGATCCAAAGAATTCATGCACAGGCTCCCAC | |
| R:CTCCTTTACCCATGAATTCTTTCTGGCTGCC | ||
| GFPHSP90-11 | F:GTGGATCCAAAGAATTCATGCACAGGCTCCCACGACGCTCCG | |
| R:CTCCTTTACCCATGAATTCTTTCTGACTGCCG |
表1 qRT-PCR和亚细胞定位引物序列
Table 1 Primer sequences of qRT-PCR and subcellular location
| 用途Useage | 基因 Gene | 引物(5′-3′)Primer |
|---|---|---|
| 实时荧光定量qRT-PCR | MdHSP90-1 | F:CGTCGATTCGGATGATTTGC;R:TGGCTCTGTTCTGGCTGTCC |
| MdHSP90-3 | F:GGAAATCAGCGACGACGAAG;R:CAAGGTGCTCCTCCCAGTCA | |
| MdHSP90-5 | F:ATGTTGCTTCTGACAGCGTGAC;R:CCATATTCCTGCTTAATCTCCTTTT | |
| MdHSP90-11 | F:CGTATTGCTCCGTTGCTTAG;R:AGTTTCTCCAGGAATGGTGTATT | |
| Actin | F:GGATTTGCTGGTGATGATGCT;R:AGTTGCTCACTATGCCGTGC | |
| 亚细胞定位 Subcellular location | GFPHSP90-1 | F:GTGGATCCAAAGAATTCATGGCCGACGTTCAGATGCAC |
| R:CTCCTTTACCCATGAATTCTCAAAATTATATTAACATTATCGAG | ||
| GFPHSP90-3 | F:GTGGATCCAAAGAATTCCCGATGTCAGCATGATTGG | |
| R:CTCCTTTACCCATGAATTCAAAAACCCGAAACATCCACAAACTC | ||
| GFPHSP90-9 | F:GTGGATCCAAAGAATTCATGCACAGGCTCCCAC | |
| R:CTCCTTTACCCATGAATTCTTTCTGGCTGCC | ||
| GFPHSP90-11 | F:GTGGATCCAAAGAATTCATGCACAGGCTCCCACGACGCTCCG | |
| R:CTCCTTTACCCATGAATTCTTTCTGACTGCCG |
| 基因名称 Gene name | 序列ID Sequence ID | 染色体 Chr | 基因组位置 Genomic location | 开放阅读框/bp ORF | 氨基酸/aa Amino acid | 蛋白质分子质量/kD mw | 等电点 pI |
|---|---|---|---|---|---|---|---|
| MdHSP90-1 | MD01G1208700 | 1 | 30 315 406 ~ 30 318 680 | 2 112 | 703 | 80.77 | 5.00 |
| MdHSP90-2 | MD03G1036600 | 3 | 2 907 372 ~ 2 910 570 | 2 100 | 699 | 80.21 | 4.95 |
| MdHSP90-3 | MD07G1279200 | 7 | 34 367 607 ~ 34 369 506 | 1 749 | 582 | 67.39 | 5.06 |
| MdHSP90-4 | MD08G1011200 | 8 | 864 457 ~ 869 106 | 2 547 | 818 | 93.59 | 4.85 |
| MdHSP90-5 | MD09G1122200 | 9 | 9 421 880 ~ 9 427 733 | 2 430 | 809 | 91.89 | 5.29 |
| MdHSP90-6 | MD11G1037400 | 11 | 3 204 284 ~ 3 207 461 | 2 100 | 699 | 80.04 | 4.95 |
| MdHSP90-7 | MD11G1152300 | 11 | 14 614 376 ~ 14 614 690 | 315 | 104 | 11.81 | 4.16 |
| MdHSP90-8 | MD13G1055300 | 13 | 3 867 643 ~ 3 871 210 | 1 143 | 380 | 43.56 | 4.89 |
| MdHSP90-9 | MD14G1110600 | 14 | 17 566 378 ~ 17 567 467 | 930 | 309 | 36.35 | 9.40 |
| MdHSP90-10 | MD16G1054100 | 16 | 3 825 148 ~ 3 830 828 | 2 385 | 794 | 89.67 | 5.02 |
| MdHSP90-11 | MD17G1113200 | 17 | 9 690 370 ~ 9 696 630 | 2 421 | 806 | 91.69 | 5.34 |
表2 苹果HSP90基因家族成员的理化性质
Table 2 Physical and chemical properties of HSP90 gene family in apple
| 基因名称 Gene name | 序列ID Sequence ID | 染色体 Chr | 基因组位置 Genomic location | 开放阅读框/bp ORF | 氨基酸/aa Amino acid | 蛋白质分子质量/kD mw | 等电点 pI |
|---|---|---|---|---|---|---|---|
| MdHSP90-1 | MD01G1208700 | 1 | 30 315 406 ~ 30 318 680 | 2 112 | 703 | 80.77 | 5.00 |
| MdHSP90-2 | MD03G1036600 | 3 | 2 907 372 ~ 2 910 570 | 2 100 | 699 | 80.21 | 4.95 |
| MdHSP90-3 | MD07G1279200 | 7 | 34 367 607 ~ 34 369 506 | 1 749 | 582 | 67.39 | 5.06 |
| MdHSP90-4 | MD08G1011200 | 8 | 864 457 ~ 869 106 | 2 547 | 818 | 93.59 | 4.85 |
| MdHSP90-5 | MD09G1122200 | 9 | 9 421 880 ~ 9 427 733 | 2 430 | 809 | 91.89 | 5.29 |
| MdHSP90-6 | MD11G1037400 | 11 | 3 204 284 ~ 3 207 461 | 2 100 | 699 | 80.04 | 4.95 |
| MdHSP90-7 | MD11G1152300 | 11 | 14 614 376 ~ 14 614 690 | 315 | 104 | 11.81 | 4.16 |
| MdHSP90-8 | MD13G1055300 | 13 | 3 867 643 ~ 3 871 210 | 1 143 | 380 | 43.56 | 4.89 |
| MdHSP90-9 | MD14G1110600 | 14 | 17 566 378 ~ 17 567 467 | 930 | 309 | 36.35 | 9.40 |
| MdHSP90-10 | MD16G1054100 | 16 | 3 825 148 ~ 3 830 828 | 2 385 | 794 | 89.67 | 5.02 |
| MdHSP90-11 | MD17G1113200 | 17 | 9 690 370 ~ 9 696 630 | 2 421 | 806 | 91.69 | 5.34 |
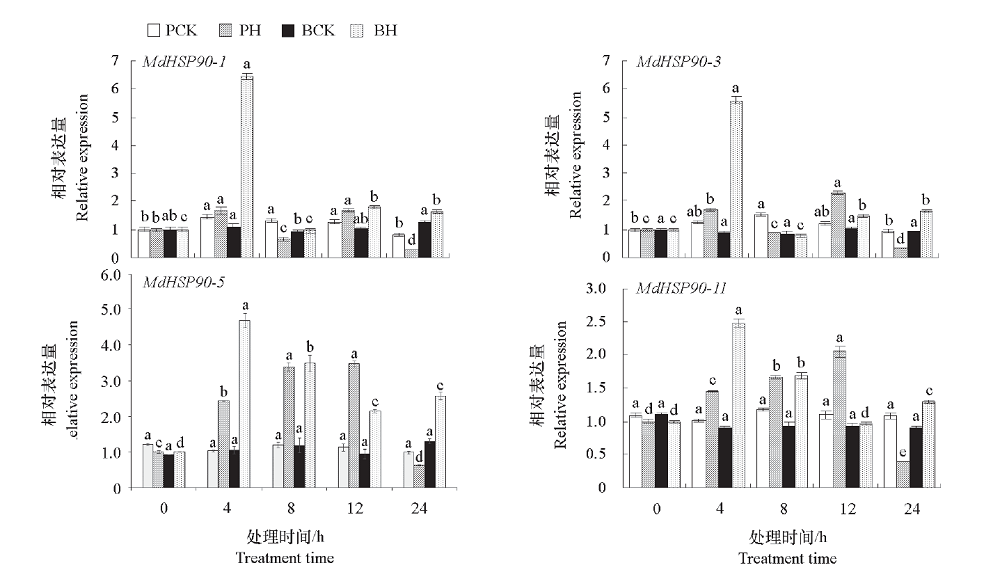
图4 苹果HSP90基因在高温胁迫下的表达分析 PCK:平邑甜茶对照;PH:平邑甜茶高温;BCK:变叶海棠对照;BH:变叶海棠高温。 不同小写字母表示同一种砧木在不同胁迫时间表达量存在显著差异(P < 0.05)。
Fig. 4 Expression analysis of HSP90 genes in apple under high temperature stress PCK:Malus hupehensis(control);PH:Malus hupehensis(high temperature);BCK:Malus toringoides(Control);BH:Malus toringoides(high temperature). Different lowercase letters in each bar indicate significant differences at different stress times in the same rootstock(P < 0.05).

图 5 苹果HSP90基因的组织表达分析 不同小写字母表示同一种砧木不同组织之间表达量存在显著差异(P < 0.05)。
Fig. 5 Tissue expression analysis of HSP90 genes in apple In the same apple rootstock,different lowercase letters indicated significant differences in the expression levels among different tissues(P < 0.05).
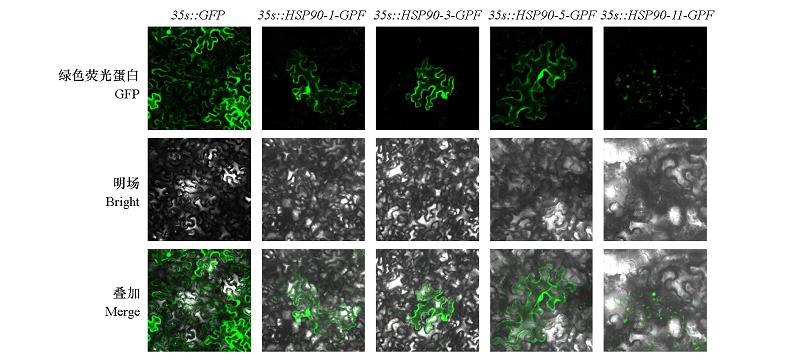
图 6 MdHSP90-1、MdHSP90-3、MdHSP90-5和MdHSP90-11蛋白在烟草细胞中的亚细胞定位
Fig. 6 Subcellular localization of MdHSP90-1、MdHSP90-3、MdHSP90-5 and MdHSP90-11proteins in tobacco
| [1] |
Al-Whaibi M H. 2011. Plant heat-shock proteins:a mini review. Journal of King Saud University science, 23:139-150.
doi: 10.1016/j.jksus.2010.06.022 URL |
| [2] | Asea A A A, Kaur P, Calderwood S K. 2016. Heat shock proteins and plants//Heat shock proteins. Switzerland: Springer International Publishing. |
| [3] |
Banilas G, Korkas E, Englezos V, Nisiotou A A, Hatzopoulos P. 2012. Genome wide analysis of the heat shock protein 90 gene family in grapevine (Vitis vinifera L.). Australian Journal of Grape and Wine Research, 18:29-38.
doi: 10.1111/ajgw.2012.18.issue-1 URL |
| [4] |
Botton A, Eccher G, Forcato C, Ferrarini A, Begheldo M, Zermiani M, Ramina A. 2011. Signalling pathways mediating the induction of apple fruitlet abscission. Plant Physiology, 155(1):185-208.
doi: 10.1104/pp.110.165779 URL |
| [5] |
Chen C, Chen H, Zhang Y, Thomas H R, Xia R. 2020. TBtools:an integrative toolkit developed for interactive analyses of big biological data. Molecular Plant, 13:8.
doi: 10.1016/j.molp.2019.12.009 URL |
| [6] | El-Gebali J, Mistry A, Bateman S R, Eddy A, Luciani S C, Potter M, Quresh L J, Richardson G A, Salazar A, Smart E L L, Sonnhammer L, Hirsh L, Paladin D, Piovesan S C E, Tosatto R D. 2019. The Pfam protein families database in 2019. Nucleic Acids Research, 47:427-432. |
| [7] |
Felsheim R F, Das A. 1992. Structure and expression of a heat-shock protein 83 gene of Pharbitis nil. Plant Physiology, 100:1764-1771.
doi: 10.1104/pp.100.4.1764 URL |
| [8] | Gupta S C, Sharma A, Mishra M, Mishra R K, Chowdhuri D K. 2010. Heat shock proteins in toxicology:how close and how far? Life Science, 86(11-12):377-384. |
| [9] |
Hsu A L, Murphy C T, Kenyon C. 2003. Regulation of aging and age-related disease by DAF-16 and heat-shock factor. Science, 300(5622):1142.
doi: 10.1126/science.1083701 URL |
| [10] |
Hu W, Hu G, Han B. 2009. Genome-wide survey and expression profiling of heat shock proteins and heat shock factors revealed overlapped and stress specific response under abiotic stresses in rice. Plant Science, 176(4):583-590.
doi: 10.1016/j.plantsci.2009.01.016 URL |
| [11] |
Ji X R, Yu Y H, Ni P Y, Zhang G H, Guo D L. 2019. Genome-wide identification of small heats hock protein(HSP20)gene family in grape and expression profile during berry development. BMC Plant Biology, 19:433.
doi: 10.1186/s12870-019-2031-4 URL |
| [12] |
Jin Z, Chandrasekaran U, Liu A. 2014. Genome-wide analysis of the Dof transcription factors in castor bean(Ricinus communis L.). Genes and Genomics, 36(4):527-537.
doi: 10.1007/s13258-014-0189-6 URL |
| [13] |
Krishna P, Gloor G. 2001. The HSP90 family of proteins in Arabidopsis thaliana. Cell Stress Chaperones, 6(3):238-246.
doi: 10.1379/1466-1268(2001)006<0238:THFOPI>2.0.CO;2 URL |
| [14] | Liu Da-li. 2006. Cloning and characterization of rHsp90 gene in rice(Oryza sativa L.)under stress[Ph. D. Dissertation]. Habin:Northeast Forestry University. (in Chinese) |
| 刘大丽. 2006. 逆境胁迫下水稻(Oryza sativa L.) rHsp90基因的克隆及功能分析[硕士论文]. 哈尔滨:东北林业大学. | |
| [15] | Liu Ling-ling. 2012. Sequence characterization and functional analysis of drought-tolerant genes ZmHSP90-1 and ZmMYB-R1 in maize[Ph. D. Dissertation]. Harbin:Northeast Agricultural University. (in Chinese) |
| 刘玲玲. 2012. 玉米耐旱相关基因ZmHSP90-1和ZmMYB-R1的序列特征分析与功能验证[硕士论文]. 哈尔滨:东北农业大学. | |
| [16] | Liu Yun-fei, Wan Jian-hong, Yang Yue-jian, Wei Yan-ping, Li Zhi-miao, Ye Qing-jing, Wang Rong-qing, Ruan Mei-ying, Yao Zhu-ping, Zhou Guo-zhi. 2014. Genome-wide identification and analysis of heat shock protein 90 in tomato. Hereditas, 36(10):1043-1052. (in Chinese) |
| 刘云飞, 万红建, 杨悦俭, 韦艳萍, 李志邈, 叶青静, 王荣青, 阮美颖, 姚祝平, 周国治. 2014. 番茄热激蛋白90的全基因组鉴定及分析. 遗传, 36(10):1043-1052. | |
| [17] |
Livak K J, Schmittgen T D. 2001. Analysis of relative gene expression data using real-time quantitative PCR and the 2-∆∆CT method. Methods, 25(4):402-408.
pmid: 11846609 |
| [18] |
Lescot M, Dhais P, Thijs G, Marchal K, Moreau Y, Van P, Rouz P, Rombauts S. 2002. PlantCARE,a database of plant cis-acting regulatory elements and a portal to tools for in silico analysis of promoter sequences. Nucleic Acids Research, 30(1):325-327.
doi: 10.1093/nar/30.1.325 URL |
| [19] |
Mukhopadhyay I, Nazir A, Saxena D K, Chowdhuri D K. 2003. Heat shock response:HSP70 in environmental monitoring. Journal of Biochemical and Molecular Toxicology, 17(5):249-54.
doi: 10.1002/(ISSN)1099-0461 URL |
| [20] |
Pearl L H, Prodromou C. 2006. Structure and mechanism of the Hsp90 molecular chaperone machinery. Annual Review of Biochemistry, 75(75):271.
doi: 10.1146/annurev.biochem.75.103004.142738 URL |
| [21] | Prasinos C, Krampis K, Samakovli D, Hatzopoulos P. 2005. Tight regulation of expression of two Arabidopsis cytosolic HSP90 genes during embryo development. Journal of Experimental Biology, 56:633-644. |
| [22] |
Prodromou C. 1997. Identification and structural characterization of the ATP/ADP binding site in the Hsp90 molecular chaperone. Cell, 90(1):65-75.
pmid: 9230303 |
| [23] |
Queitsch C, Sangster T A, Lindquist S. 2002. Hsp90 as a capacitor of phenotypic variation. Nature, 417(6889):618.
doi: 10.1038/nature749 URL |
| [24] |
Song H, Zhao R, Fan P, Wang X, Chen X, Li Y. 2009. Overexpression of AtHsp90.2,AtHsp90.5 and AtHsp90.7 in Arabidopsis thaliana enhances plant sensitivity to salt and drought stresses. Planta, 229(4):955-964.
doi: 10.1007/s00425-008-0886-y URL |
| [25] |
Song Z P, Pan F L, Yang C, Jia H F, Jiang H L, He F, Li N J, Lu X C, Zhang H Y. 2019. Genome-wide identification and expression analysis of HSP90 gene family in Nicotiana tabacum. BMC Genetics, 20:35.
doi: 10.1186/s12863-019-0738-8 URL |
| [26] | Subramanian B, Gao S H, Lercher M J, Hu S N, Chen W H. 2019. Evolview v3:a webserver for visualization,annotation,and management of phylogenetic trees. Nucleic Acids Research, 47:270-275. |
| [27] | Terasawa K, Minami M, Minami Y. 2005. Constantly updated knowledge of hsp90. Journal of Biochemistry and Molecular Biology, 137(4):443. |
| [28] |
Velasco R, Zharkikh A, Affourtit J, Dhingra A, Cestaro A, Kalyanaraman A, Fontana P, Bhatnagar S K, Troggio M, Pruss D. 2010. The genome of the domesticated apple(Malus domesticaBorkh.). Nature Genetics, 42:833-839.
doi: 10.1038/ng.654 URL |
| [29] | Wang Jing, Tan Fangjun, Liang Chengliang, Zhang Xilu, Ou Lijun, Niran Juntawong, Wang Fei, Jiao Chunyan, Zou Xuexiao, Chen Wenchao. 2020. Genome-wide identification and analysis of HSP90 gene family in pepper. Acta Horticulturae Sinica, 47(4):665-674. (in Chinese) |
| 王静, 谭放军, 梁成亮, 张西露, 欧立军, Niran Juntawong, 王飞, 焦春海, 邹学校, 陈文超. 2020. 辣椒热激蛋白HSP90家族基因鉴定及分析. 园艺学报, 47(4):665-674. | |
| [30] |
Wang W, Vinocur B, Shoseyov O, Altman A. 2004. Role of plant heat-shock proteins and molecular chaperones in the abiotic stress response. Trends Plant Science, 9(5):244-252.
doi: 10.1016/j.tplants.2004.03.006 URL |
| [31] |
Xu J Y, Xue C C, Xue D, Zhao J M, Gai J Y, Guo N, Xing H, Swati P D. 2013. Overexpression of GmHsp90s,a heat shock protein 90(HSP90)gene family cloning from soybean,decrease damage of abiotic stresses inArabidopsis thaliana. PLoS ONE, 8(7):e69810.
doi: 10.1371/journal.pone.0069810 URL |
| [32] | Yan Shuangshuang, Qiu Zhengkun, Yu Bingwei, Ming Fangyan, Chen Changming, Lei Jianjun, Cao Bihao. 2020. Advances in phytohormone auxin response to high temperature. Acta Horticulturae Sinica, 47(11):2238-2246. (in Chinese) |
| 颜爽爽, 邱正坤, 余炳伟, 明方艳, 陈长明, 雷建军, 曹必好. 2020. 植物生长素响应高温胁迫研究进展. 园艺学报, 47(11):2238-2246. | |
| [33] |
Yang Z, Zhou Y, Wang X, Gu S, Yu J, Liang G, Yan C, Xu C. 2008. Genome wide comparative phylogenetic and molecular evolutionary analysis of tubby-like protein family in Arabidopsis,rice,and poplar. Genomics, 92(4):246-53.
doi: 10.1016/j.ygeno.2008.06.001 URL |
| [34] |
Wang Yi, Li Wei, Xu Xuefeng, Qiu Changpeng, Wu Ting, Wei Qinping, Ma Fengwang, Han Zhenhai. 2019. Progress of apple rootstock breeding and its use. Horticultural Plant Journal, 5(5):183-191.
doi: 10.1016/j.hpj.2019.06.001 |
| [35] |
Zai W S, Miao L X, Xiong Z L, Zhang H L, Ma Y R, Li Y L, Chen Y B, Ye S G. 2015. Comprehensive identification and expression analysis of Hsp90s gene family in Solanum lycopersicum. Genetics and Molecular Research, 14(3):7811-7820.
doi: 10.4238/2015.July.14.7 pmid: 26214462 |
| [36] |
Zhang J, Li J, Liu B, Zhang L, Chen J, Liu M Z. 2013. Genome-wide analysis of the Populus Hsp90 gene family reveals differential expression patterns,localization,and heat stress responses. BMC Genomics, 14(1):532.
doi: 10.1186/1471-2164-14-532 URL |
| [1] | 王晓晨, 聂子页, 刘先菊, 段 伟, 范培格, 梁振昌, . 脱落酸对‘京香玉’葡萄果实单萜物质合成的影响[J]. 园艺学报, 2023, 50(2): 237-249. |
| [2] | 张 欣, 漆艳香, 曾凡云, 王艳玮, 谢培兰, 谢艺贤, 彭 军. 香蕉枯萎病菌Dicer-like基因的功能分析[J]. 园艺学报, 2023, 50(2): 279-294. |
| [3] | 任 菲, 卢苗苗, 刘吉祥, 陈信立, 刘道凤, 眭顺照, 马 婧. 蜡梅胚胎晚期丰富蛋白基因CpLEA的表达及抗性分析[J]. 园艺学报, 2023, 50(2): 359-370. |
| [4] | 于婷婷, 李 欢, 宁源生, 宋建飞, 彭璐琳, 贾竣淇, 张玮玮, 杨洪强. 苹果GRAS全基因组鉴定及其对生长素的响应分析[J]. 园艺学报, 2023, 50(2): 397-409. |
| [5] | 翟含含, 翟宇杰, 田义, 张叶, 杨丽, 温陟良, 陈海江. 桃SAUR家族基因分析及PpSAUR5功能鉴定[J]. 园艺学报, 2023, 50(1): 1-14. |
| [6] | 杨植, 张川疆, 杨芯芳, 董梦怡, 王振磊, 闫芬芬, 吴翠云, 王玖瑞, 刘孟军, 林敏娟. 枣与酸枣杂交后代果实遗传倾向及混合遗传分析[J]. 园艺学报, 2023, 50(1): 36-52. |
| [7] | 崔建, 钟雄辉, 刘泽慈, 陈登辉, 李海龙, 韩睿, 乐祥庆, 康俊根, 王超. 结球甘蓝染色体片段替换系构建[J]. 园艺学报, 2023, 50(1): 65-78. |
| [8] | 何成勇, 赵晓丽, 许腾飞, 高德航, 李世访, 王红清. 草莓病毒1山东分离物全基因组分析[J]. 园艺学报, 2023, 50(1): 153-160. |
| [9] | 罗海林, 袁雷, 翁华, 闫佳会, 郭青云, 王文清, 马新明. 蚕豆萎蔫病毒2号青海辣椒分离物的鉴定与全基因组序列克隆[J]. 园艺学报, 2023, 50(1): 161-169. |
| [10] | 忽靖宇, 阙开娟, 缪田丽, 吴少政, 王田田, 张磊, 董鲜, 季鹏章, 董家红. 侵染鸢尾的番茄斑萎病毒鉴定[J]. 园艺学报, 2023, 50(1): 170-176. |
| [11] | 韩晓蕾, 张彩霞, 刘 锴, 杨 安, 严家帝, 李武兴, 康立群, 丛佩华. 中熟苹果新品种‘中苹优蕾’[J]. 园艺学报, 2022, 49(S2): 1-2. |
| [12] | 韩晓蕾, 张彩霞, 刘 锴, 严家帝, 李武兴, 康立群, 丛佩华. 中熟苹果新品种‘苹优2号’[J]. 园艺学报, 2022, 49(S1): 1-2. |
| [13] | 王 强, 丛佩华, 刘肖烽. 晚熟苹果新品种‘华优甜娃’[J]. 园艺学报, 2022, 49(S1): 3-4. |
| [14] | 王 强, 丛佩华, 刘肖烽. 中熟苹果新品种‘华优宝蜜’[J]. 园艺学报, 2022, 49(S1): 5-6. |
| [15] | 杨 玲, 丛佩华, 王 强, 李武兴, 康立群. 中熟鲜食苹果新品种‘华丰’[J]. 园艺学报, 2022, 49(S1): 7-8. |
| 阅读次数 | ||||||
|
全文 |
|
|||||
|
摘要 |
|
|||||
版权所有 © 2012 《园艺学报》编辑部 京ICP备10030308号-2 国际联网备案号 11010802023439
编辑部地址: 北京市海淀区中关村南大街12号中国农业科学院蔬菜花卉研究所 邮编: 100081
电话: 010-82109523 E-Mail: yuanyixuebao@126.com
技术支持:北京玛格泰克科技发展有限公司