
园艺学报 ›› 2023, Vol. 50 ›› Issue (2): 279-294.doi: 10.16420/j.issn.0513-353x.2021-1081
张欣, 漆艳香, 曾凡云, 王艳玮, 谢培兰, 谢艺贤*( ), 彭军*(
), 彭军*( )
)
收稿日期:2022-06-16
修回日期:2022-09-27
出版日期:2023-02-25
发布日期:2023-03-06
通讯作者:
*(E-mail:基金资助:
ZHANG Xin, QI Yanxiang, ZENG Fanyun, WANG Yanwei, XIE Peilan, XIE Yixian*( ), PENG Jun*(
), PENG Jun*( )
)
Received:2022-06-16
Revised:2022-09-27
Online:2023-02-25
Published:2023-03-06
Contact:
*(E-mail:摘要:
香蕉枯萎病病原菌尖孢镰刀菌古巴专化型(Fusarium oxysporum f. sp. cubense)4号生理小种(Foc4)含有两个进化上高度保守的Dicer-like基因FocDCL1和FocDCL2。为了探究该基因的功能及其在RNAi中的作用机制,利用同源重组的方法获得ΔFocDCL1、ΔFocDCL2和ΔFocDCL1/2基因敲除突变体。表型分析显示,与Foc4野生型相比,突变体菌丝的营养生长无显著差异,但产孢量下降;ΔFocDCL2突变体在非生物胁迫刚果红处理后菌落明显变小且气生菌丝增多;ΔFocDCL2和ΔFocDCL1/2的致病力下降。miRNA深度测序数据显示,与Foc4野生型相比,ΔFocDCL1、ΔFocDCL2和ΔFocDCL1/2敲除突变体的miRNA长度分布和5′-端首位碱基出现频率都发生了改变,都能产生自身特异miRNA。DCL功能存在交叉和冗余,miRNA可以通过依赖独立的、交叉的或者联合的DCL发生;此外,鉴定到不依赖DCL形成的miRNA。这些结果表明FocDCL在产孢量、非生物胁迫、致病力以及小RNA发生中发挥作用。
中图分类号:
张欣, 漆艳香, 曾凡云, 王艳玮, 谢培兰, 谢艺贤, 彭军. 香蕉枯萎病菌Dicer-like基因的功能分析[J]. 园艺学报, 2023, 50(2): 279-294.
ZHANG Xin, QI Yanxiang, ZENG Fanyun, WANG Yanwei, XIE Peilan, XIE Yixian, PENG Jun. Functional Analysis of Dicer-like Genes in Fusarium oxysporum f. sp. cubense Race 4[J]. Acta Horticulturae Sinica, 2023, 50(2): 279-294.
| 引物 Primer | 序列(5′-3′) Sequence | 引物 Primer | 序列(5′-3′) Sequence |
|---|---|---|---|
| HYG-F | CTTGGCTGGAGCTAGTGGAGGT | NEO-F | TCTAGATTAACGCTTACAATTTCC |
| HYG-R DCL1-LBCK | CCCGGTCGGCATCTACTCTATTC CCAGGCTATGGTCCCAAGAA | NEO-R DCL2-NEO-LBCK | TCAGAAGAACTCGTCAAGAAGG CAGCAGATGTAATAGTCGCCG |
| DCL1-HPH-LB-R | ACCTCCACTAGCTCCAGCCAAGCAAGAGTCCGCTACAATCTCAA | DCL2-NEO--LB-R | GAAATTGTAAGCGTTAATCTAGCCTTGATGCCCTCCTTATCC |
| DCL1-HPH-RB-F | GAATAGAGTAGATGCCGACCGGGAGCGTTAGAAGCGTAGACAA | DCL2-NEO-RB-F | CCTTCTTGACGAGTTCTTCTGATTGAGAGTGCGGAGGGACTG |
| DCL1-RBCK | CAACAAACAAGACCTCCTCTC | DCL2-NEO-RBCK | GAGGGTGAGATGAACGGTGA |
| DCL1-LB-F | GCAAAGAGTCTATCGTGTGAGCC | DCL2-NEO-LB-F | CGACTTACACAAATACATCCTCCC |
| HYG-R1 | GGATGCCTCCGCTCGAAGTA | NEO-R1 | GAGCAAGGTGAGATGACAGGAG |
| HYG-F1 | CGTTGCAAGACCTGCCTGAA | NEO-F1 | CACCACTCGATCCGTCACCAAC |
| DCL1-RB-R | CGGTAAAGGATTGGGATTGTTG | DCL2-NEO-RB-R | CGTCTTTGTCTCCATCAACTTCG |
| HPT-LBCK | GACAGACGTCGCGGTGAGTT | NEO-LBCK | GAATGTCGTCAAGCGGGAAC |
| HPT-RBCK | TCTGGACCGATGGCTGTGTAG | NEO-RBCK | CGACCACCAAGCGAAACATC |
| DCL1-1784F | GTCTCTCTCTTTCTGCTGACCG | pFCC1-F1-XhoI | AATTCTCGAGAATTGATACGGCTGGCGAAG |
| DCL1-3031R | TCAAGGCTGGGATTCAACTTAC | pFCC1-R1-HindⅢ | AGAAAGCTTTCTCAACACCAAGGCCAGT |
| DCL2-LBCK | AGCATTCGTCAACTTTGCCA | pFCC1-F2-BglII | GGCAGATCTTCTCAACACCAAGGCCAGT |
| DCL2-HPH-LB-R | ACCTCCACTAGCTCCAGCCAAGTCCATCAGCACTCACATCACTC | pFCC1-R2-KpnI FCC1-qRT-F | CCAGGTACCAATTGATACGGCTGGCGAAG TCGACAGCAACGTGGAGATT |
| DCL2-HPH-RB-F | GAATAGAGTAGATGCCGACCGGGGCATCACTAAACACTCCTCCTTGT | FCC1-qRT-R DCL1-qRT-F | ACCTGTTGATCTGTTCGCGA AGAACAAGTCCTGGCTCTCC |
| DCL2-RBCK | GAGGGTGAGATGAACGGTGAC | DCL1-qRT-R | GTCGCAATCTGGAACGTCGA |
| DCL2-LB-F | ACGCTTGGAGAGAATGCGAG | DCL2-qRT-F | TCGATGGAGTTGTGGAGTCA |
| DCL2-RB-R | AGCCATCAGTCGTAAGAGCAA | DCL2-qRT-R | GCATTCTCCGCAGCTTTGGT |
| DCL2-988F | CACCTCATTCGCTCACTCTACG | qFocUBI-1F | CCAACCCTGACGATCCTCTTGTGC |
| DCL2-2373R | ATCACAACCCGACTTCCAGC | qFocUBI-1R | TACTTTCGAGTCCACTCCCGAGCTG |
表1 FocDCL单敲除和双敲除引物
Table 1 The FocDCLs single and double gene knock-out primers used in this study
| 引物 Primer | 序列(5′-3′) Sequence | 引物 Primer | 序列(5′-3′) Sequence |
|---|---|---|---|
| HYG-F | CTTGGCTGGAGCTAGTGGAGGT | NEO-F | TCTAGATTAACGCTTACAATTTCC |
| HYG-R DCL1-LBCK | CCCGGTCGGCATCTACTCTATTC CCAGGCTATGGTCCCAAGAA | NEO-R DCL2-NEO-LBCK | TCAGAAGAACTCGTCAAGAAGG CAGCAGATGTAATAGTCGCCG |
| DCL1-HPH-LB-R | ACCTCCACTAGCTCCAGCCAAGCAAGAGTCCGCTACAATCTCAA | DCL2-NEO--LB-R | GAAATTGTAAGCGTTAATCTAGCCTTGATGCCCTCCTTATCC |
| DCL1-HPH-RB-F | GAATAGAGTAGATGCCGACCGGGAGCGTTAGAAGCGTAGACAA | DCL2-NEO-RB-F | CCTTCTTGACGAGTTCTTCTGATTGAGAGTGCGGAGGGACTG |
| DCL1-RBCK | CAACAAACAAGACCTCCTCTC | DCL2-NEO-RBCK | GAGGGTGAGATGAACGGTGA |
| DCL1-LB-F | GCAAAGAGTCTATCGTGTGAGCC | DCL2-NEO-LB-F | CGACTTACACAAATACATCCTCCC |
| HYG-R1 | GGATGCCTCCGCTCGAAGTA | NEO-R1 | GAGCAAGGTGAGATGACAGGAG |
| HYG-F1 | CGTTGCAAGACCTGCCTGAA | NEO-F1 | CACCACTCGATCCGTCACCAAC |
| DCL1-RB-R | CGGTAAAGGATTGGGATTGTTG | DCL2-NEO-RB-R | CGTCTTTGTCTCCATCAACTTCG |
| HPT-LBCK | GACAGACGTCGCGGTGAGTT | NEO-LBCK | GAATGTCGTCAAGCGGGAAC |
| HPT-RBCK | TCTGGACCGATGGCTGTGTAG | NEO-RBCK | CGACCACCAAGCGAAACATC |
| DCL1-1784F | GTCTCTCTCTTTCTGCTGACCG | pFCC1-F1-XhoI | AATTCTCGAGAATTGATACGGCTGGCGAAG |
| DCL1-3031R | TCAAGGCTGGGATTCAACTTAC | pFCC1-R1-HindⅢ | AGAAAGCTTTCTCAACACCAAGGCCAGT |
| DCL2-LBCK | AGCATTCGTCAACTTTGCCA | pFCC1-F2-BglII | GGCAGATCTTCTCAACACCAAGGCCAGT |
| DCL2-HPH-LB-R | ACCTCCACTAGCTCCAGCCAAGTCCATCAGCACTCACATCACTC | pFCC1-R2-KpnI FCC1-qRT-F | CCAGGTACCAATTGATACGGCTGGCGAAG TCGACAGCAACGTGGAGATT |
| DCL2-HPH-RB-F | GAATAGAGTAGATGCCGACCGGGGCATCACTAAACACTCCTCCTTGT | FCC1-qRT-R DCL1-qRT-F | ACCTGTTGATCTGTTCGCGA AGAACAAGTCCTGGCTCTCC |
| DCL2-RBCK | GAGGGTGAGATGAACGGTGAC | DCL1-qRT-R | GTCGCAATCTGGAACGTCGA |
| DCL2-LB-F | ACGCTTGGAGAGAATGCGAG | DCL2-qRT-F | TCGATGGAGTTGTGGAGTCA |
| DCL2-RB-R | AGCCATCAGTCGTAAGAGCAA | DCL2-qRT-R | GCATTCTCCGCAGCTTTGGT |
| DCL2-988F | CACCTCATTCGCTCACTCTACG | qFocUBI-1F | CCAACCCTGACGATCCTCTTGTGC |
| DCL2-2373R | ATCACAACCCGACTTCCAGC | qFocUBI-1R | TACTTTCGAGTCCACTCCCGAGCTG |

图1 FocDCL1(左)和FocDCL2(右)敲除示意图及其突变体的PCR A、F:单基因敲除示意图,箭头指示的为引物所在的位置,HY和YG分别为HYG抗性基因前半部分和后半部分序列;B、G:突变体的上游Up和下游Down重组片段PCR扩增;C、H:突变体的内源基因PCR扩增;D、I:潮霉素B抗性基因PCR扩增;E、J:qRT-PCR检测DCL1和DCL2表达。
Fig. 1 FocDCL1(left)and FocDLC2(right)gene knockout and PCR screening the positive mutants A,F:The schematic map of single-gene knockout,the arrows indicate the position of the primers,HY and YG indicate the first and second part of HYG resistance gene,respectively;B,G:Amplification of up and down recombinant from mutants by PCR;C,H:Amplification of endogenous gene by PCR;D,I:Amplification of Hygromycin B resistance gene by PCR;E,J:qRT-PCR detection of DCL1 and DCL2.
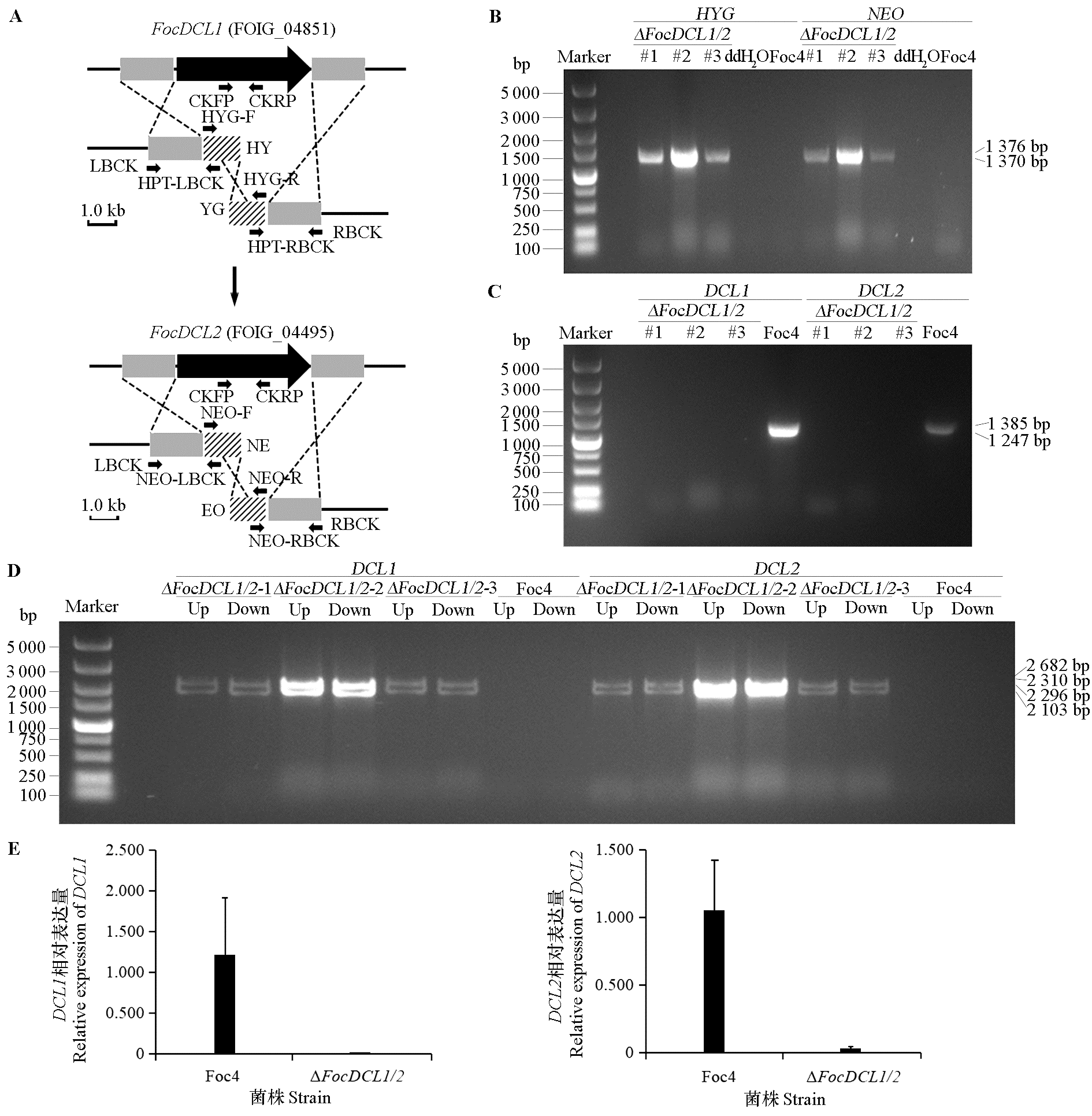
图2 ΔFocDCL1/2双敲除示意图及其突变体基因的检测 A:双敲除示意图。箭头指示的为引物位置,NE和EO分别为NEO抗性基因前半部分和后半部分序列;B:HYG和NEO的PCR扩增。C:FocDCL1和FocDCL2的PCR扩增;D:基因上、下游重组片段PCR扩增;E:qRT-PCR检测DCL1和DCL2表达。
Fig. 2 The schematic diagram of ΔFocDCL1/2 double knockout and the detection of its mutant gene A:The schematic diagram of ΔFocDCL1/2 double knockout. The arrows indicate the position of the primers,Ne and EO indicate the first and second part of NEO resistance gene,respectively;B:Amplification of HYG and NEO by PCR;C:Amplification of FocDCL1 and FocDCL2 by PCR;D:Amplification of up and down recombinant from mutants by PCR;E:qRT-PCR detection of DCL1 and DCL2.
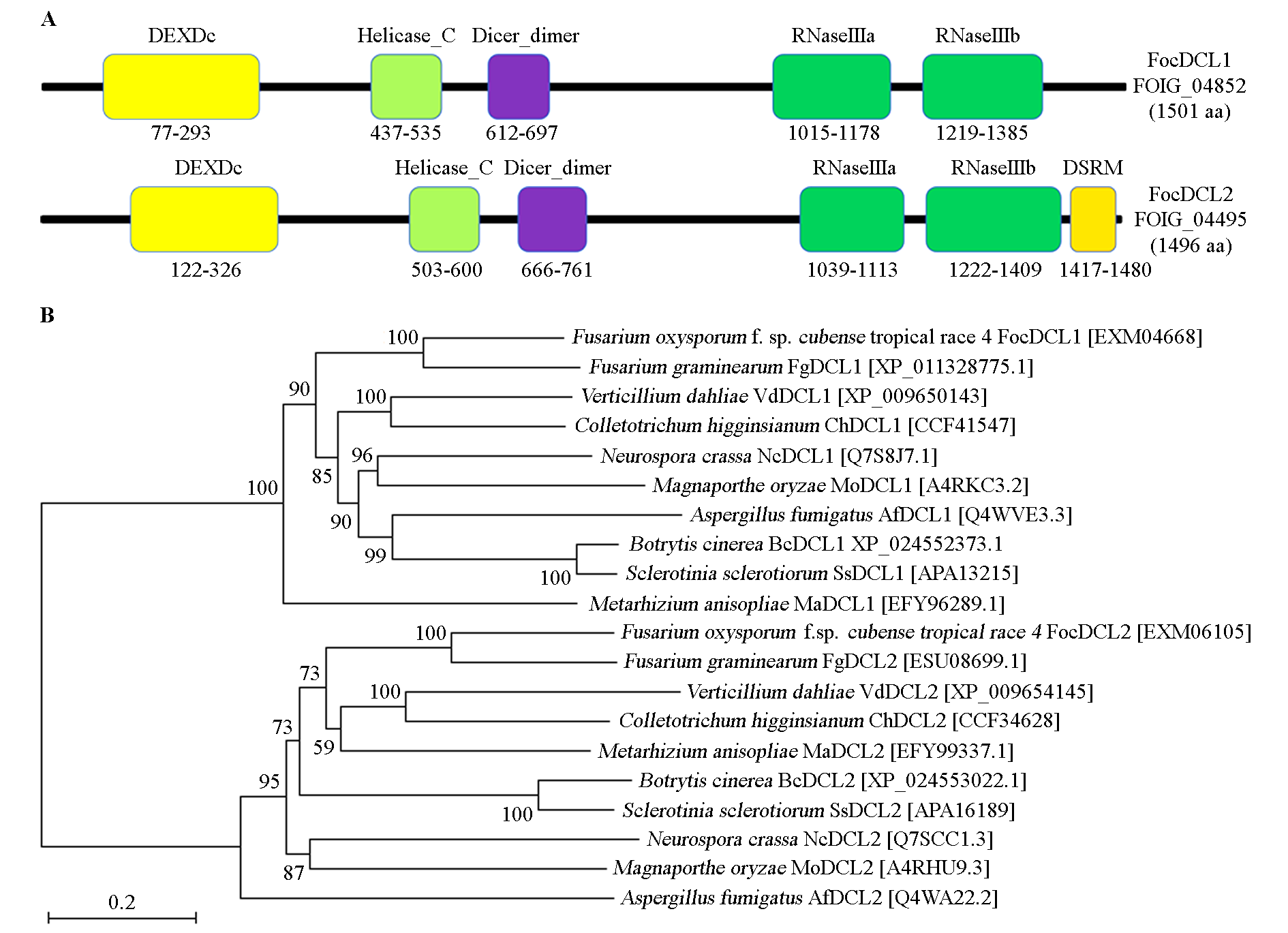
图3 FocDCL的蛋白保守结构域及系统进化树 FocDCL的系统进化树采用邻接法及1 000次重复构建。
Fig. 3 The phylogenetic tree and conserved domain of FocDCL Neighbor-joining method was used with 1 000 bootstrap replicates.

图4 FocDCL敲除突变体的表型 不同小写字母表示差异显著(P < 0.05)。下同。
Fig. 4 The phenotype of FocDCL gene knockout mutants Different lowercase letters indicate significant differences(P < 0.05). The same below.

图7 香蕉幼苗接种FocDCL敲除突变体菌株后叶片和球茎发病症状 括号内数字为病情指数。不同小写字母表示差异显著(P < 0.05)。
Fig. 7 Symptoms of banana leaves and corms after inoculation with FocDCLs knockout mutants Disease index in parentheses. Different lowercase letters indicate significant difference(P < 0.05).

图8 hpRNAi-FCC1诱导ΔFocDCL1和ΔFocDCL2突变体表型分析 紫红色的ΔFocFCC1突变体作为正对照。
Fig. 8 Phenotypic analysis of hpRNAi-FCC1 induced gene silencing in ΔFocDCL1 and ΔFocDCL2 ΔFocFCC1 with red colony phenotypes served as positive control.
| 样品 Sample ID | 总数量 Total reads | 匹配数量 Mapped reads | 已知miRNA Known-miRNA | 新的miRNA Novel-miRNA | 靶标基因 Target gene |
|---|---|---|---|---|---|
| Foc4 | 18 931 444 | 6 494 076 | 16 | 39 | 3 829 |
| ΔFocDCL1 | 8 922 109 | 2 851 863 | 10 | 33 | 1 772 |
| ΔFocDCL2 | 9 046 433 | 3 373 316 | 9 | 14 | 1 105 |
| ΔFocDCL1/2 | 19 831 731 | 11 454 808 | 6 | 11 | 818 |
表2 小RNA测序及DCL敲除突变体miRNA数据统计
Table 2 The statistics of small RNA sequence date and miRNAs of DCL knockout mutants
| 样品 Sample ID | 总数量 Total reads | 匹配数量 Mapped reads | 已知miRNA Known-miRNA | 新的miRNA Novel-miRNA | 靶标基因 Target gene |
|---|---|---|---|---|---|
| Foc4 | 18 931 444 | 6 494 076 | 16 | 39 | 3 829 |
| ΔFocDCL1 | 8 922 109 | 2 851 863 | 10 | 33 | 1 772 |
| ΔFocDCL2 | 9 046 433 | 3 373 316 | 9 | 14 | 1 105 |
| ΔFocDCL1/2 | 19 831 731 | 11 454 808 | 6 | 11 | 818 |

图10 FocDCL敲除突变体小RNA序列统计 A:unisRNA的长度分布。B:小RNA的5′末端碱基偏向性分析。C:特异miRNA统计。
Fig. 10 Statistics analysis of small RNA sequence from FocDCL knockout mutants A:The length distribution of unique reads of sRNA. B:Nucleotide frequency of the 5′ end of small RNAs;C:Statistics of specific miRNA.
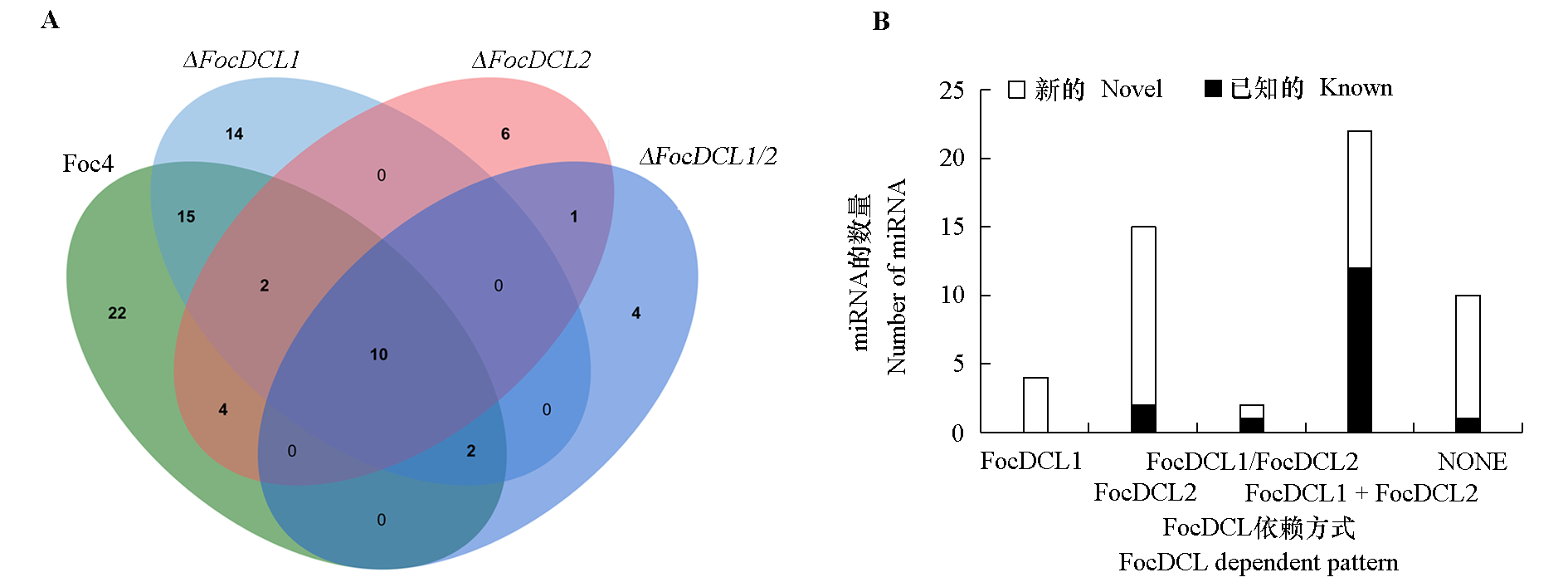
图11 FocDCL敲除突变体与Foc4野生型miRNA比对(A)及miRNA的FocDCL依赖方式鉴定(B)
Fig. 11 The miRNA comparison between FocDCL knockout mutants and Foc4(A)and identification of FocDCL-dependent miRNAs(B)
| [1] |
Allen E, Xie Z, Gustafson A M, Carrington J C. 2005. microRNA-directed phasing during trans-acting siRNA biogenesis in plants. Cell, 121 (2):207-221.
doi: 10.1016/j.cell.2005.04.004 pmid: 15851028 |
| [2] |
Carreras-Villaseñor N, Esquivel-Naranjo E U, Villalobos-Escobedo J M, Abreu-Goodger C, Herrera-Estrella A. 2013. The RNAi machinery regulates growth and development in the filamentous fungus Trichoderma atroviride. Molecular Microbiology, 89 (1):96-112.
doi: 10.1111/mmi.12261 pmid: 23672609 |
| [3] |
Chen Y, Gao Q, Huang M, Liu Y, Liu Z, Liu X, Ma Z. 2015. Characterization of RNA silencing components in the plant pathogenic fungus Fusarium graminearum. Scientific Reports, 5:12500.
doi: 10.1038/srep12500 pmid: 26212591 |
| [4] | Choi J, Kim K T, Jeon J, Wu J, Song H, Asiegbu F O, Lee Y H. 2014. funRNA:a fungi-centered genomics platform for genes encoding key components of RNAi. BMC Genomics, 15 (Suppl 9):S14. |
| [5] |
Feng H, Xu M, Liu Y, Dong R, Gao X, Huang L. 2017. Dicer-like genes are required for H2O2 and KCl stress responses,pathogenicity and small RNA generation in Valsa mali. Frontiers in Microbiology, 8:1166.
doi: 10.3389/fmicb.2017.01166 pmid: 28690605 |
| [6] |
Gaffar F Y, Imani J, Karlovsky P, Koch A, Kogel K H. 2019. Different components of the RNA interference machinery are required for conidiation,ascosporogenesis,virulence,deoxynivalenol production,and fungal inhibition by exogenous double-stranded RNA in the head blight pathogen Fusarium graminearum. Frontiers in Microbiology, 10:1662.
doi: 10.3389/fmicb.2019.01662 URL |
| [7] |
Hwang I S, Ahn I P. 2016. Multi-homologous recombination-based gene manipulation in the rice pathogen Fusarium fujikuroi. Plant Pathology Journal, 32 (3):173-181.
doi: 10.5423/PPJ.OA.12.2015.0263 pmid: 27298592 |
| [8] |
Jin Y, Zhao J H, Zhao P, Zhang T, Wang S, Guo H S. 2019. A fungal milRNA mediates epigenetic repression of a virulence gene in Verticillium dahliae. Philosophical Transactions B, 374 (1767):20180309.
doi: 10.1098/rstb.2018.0309 URL |
| [9] |
Kadotani N, Nakayashiki H, Tosa Y, Mayama S. 2004. One of the two Dicer-like proteins in the filamentous fungi Magnaporthe oryzae genome is responsible for hairpin RNA-triggered RNA silencing and related small interfering RNA accumulation. Journal of Biological Chemistry, 279 (43):44467-44474.
doi: 10.1074/jbc.M408259200 pmid: 15304480 |
| [10] | Lee H C, Li L, Gu W, Xue Z, Crosthwaite S K, Pertsemlidis A, Lewis Z A, Freitag M, Selker E U, Mello C C, Liu Y. 2010. Diverse pathways generate microRNA-like RNAs and Dicer-independent small interfering RNAs in fungi. Molecualr Cell, 38 (6):803-814. |
| [11] | Liang L Q. 2014. Function analysisi of Dicer,Argonaute and Rdrp involved in small RNAs biogenesis in Fusarium oxysporum[Ph. D. Dissertation]. Beijing: China Agricultural University. (in Chinese) |
| 梁丽琴. 2014. 尖孢镰刀菌小RNA发生相关Dicer,Argonaute和Rdrp基因功能研究[博士论文]. 北京: 中国农业大学. | |
| [12] | Luo He, Li Weijia, Li He, Zhang Zhihong. 2020. FaRGA1gene silencing changes the characteristics of flowering and runner producing in strawberry. Acta Horticulturae Sinica, 47 (12):2331-2339. (in Chinese) |
| 罗贺, 李伟佳, 李贺, 张志宏. 2020. 草莓FaRGA1基因沉默改变开花和匍匐茎抽生特性. 园艺学报, 47 (12):2331-2339. | |
| [13] |
Macrae I J, Zhou K, Li F, Repic A, Brooks A N, Cande W Z, Adams P D, Doudna J A. 2006. Structural basis for double-stranded RNA processing by Dicer. Science, 311 (5758):195-198.
doi: 10.1126/science.1121638 pmid: 16410517 |
| [14] | Mohamed A A, Mak C, Liew K W, Ho Y W. 1999. Early evaluation of banana plants at nursery stage for fusarium wilt tolerance //Molina A B,Nik Masdek N H,Liew K W. Seminar on banana Fusarium wilt management towards sustainable cultivation,Pahang,Malaysia. Montpellier:International Network for the Improvement of Banana and Plantain:174-185. |
| [15] |
Nakayashiki H, Hanada S, Nguyen B Q, Naoki K, Yukio T, Shigeyuki M. 2005. RNA silencing as a tool for exploring gene function in ascomycete fungi. Fungal Genetics and Biology, 42 (4):275-283.
doi: 10.1016/j.fgb.2005.01.002 pmid: 15749047 |
| [16] |
Nicolás F E, de Haro J P, Torres-Martínez S, Ruiz-Vázquez R M. 2007. Mutants defective in a Mucor circinelloides Dicer-like gene are not compromised in siRNA silencing but display developmental defects. Fungal Genetics and Biology, 44 (6):504-516.
doi: 10.1016/j.fgb.2006.09.003 URL |
| [17] |
Schirawski J, Mannhaupt G, Münch K, Brefort T, Schipper K, Doehlemann G, Di Stasio M, Rössel N, Mendoza-Mendoza A, Pester D, Müller O, Winterberg B, Meyer E, Ghareeb H, Wollenberg T, Münsterkötter M, Wong P, Walter M, Stukenbrock E, Güldener U, Kahmann R. 2010. Pathogenicity determinants in smut fungi revealed by genome comparison. Science, 330 (6010):1546-1548.
doi: 10.1126/science.1195330 pmid: 21148393 |
| [18] | Schultz J, Milpetz F, Bork P, Ponting C P. 1998. SMART,a simple modular architecture research tool: identification of signaling domains. Proceedings of the National Academy of Sciences of the United States of America, 95 (11):5857-5864. |
| [19] |
Shim W B, Woloshuk C P. 2001. Regulation of fumonisin B(1) biosynthesis and conidiation in Fusarium verticillioides by a cyclin-like(C-type)gene,FCC1. Applied and Environmental Microbiology, 67 (4):1607-1612.
pmid: 11282612 |
| [20] |
Siamak S B, Zheng S J. 2018. Banana fusarium wilt(Fusarium oxysporum f. sp. cubense)control and resistance,in the context of developing wilt-resistant bananas within sustainable production systems. Horticultural Plant Journal, 4 (5):208-218.
doi: 10.1016/j.hpj.2018.08.001 URL |
| [21] |
Wang M, Jin H. 2017. Spray-induced gene silencing:a powerful innovative strategy for crop protection. Trends in Microbiology, 25 (1):4-6.
doi: 10.1016/j.tim.2016.11.011 URL |
| [22] |
Wang W, Tang W. 2018. Generation of Fusarium graminearum knockout mutants by the split-marker recombination approach. Bio-protocol, e2976. doi:10.21769/BioProtoc.2976.
doi: 10.21769/BioProtoc.2976 |
| [23] |
Xue Z, Yuan H, Guo J, Liu Y. 2012. Reconstitution of an Argonaute-dependent small RNA biogenesis pathway reveals a handover mechanism involving the RNA exosome and the exonuclease QIP. Molecular Cell, 46 (3):299-310.
doi: 10.1016/j.molcel.2012.03.019 pmid: 22516970 |
| [24] |
Yang Xingyu, Xu Linbing, Wu Yuanli, Qiu Diyang, Fan Linlin, Huang Bingzhi. 2022. Genome(ABBB)identification of a hybrid banana cultivar‘Fenza 1’. Acta Horticulturae Sinica, 49 (9):1991-1997. (in Chinese)
doi: 10.16420/j.issn.0513-353x.2021-0514 |
|
杨兴玉, 许林兵, 吴元立, 邱迪洋, 范琳琳, 黄秉智. 2022. 香蕉杂交种‘粉杂1号’ABBB基因组鉴定. 园艺学报, 49 (9):1991-1997.
doi: 10.16420/j.issn.0513-353x.2021-0514 |
| [1] | 梁嘉莉, 吴启松, 陈广全, 张荣, 徐春香, 冯淑杰. 香蕉叶斑病病原菌芭蕉新拟盘多毛孢的鉴定[J]. 园艺学报, 2023, 50(2): 410-420. |
| [2] | 王喜庆, 贾云鹤, 闫闻, 付永凯, 尤海波, 李冬艳, 赵景超. 高抗枯萎病西瓜新品种‘龙盛佳力’[J]. 园艺学报, 2023, 50(2): 455-456. |
| [3] | 杨兴玉, 许林兵, 吴元立, 邱迪洋, 范琳琳, 黄秉智. 香蕉杂交种‘粉杂1号’ABBB基因组鉴定[J]. 园艺学报, 2022, 49(9): 1991-1997. |
| [4] | 张倩雯, 杨希航, 李峰, 邓颖天. miRNA调控园艺作物生长发育研究进展[J]. 园艺学报, 2022, 49(5): 1145-1161. |
| [5] | 孟臻, 张伟萍, 王莹, 李龙, 姬小雪, 董贝, 乔康. 番茄枯萎病菌RT-PCR检测技术的建立与应用[J]. 园艺学报, 2022, 49(11): 2479-2488. |
| [6] | 吕红豪, 杨丽梅, 方智远, 张扬勇, 庄木, 刘玉梅, 孙培田, 王勇, 季家磊, 李占省, 韩风庆. 抗枯萎病早熟优质春甘蓝新品种‘YR中甘21’[J]. 园艺学报, 2021, 48(9): 1839-1840. |
| [7] | 张晓艺, 洪雨慧, 张媛媛, 栾雨时. Sly-miR166b及其靶基因在番茄抗晚疫病中的作用初探[J]. 园艺学报, 2021, 48(8): 1595-1604. |
| [8] | 苏立遥, 王培育, 蒋梦琦, 黄倏祺, 薛晓东, 刘梦雨, 肖学宸, 赖春旺, 张梓浩, 陈裕坤, 赖钟雄, 林玉玲. 龙眼pri-miR319a编码短肽活性的研究[J]. 园艺学报, 2021, 48(5): 908-920. |
| [9] | 黄威剑, 李梦. 果树全基因组测序现状与展望[J]. 园艺学报, 2021, 48(4): 733-748. |
| [10] | 杨兴玉, 肖维强, 许林兵, 李华平, 黄秉智, 叶春海, 陈 彪, 陈 谷, 吕庆芳, 梁钾贤, 吴元立, 胡玲玉. 香蕉新品种‘南天红’[J]. 园艺学报, 2020, 47(S2): 2961-2962. |
| [11] | 吴元立1,黄秉智1,*,张智胜2,杨兴玉1. 香蕉枯萎病抗性离体接种鉴定方法的优化[J]. 园艺学报, 2020, 47(8): 1577-1584. |
| [12] | 李琳琳1,金 华1,刘斯超3,邹吉祥1,李天来2,*. 番茄茉莉酸缺失突变体灰霉菌侵染响应miRNA及其表达分析[J]. 园艺学报, 2020, 47(7): 1323-1334. |
| [13] | 曹云娥,吴 庆,张美君,尹 翠,高艳明,朱红艳,张文文,李建设*. 瓜类枯萎病生防菌WQ-6的筛选鉴定、发酵工艺优化及防效研究[J]. 园艺学报, 2020, 47(6): 1072-1086. |
| [14] | 刘彦英,倪珊珊,项蕾蕾,陈裕坤,赖钟雄*. 香蕉漆酶基因家族鉴定及低温胁迫下的表达分析[J]. 园艺学报, 2020, 47(5): 837-852. |
| [15] | 姚利晓,何永睿,陈善春*. microRNA 参与柑橘生长发育和抗逆的研究进展[J]. 园艺学报, 2020, 47(5): 995-1008. |
| 阅读次数 | ||||||
|
全文 |
|
|||||
|
摘要 |
|
|||||
版权所有 © 2012 《园艺学报》编辑部 京ICP备10030308号-2 国际联网备案号 11010802023439
编辑部地址: 北京市海淀区中关村南大街12号中国农业科学院蔬菜花卉研究所 邮编: 100081
电话: 010-82109523 E-Mail: yuanyixuebao@126.com
技术支持:北京玛格泰克科技发展有限公司