
Acta Horticulturae Sinica ›› 2022, Vol. 49 ›› Issue (2): 352-364.doi: 10.16420/j.issn.0513-353x.2020-1013
• Research Papers • Previous Articles Next Articles
ZHOU Zhiming, YANG Jiabao, ZHANG Cheng, ZENG Linglu, MENG Wanqiu, SUN Li( )
)
Received:2021-07-01
Revised:2021-12-23
Online:2022-02-25
Published:2022-02-28
Contact:
SUN Li
E-mail:sunlishz@126.com
CLC Number:
ZHOU Zhiming, YANG Jiabao, ZHANG Cheng, ZENG Linglu, MENG Wanqiu, SUN Li. Genome-wide Identification and Expression Analyses of Long-chain Acyl-CoA Synthetases Under Abiotic Stresses in Helianthus annuus[J]. Acta Horticulturae Sinica, 2022, 49(2): 352-364.
Add to citation manager EndNote|Ris|BibTeX
URL: https://www.ahs.ac.cn/EN/10.16420/j.issn.0513-353x.2020-1013
| 基因Gene | 正向引物(5′-3′)Forward primer | 反向引物(5′-3′)Reverse primer |
|---|---|---|
| 18S rRNA | CTACCACATCCAAGGAAGGCAG | CGACAGAAGGGACGAGTAAACC |
| HaLACS1-1 | GAGCTCGAAAACGGAACGTG | ATTGCAAGCCTCCATCGCTA |
| HaLACS1-2 | TTGGACCATGCCGAAGTTGA | AAGGAACTCGTTCCACGCAT |
| HaLACS2 | AGAAACAGACAAACCGGGCA | ACGCCTTCTTTACAGTGCCT |
| HaLACS4-1 | GGTTATGAGCATGGAGGCGT | CCACGCCAAAATTTCCGACT |
| HaLACS4-2 | GGTTATGAGCATGGAGGCGT | GGTTATGAGCATGGAGGCGT |
| HaLACS4-3 | CCTCCGATTGACGGTTTGGA | CTCAACTCCACAAGCTCGGA |
| HaLACS4-4 | CGGTGAAAGACGGAACTCCA | CGACGCCACATGAACGGATA |
| HaLACS4-5 | GAAAACAATCGTGAGCTTTGGGA | AGTTCAACGATACTTTCGCGCT |
| HaLACS6 | CACTGCCGGAGAGTTCTTCA | TGCTTCCCCGTAAGTTGTCC |
| HaLACS7 | TCATCTTCTTCCCTCCACCGA | ATCTGGTGACAAGCTTCAGCG |
| HaLACS8 | GCAGATGGATGTTGGGGGAT | CGCCAACTTCAACTGGAACG |
| HaLACS9-1 | ACTCTCGCCCTTCGGAAATC | TTTCAAATGCCCTCCCGTCT |
| HaLACS9-2 | ATTGCATTGCAGGCGTGTTT | TTTCCAACTGCTGCTTCCGT |
Table 1 Primers used for qRT-PCR
| 基因Gene | 正向引物(5′-3′)Forward primer | 反向引物(5′-3′)Reverse primer |
|---|---|---|
| 18S rRNA | CTACCACATCCAAGGAAGGCAG | CGACAGAAGGGACGAGTAAACC |
| HaLACS1-1 | GAGCTCGAAAACGGAACGTG | ATTGCAAGCCTCCATCGCTA |
| HaLACS1-2 | TTGGACCATGCCGAAGTTGA | AAGGAACTCGTTCCACGCAT |
| HaLACS2 | AGAAACAGACAAACCGGGCA | ACGCCTTCTTTACAGTGCCT |
| HaLACS4-1 | GGTTATGAGCATGGAGGCGT | CCACGCCAAAATTTCCGACT |
| HaLACS4-2 | GGTTATGAGCATGGAGGCGT | GGTTATGAGCATGGAGGCGT |
| HaLACS4-3 | CCTCCGATTGACGGTTTGGA | CTCAACTCCACAAGCTCGGA |
| HaLACS4-4 | CGGTGAAAGACGGAACTCCA | CGACGCCACATGAACGGATA |
| HaLACS4-5 | GAAAACAATCGTGAGCTTTGGGA | AGTTCAACGATACTTTCGCGCT |
| HaLACS6 | CACTGCCGGAGAGTTCTTCA | TGCTTCCCCGTAAGTTGTCC |
| HaLACS7 | TCATCTTCTTCCCTCCACCGA | ATCTGGTGACAAGCTTCAGCG |
| HaLACS8 | GCAGATGGATGTTGGGGGAT | CGCCAACTTCAACTGGAACG |
| HaLACS9-1 | ACTCTCGCCCTTCGGAAATC | TTTCAAATGCCCTCCCGTCT |
| HaLACS9-2 | ATTGCATTGCAGGCGTGTTT | TTTCCAACTGCTGCTTCCGT |
| 转录本ID Transcript ID | 基因名称 Gene name | CDS/bp | 大小/aa Size | 蛋白分子量/kD Molecular weight | 等电点 pI | 拟南芥同源基因 Arabidopsis homologous gene | |
|---|---|---|---|---|---|---|---|
| 登录号Accession No. | 名称Name | ||||||
| OTG27997 | HaLACS1-1 | 1 980 | 659 | 74.4 | 5.82 | AT2G47240 | AtLACS1 |
| OTF93791 | HaLACS1-2 | 1 986 | 661 | 74.8 | 7.23 | AT2G47240 | AtLACS1 |
| OTG12244 | HaLACS2 | 1 980 | 659 | 73.5 | 5.70 | AT1G49430 | AtLACS2 |
| OTG32156 | HaLACS4-1 | 1 986 | 661 | 73.5 | 6.07 | AT4G23850 | AtLACS4 |
| OTG33063 | HaLACS4-2 | 1 995 | 664 | 74.0 | 6.69 | AT4G23850 | AtLACS4 |
| OTG20245 | HaLACS4-3 | 1 980 | 659 | 73.5 | 6.50 | AT4G23850 | AtLACS4 |
| OTG02621 | HaLACS4-4 | 1 974 | 657 | 73.4 | 6.69 | AT4G23850 | AtLACS4 |
| OTG02629 | HaLACS4-5 | 1 983 | 660 | 73.8 | 8.49 | AT4G23850 | AtLACS4 |
| OTG11238 | HaLACS6 | 2 094 | 697 | 76.3 | 7.51 | AT3G05970 | AtLACS6 |
| OTG31091 | HaLACS7 | 2 097 | 698 | 77.0 | 6.58 | AT5G27600 | AtLACS7 |
| OTG29824 | HaLACS8 | 2 163 | 720 | 78.6 | 7.86 | AT2G04350 | AtLACS8 |
| OTG15459 | HaLACS9-1 | 2 091 | 696 | 76.2 | 6.10 | AT1G77590 | AtLACS9 |
| OTG07054 | HaLACS9-2 | 2 121 | 706 | 77.4 | 6.78 | AT1G77590 | AtLACS9 |
Table 2 Detailed information of LACS genes in sunflower genome
| 转录本ID Transcript ID | 基因名称 Gene name | CDS/bp | 大小/aa Size | 蛋白分子量/kD Molecular weight | 等电点 pI | 拟南芥同源基因 Arabidopsis homologous gene | |
|---|---|---|---|---|---|---|---|
| 登录号Accession No. | 名称Name | ||||||
| OTG27997 | HaLACS1-1 | 1 980 | 659 | 74.4 | 5.82 | AT2G47240 | AtLACS1 |
| OTF93791 | HaLACS1-2 | 1 986 | 661 | 74.8 | 7.23 | AT2G47240 | AtLACS1 |
| OTG12244 | HaLACS2 | 1 980 | 659 | 73.5 | 5.70 | AT1G49430 | AtLACS2 |
| OTG32156 | HaLACS4-1 | 1 986 | 661 | 73.5 | 6.07 | AT4G23850 | AtLACS4 |
| OTG33063 | HaLACS4-2 | 1 995 | 664 | 74.0 | 6.69 | AT4G23850 | AtLACS4 |
| OTG20245 | HaLACS4-3 | 1 980 | 659 | 73.5 | 6.50 | AT4G23850 | AtLACS4 |
| OTG02621 | HaLACS4-4 | 1 974 | 657 | 73.4 | 6.69 | AT4G23850 | AtLACS4 |
| OTG02629 | HaLACS4-5 | 1 983 | 660 | 73.8 | 8.49 | AT4G23850 | AtLACS4 |
| OTG11238 | HaLACS6 | 2 094 | 697 | 76.3 | 7.51 | AT3G05970 | AtLACS6 |
| OTG31091 | HaLACS7 | 2 097 | 698 | 77.0 | 6.58 | AT5G27600 | AtLACS7 |
| OTG29824 | HaLACS8 | 2 163 | 720 | 78.6 | 7.86 | AT2G04350 | AtLACS8 |
| OTG15459 | HaLACS9-1 | 2 091 | 696 | 76.2 | 6.10 | AT1G77590 | AtLACS9 |
| OTG07054 | HaLACS9-2 | 2 121 | 706 | 77.4 | 6.78 | AT1G77590 | AtLACS9 |
| 重复基因对 Duplicated gene pairs | 非同义替 换率Ka | 同义替换率 Ks | Ka/Ks | 重复类型 Duplicated type | 选择压力 Selective pressure | 分化时间/ Mya Divergence time |
|---|---|---|---|---|---|---|
| HaLACS9-1/HaLACS9-2 | 0.0540 | 0.5316 | 0.1016 | 片段重复Segmental | 纯化Purifying | 17.72 |
| HaLACS4-1/HaLACS4-5 | 0.0739 | 0.6713 | 0.1101 | 片段重复Segmental | 纯化Purifying | 22.38 |
| HaLACS4-1/HaLACS4-4 | 0.0905 | 0.6463 | 0.1400 | 片段重复Segmental | 纯化Purifying | 21.54 |
| HaLACS4-1/HaLACS4-3 | 0.1003 | 1.0273 | 0.0976 | 片段重复Segmental | 纯化Purifying | 34.24 |
| HaLACS4-4/HaLACS4-5 | 0.0600 | 0.3516 | 0.1706 | 串联重复Tandem | 纯化Purifying | 11.72 |
Table 3 Gene duplication events and divergence time of the LACS genes in Helianthus annuus
| 重复基因对 Duplicated gene pairs | 非同义替 换率Ka | 同义替换率 Ks | Ka/Ks | 重复类型 Duplicated type | 选择压力 Selective pressure | 分化时间/ Mya Divergence time |
|---|---|---|---|---|---|---|
| HaLACS9-1/HaLACS9-2 | 0.0540 | 0.5316 | 0.1016 | 片段重复Segmental | 纯化Purifying | 17.72 |
| HaLACS4-1/HaLACS4-5 | 0.0739 | 0.6713 | 0.1101 | 片段重复Segmental | 纯化Purifying | 22.38 |
| HaLACS4-1/HaLACS4-4 | 0.0905 | 0.6463 | 0.1400 | 片段重复Segmental | 纯化Purifying | 21.54 |
| HaLACS4-1/HaLACS4-3 | 0.1003 | 1.0273 | 0.0976 | 片段重复Segmental | 纯化Purifying | 34.24 |
| HaLACS4-4/HaLACS4-5 | 0.0600 | 0.3516 | 0.1706 | 串联重复Tandem | 纯化Purifying | 11.72 |
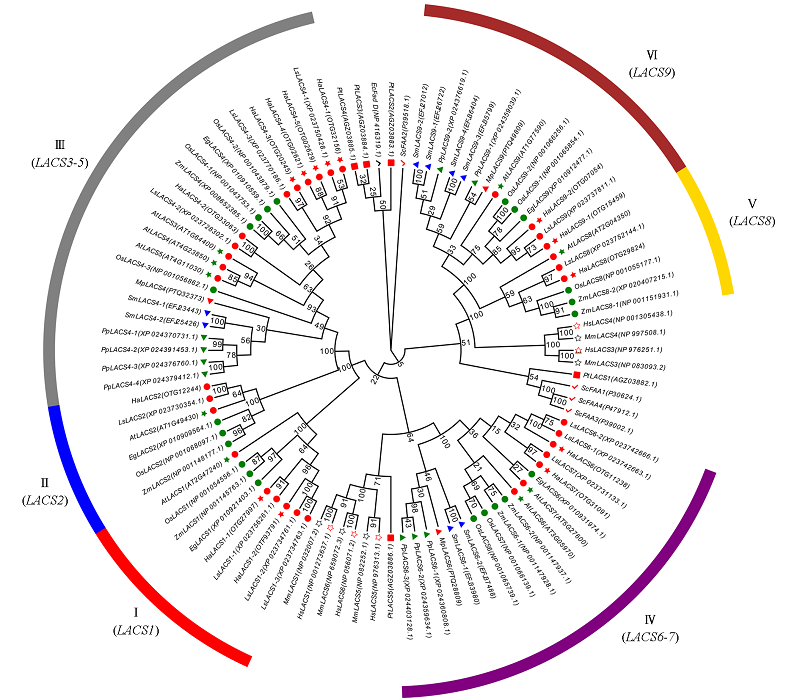
Fig. 2 Phylogenetic analysis of LACS gene family in sunflower and other species The green circles indicate monocotyledonous plants,the red circle indicates a dicot plant. Ec:Escherichia coli(prokaryote);Sc:Saccharomyces cerevisiae(fungi);Pt:Phaeodac tylumtricornutum(algae);Pp:Physcomitrella patens(moss);Sm:Selaginella moellendorffii(fern);Mp:Marchantia polymorpha(moss);At:Arabidopsis thaliana;Ha:Helianthus annuus;Ls:Lactuca sativa;Eg:Elaeis guineensis;Zm:Zea mays;Os:Oryza sativa;Hs:Homo sapiens;Mm:Mus musculus.

Fig. 4 Multiple sequence alignment of the HaLACS amino acid sequences with other proteins Ec:Escherichia coli;At:Arabidopsis thaliana;Sc:Selaginella moellendorffii;Ha:Helianthus annuus.
| [1] |
Aznar-Moreno J A, Calerón M V, Martínez-Force E, Garcés R, Mullen R, Gidda S K, Salas J J. 2014. Sunflower(Helianthus annuus L.)long-chain acyl-coenzyme A synthetases expressed at high levels in developing seeds. Physiologia Plantarum, 150 (3):363-373.
doi: 10.1111/ppl.2014.150.issue-3 URL |
| [2] |
Bernard A, Joubès J. 2013. Arabidopsis cuticular waxes:advances in synthesis,export and regulation. Progress in Lipid Research, 52 (1):110-129.
doi: 10.1016/j.plipres.2012.10.002 pmid: 23103356 |
| [3] |
Chen C J, Chen H, Zhang Y, Thomas H R, Frank M H, He Y H, Xia R. 2020. TBtools:an integrative toolkit developed for interactive analyses of big biological data. Molecular Plant, 13 (8):1194-1202.
doi: 10.1016/j.molp.2020.06.009 URL |
| [4] |
Dimopoulos N, Tindjau R, Wong D C J, Matzat T, Haslam T, Song C, Gambetta G A, Kunst L, Castellarin S D. 2020. Drought stress modulates cuticular wax composition of the grape berry. Journal of Experimental Botany, 71 (10):3126-3141.
doi: 10.1093/jxb/eraa046 pmid: 31985780 |
| [5] |
Ding L N, Gu S L, Zhu F G, Ma Z Y, Li J, Li M, Zheng W, Tan X L. 2020. Long-chain acyl-CoA synthetase 2 is involved in seed oil production in Brassica napus. BMC Plant Biology, 20 (1):21-34.
doi: 10.1186/s12870-020-2240-x pmid: 31931712 |
| [6] |
Etchegaray A, Ralf D, Paul C E, Geoffrey T, von Döhren H. 1998. Group specific antibodies against the putative AMP-binding domain signature SGTTGXPKG in peptide synthetases and related enzymes. Biochemistry and Molecular Biology International, 44 (2):235-243.
pmid: 9530507 |
| [7] |
Flagel L E, Wendel J F. 2009. Gene duplication and evolutionary novelty in plants. New Phytologist, 183 (3):557-564.
doi: 10.1111/j.1469-8137.2009.02923.x pmid: 19555435 |
| [8] |
Fulda M, Heinz E, Wolter F P. 1994. The fadD gene of Escherichia coli K 12 is located close to rnd at 39.6 min of the chromosomal-map and is a new member of the AMP-binding protein family. Molecular and General Genetics, 242 (3):241-249.
pmid: 8107670 |
| [9] |
Fulda M, Shockey J, Werber M, Wolter F P, Heinz E. 2002. Two long-chain acyl-CoA synthetases from Arabidopsis thaliana involved in peroxisomal fatty acid b-oxidation. The Plant Journal, 32 (1):93-103.
doi: 10.1046/j.1365-313X.2002.01405.x URL |
| [10] |
Iijima H, Fujino T, Minekura H, Suzuki H, Kang M J, Yamamoto T. 1996. Biochemical studies of two rat Acyl-CoA synthetases,ACS1 and ACS2. European Journal of Biochemistry, 242 (2):186-190.
doi: 10.1111/ejb.1996.242.issue-2 URL pmid: 8973631 |
| [11] |
Jessen D, Olbrich A, Knüfer J, Krüger A, Hoppert M, Polle A, Fulda M. 2011. Combined activity of LACS1 and LACS4 is required for proper pollen coat formation in Arabidopsis. The Plant Journal, 68 (4):715-726.
doi: 10.1111/j.1365-313X.2011.04722.x pmid: 21790813 |
| [12] |
Kosma D K, Bourdenx B, Bernard A, Parsons E P, Lv S Y, Joubès J, Jenks M A. 2009. The impact of water deficiency on leaf cuticle lipids of Arabidopsis. Plant Physiology, 151 (4):1918-1929.
doi: 10.1104/pp.109.141911 URL |
| [13] |
Li Xiao-cui, Kang Kai-cheng, Huang Xian-zhong, Fan Yong-bin, Song Miao-miao, Huang Yun-jie, Ding Jia-jia. 2020. Genome-wide identification,phylogenetic analysis and expression profiling of the MKK gene family in Arabidopsis pumila. Hereditas(Beijing), 42:403-421. (in Chinese)
doi: 10.16288/j.yczz.19-388 pmid: 32312709 |
|
李晓翠, 康凯程, 黄先忠, 范永斌, 宋苗苗, 黄韵杰, 丁佳佳. 2020. 小拟南芥MKK基因家族全基因组鉴定及进化和表达分析. 遗传, 42:403-421.
pmid: 32312709 |
|
| [14] |
Lü S Y, Song T, Kosma D K, Parsons E P, Rowland O, Jenks M A. 2009. Arabidopsis CER8 encodes LONG-CHAIN ACYL-COA SYNTHETASE 1 (LACS1) that has overlapping functions with LACS2 in plant wax and cutin synthesis. The Plant Journal, 59 (4):553-564.
doi: 10.1111/tpj.2009.59.issue-4 URL |
| [15] |
Mashek D G, Li L O, Coleman R A. 2007. Long-chain Acyl-CoA synthetases and fatty acid channeling. Future Lipidology, 2 (4):465-476.
doi: 10.2217/17460875.2.4.465 URL |
| [16] |
Schnurr J, Shockey J, Browse J. 2004. The Acyl-CoA synthetase encoded by LACS2 is essential for normal cuticle development in Arabidopsis. The Plant Cell, 16 (3):629-642.
doi: 10.1105/tpc.017608 URL |
| [17] |
Shockey J M, Fulda M S, Browse J A. 2002. Arabidopsis contains nine long-chain acyl-coenzyme a synthetase genes that participate in fatty acid and glycerolipid metabolism. Plant Physiology, 129 (4):1710-1722.
pmid: 12177484 |
| [18] |
Suh M C, Samuels A L, Jetter R, Kunst L, Pollard M, Ohlrogge J, Beisson F. 2005. Cuticular lipid composition,surface structure,and gene expression in Arabidopsis stem epidermis. Plant Physiology, 139 (4):1649-1665.
doi: 10.1104/pp.105.070805 URL |
| [19] |
Vatansever R, Koc I, Ozyigit I I, Sen U, Uras M E, Anjum N A, Pereira E, Filiz E. 2016. Genome-wide identification and expression analysis of sulfate transporter(SULTR)genes in potato(Solanum tuberosum L.). Planta, 244 (6):1167-1183.
pmid: 27473680 |
| [20] |
Wang X L, Li X B. 2009. The GhACS1 gene encodes an acyl-CoA synthetase which is essential for normal microsporogenesis in early anther development of cotton. The Plant Journal, 57 (3):473-486.
doi: 10.1111/tpj.2009.57.issue-3 URL |
| [21] |
Xiao Y, Li X T, Yao L H, Xu D X, Li Yang, Zhang X F, Li Z, Xiao Q L, Ni Y, Guo Y J. 2020. Chemical profiles of cuticular waxes on various organs of Sorghum bicolor and their antifungal activities. Plant Physiology and Biochemistry, 155:596-604.
doi: S0981-9428(20)30408-3 pmid: 32846395 |
| [22] |
Xiao Z C, Li N N, Wang S F, Sun J J, Zhang L Y, Zhang C, Yang H, Zhao H Y, Yang B, Wei L J, Du H, Qu C M, Lu K, Li J N. 2019. Genome-wide identification and comparative expression profile analysis of the long-chain Acyl-CoA synthetase(LACS)gene family in two different oil content cultivars of Brassica napus. Biochemical Genetics, 57 (6):781-800.
doi: 10.1007/s10528-019-09921-5 URL |
| [23] | Yi Ting, Zhang Zhishuo, Tang Bingqian, Xie Lingling, Zou Xuexiao. 2020. Identification and expression analysis of the KCS gene family in pepper. Acta Horticulturae Sinica, 47 (2):370-380. (in Chinese) |
| 易婷, 张志硕, 汤冰倩, 谢玲玲, 邹学校. 2020. 辣椒β-酮脂酰辅酶A合酶基因家族的鉴定与表达分析. 园艺学报, 47 (2):370-380. | |
| [24] |
Zhang C L, Mao K, Zhou L J, Wang G L, Zhang Y L, Li Y Y, Hao Y J. 2018. Genome-wide identification and characterization of apple long-chain Acyl-CoA synthetases and expression analysis under different stresses. Plant Physiology and Biochemistry, 132:320-332.
doi: 10.1016/j.plaphy.2018.09.004 URL |
| [25] |
Zhao L F, Haslam T M, Sonntag A, Molina I, Kunst L. 2019. Functional overlap of long-chain Acyl-CoA synthetases in Arabidopsis. Plant and Cell Physiology, 60 (5):1041-1054.
doi: 10.1093/pcp/pcz019 URL |
| [26] |
Zhao L F, Katavic V, Li F L, Haughn G W, Kunst L. 2010. Insertional mutant analysis reveals that long-chain acyl-CoA synthetase 1(LACS1),but not LACS8,functionally overlaps with LACS9 in Arabidopsis seed oil biosynthesis. The Plant Journal, 64 (6):1048-1058.
doi: 10.1111/tpj.2010.64.issue-6 URL |
| [27] | Zhu X D, Wang M Q, Li X P, Jiu S T, Wang C, Fang J G. 2017. Genome-wide analysis of the sucrose synthase gene family in grape(Vitis vinifera):structure,evolution,and expression profiles. Genes(Basel), 8 (4):111-135. |
| [1] | WANG Xiaochen, NIE Ziye, LIU Xianju, DUAN Wei, FAN Peige, and LIANG Zhenchang, . Effects of Abscisic Acid on Monoterpene Synthesis in‘Jingxiangyu’Grape Berries [J]. Acta Horticulturae Sinica, 2023, 50(2): 237-249. |
| [2] | ZHAI Hanhan, ZHAI Yujie, TIAN Yi, ZHANG Ye, YANG Li, WEN Zhiliang, CHEN Haijiang. Genome-wide Identification of Peach SAUR Gene Family and Characterization of PpSAUR5 Gene [J]. Acta Horticulturae Sinica, 2023, 50(1): 1-14. |
| [3] | XU Xiaoping, CAO Qingying, CAI Roudi, GUAN Qingxu, ZHANG Zihao, CHEN Yukun, XU HAN, LIN Yuling, LAI Zhongxiong. Gene Cloning and Expression Analysis of miR408 and Its Target DlLAC12 in Globular Embryo Development and Abiotic Stress in Dimocarpus longan [J]. Acta Horticulturae Sinica, 2022, 49(9): 1866-1882. |
| [4] | JIA Xin, ZENG Zhen, CHEN Yue, FENG Hui, LÜ Yingmin, ZHAO Shiwei. Cloning and Expression Analysis of RcDREB2A Gene in Rosa chinensis‘Old Blush’ [J]. Acta Horticulturae Sinica, 2022, 49(9): 1945-1956. |
| [5] | ZHANG Qiuyue, LIU Changlai, YU Xiaojing, YANG Jiading, FENG Chaonian. Screening of Reference Genes for Differentially Expressed Genes in Pyrus betulaefolia Plant Under Salt Stress by qRT-PCR [J]. Acta Horticulturae Sinica, 2022, 49(7): 1557-1570. |
| [6] | MA Weifeng, LI Yanmei, MA Zonghuan, CHEN Baihong, MAO Juan. Identification of Apple POD Gene Family and Functional Analysis of MdPOD15 Gene [J]. Acta Horticulturae Sinica, 2022, 49(6): 1181-1199. |
| [7] | LI Yamei, MA Fuli, ZHANG Shanqi, HUANG Jinqiu, CHEN Mengting, ZHOU Junyong, SUN Qibao, SUN Jun. Optimization of Jujube Callus Transformation System and Application of ZjBRC1 in Regulating ZjYUCCA Expression [J]. Acta Horticulturae Sinica, 2022, 49(4): 749-757. |
| [8] | WANG Ying, AI Penghui, LI Shuailei, KANG Dongru, LI Zhongai, WANG Zicheng. Identification and Expression Analysis of Genes Related to DNA Methylation in Chrysanthemum × morifolium and C. nankingense [J]. Acta Horticulturae Sinica, 2022, 49(4): 827-840. |
| [9] | ZHANG Rui, ZHANG Xiayi, ZHAO Ting, WANG Shuangcheng, ZHANG Zhongxing, LIU Bo, ZHANG De, WANG Yanxiu. Transcriptome Analysis of the Molecular Mechanism of Saline-alkali Stress Response in Malus halliana Leaves [J]. Acta Horticulturae Sinica, 2022, 49(2): 237-251. |
| [10] | QIAO Jun, WANG Liying, LIU Jing, LI Suweng. Expression Analysis of Genes Related to Photosensitive Color Under the Caylx in Eggplant Based on Transcriptome Sequencing [J]. Acta Horticulturae Sinica, 2022, 49(11): 2347-2356. |
| [11] | HOU Tianze, YI Shuangshuang, ZHANG Zhiqun, WANG Jian, LI Chonghui. Selection and Validation of Reference Genes for RT-qPCR in Phalaenopsis- type Dendrobium Hybrid [J]. Acta Horticulturae Sinica, 2022, 49(11): 2489-2501. |
| [12] | ZHOU Tie, PAN Bin, LI Feifei, MA Xiaochuan, TANG Mengjing, LIAN Xuefei, CHANG Yuanyuan, CHEN Yuewen, LU Xiaopeng. Effects of Drought Stress at Enlargement Stage on Fruit Quality Formation of Satsuma Mandarin and the Law of Water Absorption and Transportation in Tree After Re-watering [J]. Acta Horticulturae Sinica, 2022, 49(1): 11-22. |
| [13] | XIE Siyi, ZHOU Chengzhe, ZHU Chen, ZHAN Dongmei, CHEN Lan, WU Zuchun, LAI Zhongxiong, GUO Yuqiong. Genome-wide Identification and Expression Analysis of CsTIFY Transcription Factor Family Under Abiotic Stress and Hormone Treatments in Camellia sinensis [J]. Acta Horticulturae Sinica, 2022, 49(1): 100-116. |
| [14] | HE Yan, SUN Yanli, ZHAO Fangfang, DAI Hongjun. Effect of Exogenous Brassinolides Treatment on Sugar Metabolism of Merlot Grape Berries [J]. Acta Horticulturae Sinica, 2022, 49(1): 117-128. |
| [15] | LI Maofu, YANG Yuan, WANG Hua, FAN Youwei, SUN Pei, JIN Wanmei. Identification and Analysis of Self Incompatibility S-RNase in Rose [J]. Acta Horticulturae Sinica, 2022, 49(1): 157-165. |
| Viewed | ||||||
|
Full text |
|
|||||
|
Abstract |
|
|||||
Copyright © 2012 Acta Horticulturae Sinica 京ICP备10030308号-2 国际联网备案号 11010802023439
Tel: 010-82109523 E-Mail: yuanyixuebao@126.com
Support by: Beijing Magtech Co.Ltd