
Acta Horticulturae Sinica ›› 2022, Vol. 49 ›› Issue (7): 1557-1570.doi: 10.16420/j.issn.0513-353x.2021-0376
• Research Notes • Previous Articles Next Articles
ZHANG Qiuyue1, LIU Changlai1,2,*( ), YU Xiaojing1, YANG Jiading1, FENG Chaonian1,*(
), YU Xiaojing1, YANG Jiading1, FENG Chaonian1,*( )
)
Received:2022-03-25
Revised:2022-04-27
Online:2022-07-25
Published:2022-07-29
Contact:
LIU Changlai,FENG Chaonian
E-mail:clc2012@njfu.edu.cn;fcn@njfu.edu.cn
CLC Number:
ZHANG Qiuyue, LIU Changlai, YU Xiaojing, YANG Jiading, FENG Chaonian. Screening of Reference Genes for Differentially Expressed Genes in Pyrus betulaefolia Plant Under Salt Stress by qRT-PCR[J]. Acta Horticulturae Sinica, 2022, 49(7): 1557-1570.
Add to citation manager EndNote|Ris|BibTeX
URL: https://www.ahs.ac.cn/EN/10.16420/j.issn.0513-353x.2021-0376
| 引物名称 Primer names | 基因名称 Gene name | 基因编号 Gene ID | 片段长度/bp Gene length | 来源 Source | 变异系数/% CV | 引物序列 Primer sequence |
|---|---|---|---|---|---|---|
| EF1a-1 | EF1a-1 | Chr3.g19898 | 168 | 本研究设计 Designed in this study | 18.0 | F:CATCGAGAGGTTCGAGAAGG R:CCGGGAGCATCAATAACAGT |
| EF1a-2A | EF1a-2 | Chr4.g38173 | 167 | 蒲小秋 等, | 26.0 | F:GGTGTGAAGCAGATGATTTG R:TCACCCTCAAACCCAGATAT |
| EF1a-2B | EF1a-2 | Chr4.g38173 | 201 | 本研究设计 Designed in this study | 26.0 | F:AGGTCCACCAACCTTGACTG R:TGGACCAAAAGTGACAACCA |
| EF2 | EF2 | Chr5.g06899 | 179 | 本研究设计 Designed in this study | 8.7 | F:CCCAAGAGATGATCCCAAGA R:ACCCAGCAACAACAGAATCC |
| Actin2 | Actin2 | Chr15.g01351 | 101 | 蒲小秋 等, | 13.8 | F:CTTCAATGTGCCTGCCATGT R:TCACACCATCACCAGAGTCC |
| GAPDH-1 | GAPDH-1 | Chr16.g30426 | 171 | 本研究设计 Designed in this study | 17.9 | F:GTTCGTTGTTGGTGTGAACG R:GTCTTTTGGGTGGCAGTGAT |
| GAPDH-2 | GAPDH-2 | Chr13.g23532 | 123 | 张雪 等, | 11.2 | F:GAGGGTCTCATGACCACAGT R:TCCAGTGCTGCTAGGAATGA |
| TUBB-A | TUBB | Chr5.g06472 | 212 | 张雪 等, | 20.6 | F:CTGCTGTGTTCCGTGGTAAG R:CTGCTCGCTAACTCTCCTGA |
| TUBB-B | TUBB | Chr5.g06472 | 237 | 本研究设计 Designed in this study | 20.6 | F:ACCCGATAACTTCGTGTTCG R:AACATCATTCGATCCGGGTA |
| UBQE | UBQ | Chr4.g40121 | 140 | 本研究设计 Designed in this study | 8.8 | F:GGCAGAACTGCCTGCTAATC R:CGGTTTTGCTCGATAAGCTC |
| HKT | HKT | Chr16.g29024 | 197 | 本研究设计 Designed in this study | 63.9 | F:TGGGCTACTGTCATTTGCTG R:AACGGATTCACCGCTATGTC |
Table 1 Primer sequences of candidate reference genes and target gene
| 引物名称 Primer names | 基因名称 Gene name | 基因编号 Gene ID | 片段长度/bp Gene length | 来源 Source | 变异系数/% CV | 引物序列 Primer sequence |
|---|---|---|---|---|---|---|
| EF1a-1 | EF1a-1 | Chr3.g19898 | 168 | 本研究设计 Designed in this study | 18.0 | F:CATCGAGAGGTTCGAGAAGG R:CCGGGAGCATCAATAACAGT |
| EF1a-2A | EF1a-2 | Chr4.g38173 | 167 | 蒲小秋 等, | 26.0 | F:GGTGTGAAGCAGATGATTTG R:TCACCCTCAAACCCAGATAT |
| EF1a-2B | EF1a-2 | Chr4.g38173 | 201 | 本研究设计 Designed in this study | 26.0 | F:AGGTCCACCAACCTTGACTG R:TGGACCAAAAGTGACAACCA |
| EF2 | EF2 | Chr5.g06899 | 179 | 本研究设计 Designed in this study | 8.7 | F:CCCAAGAGATGATCCCAAGA R:ACCCAGCAACAACAGAATCC |
| Actin2 | Actin2 | Chr15.g01351 | 101 | 蒲小秋 等, | 13.8 | F:CTTCAATGTGCCTGCCATGT R:TCACACCATCACCAGAGTCC |
| GAPDH-1 | GAPDH-1 | Chr16.g30426 | 171 | 本研究设计 Designed in this study | 17.9 | F:GTTCGTTGTTGGTGTGAACG R:GTCTTTTGGGTGGCAGTGAT |
| GAPDH-2 | GAPDH-2 | Chr13.g23532 | 123 | 张雪 等, | 11.2 | F:GAGGGTCTCATGACCACAGT R:TCCAGTGCTGCTAGGAATGA |
| TUBB-A | TUBB | Chr5.g06472 | 212 | 张雪 等, | 20.6 | F:CTGCTGTGTTCCGTGGTAAG R:CTGCTCGCTAACTCTCCTGA |
| TUBB-B | TUBB | Chr5.g06472 | 237 | 本研究设计 Designed in this study | 20.6 | F:ACCCGATAACTTCGTGTTCG R:AACATCATTCGATCCGGGTA |
| UBQE | UBQ | Chr4.g40121 | 140 | 本研究设计 Designed in this study | 8.8 | F:GGCAGAACTGCCTGCTAATC R:CGGTTTTGCTCGATAAGCTC |
| HKT | HKT | Chr16.g29024 | 197 | 本研究设计 Designed in this study | 63.9 | F:TGGGCTACTGTCATTTGCTG R:AACGGATTCACCGCTATGTC |
| 引物名称 Primer name | 斜率(K) Slope | 决定系数 R2 | 扩增效率/% Amplification efficiency |
|---|---|---|---|
| EF1a-1 | -3.43 | 0.998 | 95.66 |
| EF1a-2A | -3.15 | 0.999 | 107.96 |
| EF1a-2B | -3.54 | 0.999 | 91.82 |
| EF2 | -3.43 | 0.998 | 95.66 |
| Actin2 | -3.43 | 0.998 | 95.66 |
| GAPDH-1 | -3.35 | 0.999 | 98.77 |
| GAPDH-2 | -3.21 | 0.999 | 104.95 |
| TUBB-A | -3.32 | 0.996 | 100.16 |
| TUBB-B | -3.05 | 0.989 | 112.99 |
| UBQE | -3.52 | 0.999 | 92.49 |
Table 2 The vector characteristics of candidate internal reference genes and amplification efficiency of primer pairs
| 引物名称 Primer name | 斜率(K) Slope | 决定系数 R2 | 扩增效率/% Amplification efficiency |
|---|---|---|---|
| EF1a-1 | -3.43 | 0.998 | 95.66 |
| EF1a-2A | -3.15 | 0.999 | 107.96 |
| EF1a-2B | -3.54 | 0.999 | 91.82 |
| EF2 | -3.43 | 0.998 | 95.66 |
| Actin2 | -3.43 | 0.998 | 95.66 |
| GAPDH-1 | -3.35 | 0.999 | 98.77 |
| GAPDH-2 | -3.21 | 0.999 | 104.95 |
| TUBB-A | -3.32 | 0.996 | 100.16 |
| TUBB-B | -3.05 | 0.989 | 112.99 |
| UBQE | -3.52 | 0.999 | 92.49 |
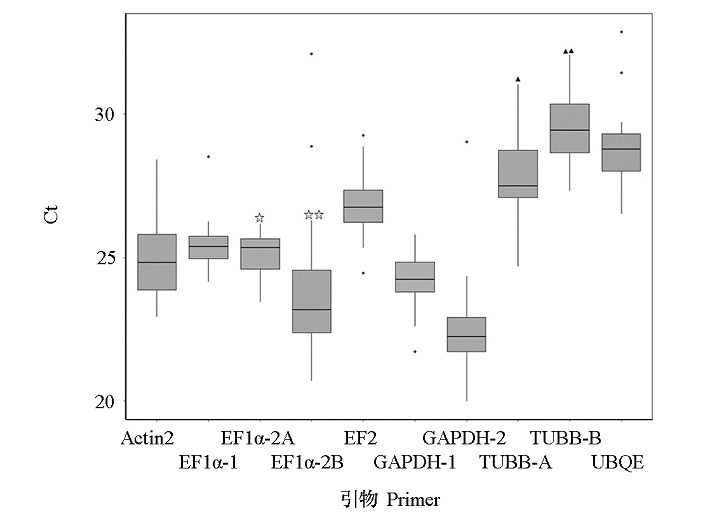
Fig. 4 Ct value distribution of amplification products by primer pairs of candidate internal reference genes in 24 leaf samples☆ and ▲ represent the significant difference between PCR products using the primers of EF1α-2A and EF1α-2B,TUBB-A and TUBB-B,at P < 0.01 level,respectively. The little black spot represent the discrete value.
| 引物 Primer | 几何平均值 Geo mean | 算术平均值 AM | 最小值 Min | 最大值 Max | 相关系数 r | 标准偏差 SD | 变异系数/% CV |
|---|---|---|---|---|---|---|---|
| GAPDH-1 | 24.24 | 24.26 | 21.71 | 25.79 | 0.870 | 0.75 | 3.09 |
| EF2 | 26.83 | 26.85 | 24.46 | 29.27 | 0.806 | 0.89 | 3.32 |
| UBQE | 28.81 | 28.84 | 26.52 | 32.87 | 0.697 | 0.86 | 2.98 |
| EF1α-1 | 25.39 | 25.40 | 24.15 | 28.52 | 0.688 | 0.56 | 2.21 |
| EF1α-2A | 25.18 | 25.19 | 23.44 | 26.18 | 0.640 | 0.57 | 2.27 |
| TUBB-B | 29.56 | 29.58 | 27.32 | 32.08 | 0.817 | 1.02 | 3.45 |
| TUBB-A | 27.79 | 27.82 | 24.68 | 31.04 | 0.890 | 1.04 | 3.72 |
| GAPDH-2 | 22.47 | 22.53 | 19.99 | 29.04 | 0.912 | 1.06 | 4.69 |
| Actin2 | 25.01 | 25.05 | 22.93 | 28.41 | 0.792 | 1.27 | 5.05 |
| EF1α-2B | 23.71 | 23.83 | 20.70 | 32.10 | 0.648 | 1.75 | 7.34 |
Table 3 Analysis results of expression stability of candidate reference genes by BestKeeper
| 引物 Primer | 几何平均值 Geo mean | 算术平均值 AM | 最小值 Min | 最大值 Max | 相关系数 r | 标准偏差 SD | 变异系数/% CV |
|---|---|---|---|---|---|---|---|
| GAPDH-1 | 24.24 | 24.26 | 21.71 | 25.79 | 0.870 | 0.75 | 3.09 |
| EF2 | 26.83 | 26.85 | 24.46 | 29.27 | 0.806 | 0.89 | 3.32 |
| UBQE | 28.81 | 28.84 | 26.52 | 32.87 | 0.697 | 0.86 | 2.98 |
| EF1α-1 | 25.39 | 25.40 | 24.15 | 28.52 | 0.688 | 0.56 | 2.21 |
| EF1α-2A | 25.18 | 25.19 | 23.44 | 26.18 | 0.640 | 0.57 | 2.27 |
| TUBB-B | 29.56 | 29.58 | 27.32 | 32.08 | 0.817 | 1.02 | 3.45 |
| TUBB-A | 27.79 | 27.82 | 24.68 | 31.04 | 0.890 | 1.04 | 3.72 |
| GAPDH-2 | 22.47 | 22.53 | 19.99 | 29.04 | 0.912 | 1.06 | 4.69 |
| Actin2 | 25.01 | 25.05 | 22.93 | 28.41 | 0.792 | 1.27 | 5.05 |
| EF1α-2B | 23.71 | 23.83 | 20.70 | 32.10 | 0.648 | 1.75 | 7.34 |
| 评价方法 Method | 等级Ranking order(Better--Good--Average) | |||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| 1 | 2 | 3 | 4 | 5 | 6 | 7 | 8 | 9 | 10 | |
| delta Ct | GΑPDH-1 | TUBB-Α | TUBB-B | EF2 | EF1α-1 | EF1α-2Α | UBQE | GΑPDH-2 | Αctin2 | EF1α-2B |
| BestKeeper | EF1α-1 | EF1α-2Α | GΑPDH-1 | UBQE | EF2 | TUBB-B | TUBB-Α | GΑPDH-2 | Αctin2 | EF1α-2B |
| NormFinder | GΑPDH-1 | TUBB-Α | EF2 | TUBB-B | EF1α-1 | EF1α-2Α | UBQE | GΑPDH-2 | Αctin2 | EF1α-2B |
| geNorm | GΑPDH-1 TUBB-Α | TUBB-B | EF1α-2Α | EF1α-1 | EF2 | Αctin2 | GΑPDH-2 | UBQE | EF1α-2B | |
| 综合评价 Comprehensive | GΑPDH-1 | TUBB-Α | EF1α-1 | TUBB-B | EF1α-2Α | EF2 | UBQE | GΑPDH-2 | Αctin2 | EF1α-2B |
Table 4 Expression stability of candidate reference genes analysed by RefFinder
| 评价方法 Method | 等级Ranking order(Better--Good--Average) | |||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| 1 | 2 | 3 | 4 | 5 | 6 | 7 | 8 | 9 | 10 | |
| delta Ct | GΑPDH-1 | TUBB-Α | TUBB-B | EF2 | EF1α-1 | EF1α-2Α | UBQE | GΑPDH-2 | Αctin2 | EF1α-2B |
| BestKeeper | EF1α-1 | EF1α-2Α | GΑPDH-1 | UBQE | EF2 | TUBB-B | TUBB-Α | GΑPDH-2 | Αctin2 | EF1α-2B |
| NormFinder | GΑPDH-1 | TUBB-Α | EF2 | TUBB-B | EF1α-1 | EF1α-2Α | UBQE | GΑPDH-2 | Αctin2 | EF1α-2B |
| geNorm | GΑPDH-1 TUBB-Α | TUBB-B | EF1α-2Α | EF1α-1 | EF2 | Αctin2 | GΑPDH-2 | UBQE | EF1α-2B | |
| 综合评价 Comprehensive | GΑPDH-1 | TUBB-Α | EF1α-1 | TUBB-B | EF1α-2Α | EF2 | UBQE | GΑPDH-2 | Αctin2 | EF1α-2B |
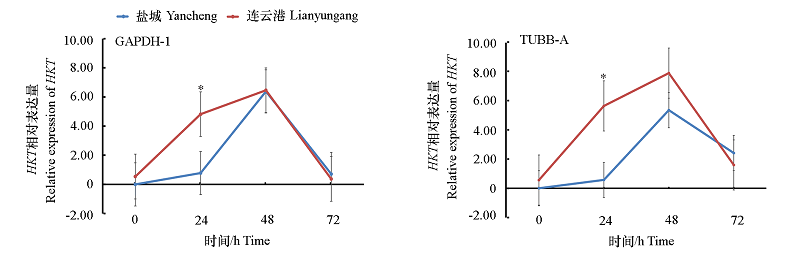
Fig. 6 Relative expression of HKT gene measured by using GAPDH-1 and TUBB-A as primer pairs of internal reference genes respectively* represent HKT expression significant difference of Pyrus betulaefolia Bunge. from Lianyungang and Yancheng at P < 0.05.
| [1] |
Andersen C L, Jensen J L, Ørntoft T F. 2004. Normalization of real-time quantitative reverse transcription-PCR data:a model-based variance estimation approach to identify genes suited for normalization. Applied to Bladder and Colon Cancer Data Sets. Cancer Research, 64 (15):5245.
doi: 10.1158/0008-5472.CAN-04-0496 URL |
| [2] |
Butterfield D A, Hardas S S, Lange M L B. 2010. Oxidatively modified glyceraldehyde-3-phosphate dehydrogenase(GAPDH)and Alzheimer's Disease:many pathways to neurodegeneration. Journal of Alzheimer's Disease, 20:369-393.
doi: 10.3233/JAD-2010-1375 pmid: 20164570 |
| [3] |
Chen J, Li X, Wang D, Li L, Zhou H, Liu Z, Wu J, Wang P, Jiang X, Fabrice M R, Zhang S, Wu J. 2015. Identification and testing of reference genes for gene expression analysis in pollen of Pyrus bretschneideri. Scientia Horticulturae, 190:43-56.
doi: 10.1016/j.scienta.2015.04.010 URL |
| [4] | Chen Yangyang, Wu Xiao, Gu Chao, Yin Hao, Zhang Shaoling. 2018. Selection of reference genes in qRT-PCR of pear‘Dangshansuli’. China Fruits,(1):16-22. (in Chinese) |
| 陈杨杨, 吴潇, 谷超, 殷豪, 张绍铃. 2018. ‘砀山酥梨’实时荧光定量PCR内参基因的筛选. 中国果树,(1):16-22 | |
| [5] |
Dominguez R, Holmes K C. 2011. Actin structure and function. Annual Review of Biophysics, 40 (1):169-186.
doi: 10.1146/annurev-biophys-042910-155359 URL |
| [6] |
Dong X, Wang Z, Tian L, Zhang Y, Qi D, Huo H, Xu J, Li Z, Liao R, Shi M, Wahocho S A, Liu C, Zhang S, Tian Z, Cao Y. 2019. De novo assembly of a wild pear(Pyrus betuleafolia)genome. Plant Biotechnology Journal, 18 (2):581-595.
doi: 10.1111/pbi.13226 URL |
| [7] | Dubbelhuis P F, Meijer A J. 2002. Chapter 14 - Amino acid-dependent signal transduction. Sensing. Amsterdam:Elsevier:207-219. |
| [8] | Gao Haiming, Zhao Yanyan. 2020. Application progress of next generation and the third generation sequencing technology in genetic diagnosis. Current Biotechnology, 10 (6):646-654. (in Chinese) |
| 高海明, 赵彦艳. 2020. 二代及三代测序技术在遗传学诊断中的应用进展. 生物技术进展, 10 (6):646-654. | |
| [9] | Hou Lei, Chen Longjun. 2011. Effects of salt stress on epidermal cell expansion in leaves of Arabidopsis thaliana. Journal of Anhui Agricultural Sciences, 39 (13):7615-7616. (in Chinese) |
| 侯蕾, 陈龙俊. 2011. 盐胁迫对拟南芥叶片和下表皮细胞大小的影响. 安徽农业科学, 39 (13):7615-7616. | |
| [10] | Hu Lisong, Wu Siting, Duan Xingshuai, Cen Yi, Su Yuefeng, Fan Rui, Wu Baoduo, Hao Chaoyun. 2020. The identification of internal control genes based on genome and transcriptome data in black pepper(Piper nigrum L.). Chinese Journal of Tropical Crops, 41 (10):2120-2129. (in Chinese) |
| 胡丽松, 吴思婷, 段兴帅, 岑怡, 苏岳峰, 范睿, 伍宝朵, 郝朝运. 2020. 基于基因组和转录组数据的胡椒内参基因鉴定. 热带作物学报, 41 (10):2120-2129. | |
| [11] | Jiang Lei, Li Huanyong, Zhang Qin, Zhang Huilong, Qiao Yanhui, Zhang Huaxin, Yang Xiuyan. 2020. Effects of arbuscular mycorrhiza fungi on the growth and physiological metabolism of Pyrus betulaefolia Bunge seedlings under saline-alkaline stress. Journal of Nanjing Forestry University(Natural Science Edition), 44 (6):152-160. (in Chinese) |
| 姜磊, 李焕勇, 张芹, 张会龙, 乔艳辉, 张华新, 杨秀艳. 2020. AM真菌对盐碱胁迫下杜梨幼苗生长与生理代谢的影响. 南京林业大学学报(自然科学版), 44 (6):152-160. | |
| [12] |
Lehmann S G, Bourgoin-Voillard S, Seve M, Rachidi W, Ayala A. 2017. Tubulin Beta-3 Chain as a new candidate protein biomarker of human skin aging:a preliminary study. Oxidative Medicine and Cellular Longevity,doi: 0.1155/2017/5140360.
doi: 0.1155/2017/5140360 URL |
| [13] |
Li H, Lin J, Yang Q, Li X, Chang Y. 2017. Comprehensive analysis of differentially expressed genes under salt stress in pear(Pyrus betulaefolia)using RNA-Seq. Plant Growth Regulation, 82 (3):409-420.
doi: 10.1007/s10725-017-0266-3 URL |
| [14] | Li Yongping, Ye Xinru, Wang Bin, Chen Meidong, Liu Jianting, Zhu Haisheng, Wen Qingfang. 2021. Cloning and selection evaluation of reference gene for quantitative real-time PCR in Hibiscus esculentus L. Journal of Nuclear Agricultural Sciences, 35 (1):60-71. (in Chinese) |
|
李永平, 叶新如, 王彬, 陈敏氡, 刘建汀, 朱海生, 温庆放. 2021. 黄秋葵实时荧光定量PCR内参基因的克隆与筛选评价. 核农学报, 35 (1):60-71.
doi: 10.11869/j.issn.100-8551.2021.01.0060 |
|
| [15] | Liu Xiaofei, Yu Bo, Huang Lili, Sun Yingbo. 2020. Screening and validation of reference genes of Camellia azalea by quantitative real-time PCR. Guangdong Agricultural Sciences, 47 (12):203-211. (in Chinese) |
| 刘小飞, 于波, 黄丽丽, 孙映波. 2020. 杜鹃红山茶实时定量PCR内参基因筛选及验证. 广东农业科学, 47 (12):203-211. | |
| [16] | Ma Lina, Yang Jinbo, Ding Yifei, Li Yingkang. 2019. Research progress on three generations sequencing technology and its application. China Animal Husbandry & Veterinary Medicine, 46 (8):2246-2256. (in Chinese) |
| 马丽娜, 杨进波, 丁逸菲, 李颖康. 2019. 三代测序技术及其应用研究进展. 中国畜牧兽医, 46 (8):2246-2256. | |
| [17] | Mou Lifei, Yu Xiaojing, Zhang Qiuyue, Feng Chaonian. 2020. Salt tolerance of four half-sib families of Pyrus betulaefolia Bunge from coastal areas. Journal of Nanjing Forestry University(Natural Science Edition), 44 (5):157-166. (in Chinese) |
| 缪李飞, 于晓晶, 张秋悦, 封超年. 2020. 4个杜梨半同胞家系苗期耐盐性分析. 南京林业大学学报(自然科学版), 44 (5):157-166. | |
| [18] |
Pfaffl M W, Tichopad A, Prgomet C, Neuvians T P. 2004. Determination of stable housekeeping genes,differentially regulated target genes and sample integrity:BestKeeper-Excel-based tool using pair-wise correlations. Biotechnology Letters, 26 (6):509-515.
doi: 10.1023/B:BILE.0000019559.84305.47 URL |
| [19] | Pu Xiaoqiu, Tian Jia, Li Jiang, Zhang Yan, Li Peng, Tan Weiming, Jing Chunzhi. 2020. Analysis on expression stability of internal reference genes at cell division stage of pear fruits. Nonwood Forest Research, 38 (1):66-74. (in Chinese) |
| 蒲小秋, 田嘉, 李疆, 张艳, 李鹏, 覃伟铭, 井春芝. 2020. 梨果实细胞分裂期内参基因表达稳定性分析. 经济林研究, 38 (1):66-74. | |
| [20] | Qiao Yonggang, Wang Yongfei, Cao Yaping, He Jiaxin, Jia Mengjun, Li Zheng, Zhang Xinrui, Song Yun. 2020. Reference genes selection and related genes expression analysis under low and high temperature stress in Taraxacum officinale. Acta Horticulturae Sinica, 47 (6):1153-1164. (in Chinese) |
| 乔永刚, 王勇飞, 曹亚萍, 贺嘉欣, 贾孟君, 李政, 张鑫瑞, 宋芸. 2020. 药用蒲公英低温和高温胁迫下内参基因筛选与相关基因表达分析. 园艺学报, 47 (6):1153-1164. | |
| [21] |
Roger A J, Sandblom O, Doolittle W F, Philippe H. 1999. An evaluation of elongation factor 1 alpha as a phylogenetic marker for eukaryotes. Molecular Biology and Evolution, 16 (2):218-233.
pmid: 10028289 |
| [22] |
Silver N, Best S, Jiang J, Thein S L. 2006. Selection of housekeeping genes for gene expression studies in human reticulocytes using real-time PCR. BMC Molecular Biology, 7 (1):33.
doi: 10.1186/1471-2199-7-33 URL |
| [23] | Stone S L. 2016. Chapter 26-ubiquitination of plant transcription factors, Boston: Academic Press:395-409. |
| [24] |
Umadevi P, Suraby E J, Anandaraj M, Nepolean T. 2019. Identification of stable reference gene for transcript normalization in black pepper-Phytophthora capsici pathosystem. Physiology and Molecular Biology of Plants, 25 (4):945-952.
doi: 10.1007/s12298-019-00653-9 pmid: 31402818 |
| [25] | Vandesompele J, De Preter K, Pattyn F, Poppe B, Van Roy N, De Paepe A, Speleman F. 2002. Accurate normalization of real-time quantitative RT-PCR data by geometric averaging of multiple internal control genes. Genome Biology, 3 (7):h31-h34. |
| [26] |
Wang Hao, Cai Qizhong, Liu Lu, Yang Quan, Zhou Liangyun. 2021. Reference gene screening for real-time quantitative PCR in Polygonum multiflorum. China Journal of Chinese Materia Medica, 46 (1):80-85. (in Chinese)
doi: 10.19540/j.cnki.cjcmm.20201024.103 pmid: 33645055 |
|
王浩, 蔡启忠, 刘露, 杨全, 周良云. 2021. 何首乌实时荧光定量PCR内参基因筛选. 中国中药杂志, 46 (1):80-85.
pmid: 33645055 |
|
| [27] |
Wu H. 2018. Plant salt tolerance and Na+ sensing and transport. The Crop Journal, 6 (3):215-225.
doi: 10.1016/j.cj.2018.01.003 URL |
| [28] |
Xie F, Xiao P, Chen D, Xu L, Zhang B. 2012. miRDeepFinder:a miRNA analysis tool for deep sequencing of plant small RNAs. Plant Mol Biol, 80:75-84.
doi: 10.1007/s11103-012-9885-2 URL |
| [29] | Yang Ting, Xue Zhenzhen, Li Na, Lang Xiaoan, Li Lingfei, Zhong Chunmei. 2021. Reference genes selection and validation in Begonia masoniana leaves of different developmental stages. Acta Horticulturae Sinica, 48 (11):2251-2261. (in Chinese) |
| 杨婷, 薛珍珍, 李娜, 郎校安, 李凌飞, 钟春梅. 2021. 铁十字秋海棠斑叶发育过程内参基因筛选及验证. 园艺学报, 48 (11):2251-2261. | |
| [30] | Ye Bihuan, Song Qiyan, Chen Youwu, Hu Chuanjiu, Du Guojian, Liao Rongjun, Li Haibo. 2020. Selection and validation of internal reference genes for qPCR in Polygonatum cyrtonema tubers at different development stages and in response to abiotic stress. China Journal of Chinese Materia Medica, 45 (24):5967-5975. (in Chinese) |
| 杨阳, 叶碧欢, 宋其岩, 陈友吾, 胡传久, 杜国坚, 廖荣俊, 李海波. 2020. 多花黄精块茎发育和胁迫条件下qPCR内参基因的筛选与验证. 中国中药杂志, 45 (24):5967-5975. | |
| [31] | Zhang Xue, Wang Li, Ju Fei, Yang Shengjun. 2019. Reference gene screening for real-time quantitative PCR in red pear(Pyrus pyrifolia). Journal of Agricultural Biotechnology, 27 (2):361-370. (in Chinese) |
| 张雪, 王荔, 瞿飞, 杨胜俊. 2019. 红梨实时荧光定量PCR内参基因的筛选. 农业生物技术学报, 27 (2):361-370. | |
| [32] | Zhang Zhengrui, Zhang Yaohua, Wang Qiushi, Yu Hui, Yang Suxin. 2020. Screening and validation of reference genes for real-time quantitative PCR in soybean. Plant Physiology Communications, 56 (9):1963-1973. (in Chinese) |
| 张芷睿, 张耀华, 王秋实, 于慧, 杨素欣. 2020. 大豆实时荧光定量PCR内参基因的筛选与验证. 植物生理学报, 56 (9):1963-1973. |
| [1] | WANG Hong, YANG Wangli, LIN Jing, YANG Qingsong, LI Xiaogang, SHENG Baolong, CHANG Youhong. Comparative Metabolic and Transcriptomic Analysis of Ripening Fruit in Pear Cultivars of‘Sucui 1’‘Cuiguan’and‘Huasu’ [J]. Acta Horticulturae Sinica, 2022, 49(3): 493-508. |
| [2] | HOU Tianze, YI Shuangshuang, ZHANG Zhiqun, WANG Jian, LI Chonghui. Selection and Validation of Reference Genes for RT-qPCR in Phalaenopsis- type Dendrobium Hybrid [J]. Acta Horticulturae Sinica, 2022, 49(11): 2489-2501. |
| [3] | ZHANG Zhiqiang, LU Shixiong, MA Zonghuan, LI Yanbiao, GAO Caixia, CHEN Baihong, MAO Juan. Bioinformatic Identification and Expression Analysis of Candidate Genes of LIM Protein Family in Strawberry Under Abiotic Stresses [J]. Acta Horticulturae Sinica, 2021, 48(8): 1485-1503. |
| [4] | ZHU Junfei, LI Xin, DONG Kangting, TANG Zhifei, BIAN Xiuju, WANG Lihong, LI Huibin, SUN Xinbo. Effect of Creeping Bentgrass Small Heat Shock Protein AsHSP26.8a on Transgenic Photosynthesis in Arabidopsis thaliana [J]. Acta Horticulturae Sinica, 2021, 48(8): 1619-1625. |
| [5] | JIA Bing, GUO Guoling, WANG Youyu, YE Zhenfeng, LIU Li, LIU Pu, HENG Wei, ZHU Liwu. Studies on the Regreening Mechanism of the Iron-deficiency Chlorosis Leaves induced by GA3 in‘Whangkeumbae’(Pyrus pyrifolia Nakai) [J]. Acta Horticulturae Sinica, 2021, 48(2): 254-264. |
| [6] | MA Lulin, DUAN Qing, CUI Guangfen, DU Wenwen, JIA Wenjie, WANG Xiangning, WANG Jihua, CHEN Fadi. Selection and Validation of Reference Genes for qRT-PCR Analysis of the Correlated Genes in Flower Pigments Biosynthesis Pathway of Anemone obtusiloba [J]. Acta Horticulturae Sinica, 2021, 48(2): 377-388. |
| [7] | JIANG Mengqi, XUE Xiaodong, SU Liyao, CHEN Yan, ZHANG Shuting, LI Xiaofei, WANG Peiyu, ZHANG Zihao, LAI Zhongxiong, LIN Yuling. Genome-wide Identification and Expression Analysis of TCP Family in Dimocarpus longan [J]. Acta Horticulturae Sinica, 2021, 48(12): 2481-2496. |
| [8] | YANG Ting, XUE Zhenzhen, LI Na, LANG Xiaoan, LI Lingfei, ZHONG Chunmei. Reference Genes Selection and Validation in Begonia masoniana Leaves of Different Developmental Stages [J]. Acta Horticulturae Sinica, 2021, 48(11): 2251-2261. |
| [9] | CHENG Shaoyu,Xuan Lingjuan,DONG Bin,GU Cuihua,SHEN Yamei*,ZHANG Mingru,DAI Mengyi,WANG Zhuowei,ZHANG Yingjia,and LU Danying. Identification of Differential Metabolic Pathways and Key Regulatory Genes in the Two Flower Bud Differentiation Processes of Magnolia liliiflora [J]. ACTA HORTICULTURAE SINICA, 2020, 47(8): 1490-1504. |
| [10] | MAO Juan,LU Shixiong,WANG Ping,MA Zonghuan,LIU Tao,HE Honghong,NAI Guojie,ZHANG Zhiqiang,and CHEN Baihong*. Analysis of Gene Characteristic and Expression Pattern of LIM Family in Grape [J]. ACTA HORTICULTURAE SINICA, 2020, 47(7): 1277-1288. |
| [11] | ZHOU Tao1,2,WANG Juan2,WANG Lulu1,WANG Baike2,HU Jiahui1,2,LAN Haiyan1,*,and YU Qinghui2,*. Cloning and Prokaryotic Expression Analysis of a Transcription Factor Gene SlWRKY16 in Tomato [J]. ACTA HORTICULTURAE SINICA, 2020, 47(7): 1312-1322. |
| [12] | QIAO Yonggang*,WANG Yongfei,CAO Yaping,HE Jiaxin,JIA Mengjun,LI Zheng,ZHANG Xinrui,and SONG Yun. Reference Genes Selection and Related Genes Expression Analysis under Low and High Temperature Stress in Taraxacum officinale [J]. ACTA HORTICULTURAE SINICA, 2020, 47(6): 1153-1164. |
| [13] | FU Yajuan1,2,CHEN Xiating1,QIAO Jie1,2,WANG Jing1,2,LI Wenjing1,and HOU Xiaoqiang1,2,*. Molecular Cloning and Expression Characterization of Cyclophilin Gene(DoCyP)in Dendrobium officinale [J]. ACTA HORTICULTURAE SINICA, 2020, 47(3): 581-589. |
| [14] | LIU Tao,WANG Pingping,HE Honghong,LIANG Guoping,LU Shixiong,CHEN Baihong,and MAO Juan*. Identification and Expression Analysis of CIPK Gene Family in Strawberry [J]. ACTA HORTICULTURAE SINICA, 2020, 47(1): 127-142. |
| [15] | LU Shixiong,WANG Ping,HE Honghong,LIANG Guoping,MA Zonghuan,QIAO Yali,WU Yuxia,CHEN Baihong*,and MAO Juan*. Bioinformatics Identification and Expression Analysis of Grape Trihelix Transcription Factor Family [J]. ACTA HORTICULTURAE SINICA, 2019, 46(7): 1257-1269. |
| Viewed | ||||||
|
Full text |
|
|||||
|
Abstract |
|
|||||
Copyright © 2012 Acta Horticulturae Sinica 京ICP备10030308号-2 国际联网备案号 11010802023439
Tel: 010-82109523 E-Mail: yuanyixuebao@126.com
Support by: Beijing Magtech Co.Ltd