
Acta Horticulturae Sinica ›› 2022, Vol. 49 ›› Issue (2): 365-377.doi: 10.16420/j.issn.0513-353x.2021-0574
• Research Papers • Previous Articles Next Articles
DENG Jiao1,2, SU Mengyue1, LIU Xuelian1, OU Kefang3, HU Zhengrong1, YANG Pingfang1,*( )
)
Received:2021-06-18
Revised:2021-09-06
Online:2022-02-25
Published:2022-02-28
Contact:
YANG Pingfang
E-mail:yangpf@hubu.edu.cn
CLC Number:
DENG Jiao, SU Mengyue, LIU Xuelian, OU Kefang, HU Zhengrong, YANG Pingfang. Transcriptome Analysis Revealed the Formation Mechanism of Floral Color of Lotus‘Dasajin’with Bicolor Petal[J]. Acta Horticulturae Sinica, 2022, 49(2): 365-377.
Add to citation manager EndNote|Ris|BibTeX
URL: https://www.ahs.ac.cn/EN/10.16420/j.issn.0513-353x.2021-0574
| 样本 Sample | 原始序列 Raw reads | 过滤后序列 Clean reads | 过滤后的碱基/Gb Clean bases | 错误率/% Error rate | Q20/% | Q30/% | GC/% |
|---|---|---|---|---|---|---|---|
| 红色1 Red 1 | 91 323 664 | 90 563 834 | 13.58 | 0.03 | 97.97 | 93.90 | 45.08 |
| 红色2 Red 2 | 73 024 058 | 72 316 724 | 10.85 | 0.03 | 97.99 | 93.95 | 45.13 |
| 红色3 Red 3 | 56 698 476 | 56 146 640 | 8.42 | 0.03 | 97.94 | 93.82 | 45.11 |
| 白色1 White 1 | 59 612 084 | 59 030 858 | 8.85 | 0.02 | 98.11 | 94.25 | 45.19 |
| 白色2 White 2 | 54 401 662 | 53 731 038 | 8.06 | 0.03 | 97.93 | 93.81 | 45.28 |
| 白色3 White 3 | 49 534 162 | 49 072 084 | 7.36 | 0.03 | 98.05 | 94.09 | 45.28 |
Table 1 The information form of quality of transcription data of red and white petals
| 样本 Sample | 原始序列 Raw reads | 过滤后序列 Clean reads | 过滤后的碱基/Gb Clean bases | 错误率/% Error rate | Q20/% | Q30/% | GC/% |
|---|---|---|---|---|---|---|---|
| 红色1 Red 1 | 91 323 664 | 90 563 834 | 13.58 | 0.03 | 97.97 | 93.90 | 45.08 |
| 红色2 Red 2 | 73 024 058 | 72 316 724 | 10.85 | 0.03 | 97.99 | 93.95 | 45.13 |
| 红色3 Red 3 | 56 698 476 | 56 146 640 | 8.42 | 0.03 | 97.94 | 93.82 | 45.11 |
| 白色1 White 1 | 59 612 084 | 59 030 858 | 8.85 | 0.02 | 98.11 | 94.25 | 45.19 |
| 白色2 White 2 | 54 401 662 | 53 731 038 | 8.06 | 0.03 | 97.93 | 93.81 | 45.28 |
| 白色3 White 3 | 49 534 162 | 49 072 084 | 7.36 | 0.03 | 98.05 | 94.09 | 45.28 |
| 样本 Sample | FPKM | ||||
|---|---|---|---|---|---|
| 0 ~ 1 | 1 ~ 3 | 3 ~ 15 | 15 ~ 16 | > 60 | |
| 红色1 Red 1 | 11 710(41.43%) | 2 957(10.46%) | 6 609(23.38%) | 5 017(17.75%) | 1 973(6.98%) |
| 红色2 Red 2 | 11 028(39.02%) | 3 038(10.75%) | 6 778(23.98%) | 5 313(18.80%) | 2 109(7.46%) |
| 红色3 Red 3 | 11 077(39.19%) | 2 986(10.56%) | 6 788(24.01%) | 5 303(18.76%) | 2 112(7.47%) |
| 白色1 White 1 | 11 653(41.23%) | 3 034(10.73%) | 6 680(23.63%) | 4 771(16.88%) | 2 128(7.53%) |
| 白色2 White 2 | 11 484(40.63%) | 2 992(10.59%) | 6 705(23.72%) | 5 008(17.72%) | 2 077(7.35%) |
| 白色3 White 3 | 10 976(38.83%) | 2 961(10.48%) | 6 818(24.12%) | 5 372(19.01%) | 2 139(7.57%) |
Table 2 Statistical table of the number of genes in different FPKM interval of red and white petals
| 样本 Sample | FPKM | ||||
|---|---|---|---|---|---|
| 0 ~ 1 | 1 ~ 3 | 3 ~ 15 | 15 ~ 16 | > 60 | |
| 红色1 Red 1 | 11 710(41.43%) | 2 957(10.46%) | 6 609(23.38%) | 5 017(17.75%) | 1 973(6.98%) |
| 红色2 Red 2 | 11 028(39.02%) | 3 038(10.75%) | 6 778(23.98%) | 5 313(18.80%) | 2 109(7.46%) |
| 红色3 Red 3 | 11 077(39.19%) | 2 986(10.56%) | 6 788(24.01%) | 5 303(18.76%) | 2 112(7.47%) |
| 白色1 White 1 | 11 653(41.23%) | 3 034(10.73%) | 6 680(23.63%) | 4 771(16.88%) | 2 128(7.53%) |
| 白色2 White 2 | 11 484(40.63%) | 2 992(10.59%) | 6 705(23.72%) | 5 008(17.72%) | 2 077(7.35%) |
| 白色3 White 3 | 10 976(38.83%) | 2 961(10.48%) | 6 818(24.12%) | 5 372(19.01%) | 2 139(7.57%) |
| 基因编号 Gene ID | 注释 Description | FPKM | FC | 校正的p值 padj | 变化 Change | |
|---|---|---|---|---|---|---|
| 红色Red | 白色White | |||||
| 104585760 | 吲哚-3-丙酮酸单氧酶YUCCA10 Indole-3-pyruvate monooxygenase YUCCA10 | 379.86 | 1 057.48 | 0.36 | 0.0061 | 下调Down |
| LOC104586332△ | 类囊体腔内15.0 kD蛋白2,类叶绿体 Thylakoid lumenal 15.0 kD protein 2, chloroplastic-like | 25.56 | 119.73 | 0.21 | 0.0146 | 下调Down |
| LOC104586359 | 包含 BTB/POZ 结构域蛋白 BTB/POZ domain-containing protein | 381.38 | 980.36 | 0.39 | 0.0204 | 下调Down |
| LOC104587048 | 酯酶 Esterase | 76.90 | 292.68 | 0.26 | 0.0023 | 下调Down |
| LOC104587342 | UDP-D-芹菜糖/UDP-D-木糖合酶1 UDP-D-apiose/UDP-D-xylose synthase 1 | 617.36 | 1 595.39 | 0.39 | 0.0118 | 下调Down |
| LOC104587387 | 未知 Unkown | 188.34 | 586.55 | 0.32 | 0.0023 | 下调Down |
| LOC104587394 | 包含 BTB/POZ 结构域蛋白 BTB/POZ domain-containing protein | 200.87 | 751.03 | 0.27 | 5.61E-05 | 下调Down |
| LOC104587400 | 未知 Unkown | 264.94 | 812.91 | 0.33 | 0.0481 | 下调Down |
| LOC104587793 | 未知 Unkown | 194.34 | 605.49 | 0.32 | 0.0023 | 下调Down |
| LOC104587865 | 未知 Unkown | 35.16 | 149.94 | 0.23 | 0.0146 | 下调Down |
| LOC104587866△ | 叶绿体运动蛋白CHUP1 Protein CHUP1,chloroplastic-like | 12.66 | 89.50 | 0.14 | 0.0053 | 下调Down |
| LOC104588342△ | 光系统I反应中心亚基 psaK,叶绿体 Photosystem I reaction center subunit psaK,chloroplastic | 188.65 | 553.88 | 0.34 | 0.0236 | 下调Down |
| LOC104588655△ | 组成型活性ROPs 2 的交互器,叶绿体 Interactor of constitutive active ROPs 2,chloroplastic | 62.64 | 422.51 | 0.15 | 2.18E-05 | 下调Down |
| LOC104589017 | GATA 锌指蛋白 GATA zinc finger protein | 974.90 | 2 752.10 | 0.35 | 0.0023 | 下调Down |
| LOC104590162△ | 氧进化增强蛋白3-1,叶绿体 Oxygen-evolving enhancer protein 3-1,chloroplastic | 1 231.52 | 2 756.21 | 0.45 | 0.0390 | 下调Down |
| LOC104590347 | ABC转运体G家族成员22 ABC transporter G family member 22 | 558.88 | 2 026.67 | 0.28 | 0.0263 | 下调Down |
| LOC104590803 | 轴向调节因子YABBY 2 Axial regulator YABBY 2 | 613.91 | 1 645.85 | 0.37 | 0.0054 | 下调Down |
| LOC104591558 | 豆肽链内切酶 Vignain | 1 308.09 | 6 381.86 | 0.20 | 0.0231 | 下调Down |
| LOC104591924△ | 甘油醛-3-磷酸脱氢酶A,叶绿体 Glyceraldehyde-3-phosphate dehydrogenase A,chloroplastic | 303.68 | 1 306.44 | 0.23 | 5.28E-05 | 下调Down |
| LOC104592073△ | 催泪因子合成酶,叶绿体 Lachrymatory-factor synthase | 140.18 | 437.15 | 0.32 | 0.0059425 | 下调Down |
| LOC104592306 | 赤霉素3-β-双加氧酶 1 Gibberellin 3-beta-dioxygenase 1 | 44.02 | 212.18 | 0.21 | 0.0009824 | 下调Down |
| LOC104592455 | 转录因子YABBY 5-like Axial regulator YABBY 5-like | 8.59 | 913.39 | 0.01 | 1.66E-05 | 下调Down |
| LOC104592502 | 14 kD富含脯氨酸的蛋白质DC2.15 14 kD proline-rich protein DC2.15 | 206.09 | 672.66 | 0.31 | 0.0056149 | 下调Down |
| LOC104593007△ | 光系统II核心复合蛋白psbY,叶绿体 Photosystem II core complex proteins psbY,chloroplastic | 179.71 | 547.87 | 0.33 | 0.039928 | 下调Down |
| LOC104593587 | α-L-阿拉伯呋喃糖酶 1 Alpha-L-arabinofuranosidase 1-like | 3 860.50 | 12 376.06 | 0.31 | 0.0016702 | 下调Down |
| LOC104594041 | S型阴离子通道SLAH1 S-type anion channel SLAH1 | 254.66 | 75.28 | 3.38 | 0.013928 | 上调Up |
| LOC104594703 | 谷胱甘肽 S-转移酶 L3 Glutathione S-transferase L3 | 18.45 | 125.60 | 0.15 | 0.0047725 | 下调Down |
| LOC104594809 | 过氧化物酶 21 Peroxidase 21 | 1 697.61 | 626.18 | 2.71 | 0.01402 | |
| LOC104594846 | α碳酸酐酶8 Alpha carbonic anhydrase 8 | 434.02 | 1 641.06 | 0.26 | 0.031317 | 下调Down |
| LOC104595273 | 赤霉素3-β-双加氧酶 1 Gibberellin 3-beta-dioxygenase 1 | 46.11 | 251.33 | 0.18 | 0.019816 | 下调Down |
| LOC104595276△ | 丙酮酸钠协同转运蛋白BASS2 sodium/pyruvate cotransporter BASS2 | 2 043.27 | 4 692.64 | 0.44 | 0.030206 | 下调Down |
| LOC104595371 | 包含EG45结构域蛋白 EG45-like domain containing protein | 1 011.13 | 367.72 | 2.75 | 0.032524 | 上调Up |
| LOC104595406 | 轴向调节因子YABBY 5-like Axial regulator YABBY 5-like | 33.82 | 1 727.01 | 0.02 | 6.39E-09 | 下调Down |
| LOC104596121 | 含重复WD 结构域的蛋白 RUP2 WD repeat-containing protein RUP2 | 97.23 | 387.20 | 0.25 | 0.0002 | 下调Down |
| LOC104596470 | 谷胱甘肽S-转移酶 GSTU17 Glutathione S-transferase U17 | 117.27 | 509.20 | 0.23 | 0.0390 | 下调Down |
| LOC104596509 | 亚油酸酯9S-脂氧合酶 5 Probable linoleate 9S-lipoxygenase 5 | 556.77 | 2 032.33 | 0.27 | 5.28E-05 | 下调Down |
| LOC104597338 | 未知 Unkown | 524.39 | 2 025.64 | 0.26 | 0.0111 | 下调Down |
| LOC104597400 | 蔗糖—肌醇半乳糖基转移酶2 Galactinol-sucrose galactosyltransferase 2 | 943.20 | 3 133.75 | 0.30 | 0.0020 | 下调Down |
| LOC104597497 | 细胞色素P450 93A3 Cytochrome P450 93A3 | 920.50 | 383.45 | 2.40 | 0.0483 | 上调Up |
| LOC104597628 | 种子胰凝乳蛋白酶抑制剂TI5-72 Seed trypsin/chymotrypsin inhibitor TI5-72 | 259.84 | 1 005.47 | 0.26 | 0.0061 | 下调Down |
| LOC104597911△ | 亚复合体b4的光合NDH亚基,叶绿体 Photosynthetic NDH subunit of subcomplex B 4, chloroplastic | 4.87 | 104.81 | 0.05 | 5.75E-06 | 下调Down |
| LOC104597930 | 含 NAC 结构域蛋白NAC100 NAC domain-containing protein 100 | 112.47 | 461.16 | 0.24 | 0.0072 | 下调Down |
| LOC104597931 | 转录因子 TCP5 Transcription factor TCP5 | 262.31 | 58.19 | 4.51 | 0.0004 | 上调Up |
| LOC104598813 | β-D-木糖酶 Beta-D-xylosidase | 1 221.24 | 5 693.74 | 0.21 | 0.0146 | 下调Down |
| LOC104598896 | 内切葡聚糖酶17 Endoglucanase 17 | 529.67 | 1 602.49 | 0.33 | 0.0139 | 下调Down |
| LOC104599017 | 支架蛋白 Cullin | 70.23 | 222.45 | 0.32 | 0.0271 | 下调Down |
| LOC104599122 | 木葡聚糖内糖基转移酶/水解酶 2 Xyloglucan endotransglucosylase/hydrolase 2 | 61.05 | 220.88 | 0.28 | 0.0111 | 下调Down |
| LOC104599774△ | 肽基脯氨酸顺反异构酶FKBP13,叶绿体 Peptidyl-prolyl cis-trans isomerase FKBP13, chloroplastic | 100.34 | 697.49 | 0.14 | 3.10E-10 | 下调Down |
| LOC104599896 | 脂质转移蛋白 Patellin | 29.09 | 160.76 | 0.18 | 0.0016 | 下调Down |
| LOC104600529 | 类异胡豆苷合成酶4基因SSL4 Protein STRICTOSIDINE SYNTHASE-LIKE 4-like | 1 001.83 | 2 658.36 | 0.38 | 0.0054 | 下调Down |
| LOC104600691 | β-果糠糖苷酶,不溶同工酶1 Beta-fructofuranosidase,insoluble isoenzyme 1 | 218.91 | 48.72 | 4.49 | 0.0009 | 上调Up |
| LOC104600849 | 转录因子 EMB1444 Transcription factor EMB1444 | 25.06 | 108.77 | 0.23 | 0.0393 | 下调Down |
| LOC104600890 | 未知 Unkown | 11.48 | 70.98 | 0.16 | 0.0474 | 下调Down |
| LOC104601317 | 普遍应激蛋白 PHOS34 Universal stress protein PHOS34-like | 4.66 | 46.12 | 0.10 | 0.0483 | 下调Down |
| LOC104601584 | 钠转运体 HKT1 Sodium transporter HKT1 | 349.98 | 1 418.70 | 0.25 | 6.66E-06 | 下调Down |
| LOC104601751 | 类枯草菌素蛋白酶SBT1.9 Subtilisin-like protease SBT1.9 | 82.89 | 385.71 | 0.21 | 0.0438 | 下调Down |
| LOC104601926 | 生长素外排载体成分1 Auxin efflux carrier component 1 | 240.85 | 721.72 | 0.33 | 0.0054 | 下调Down |
| LOC104602271 | 21 kD 蛋白 21 kD protein | 1 519.82 | 4 115.71 | 0.37 | 0.0024 | 下调Down |
| LOC104602296 | β-果糠糖苷酶,可溶性同工酶I Beta-fructofuranosidase,soluble isoenzyme I | 3 037.56 | 10 350.37 | 0.29 | 3.83E-05 | 下调Down |
| LOC104602308 | 14 kD富含脯氨酸的蛋白质DC2.15 14 kD proline-rich protein DC2.15 | 14.11 | 86.45 | 0.16 | 0.0231 | 下调Down |
| LOC104602311 | 14 kD富含脯氨酸的蛋白质DC2.15 14 kD proline-rich protein DC2.15 | 2 417.69 | 8 525.97 | 0.28 | 5.44E-05 | 下调Down |
| LOC104603231 | 天冬氨酸蛋白酶家族蛋白 Aspartyl protease family protein | 3.11 | 130.60 | 0.02 | 0.0023 | 下调Down |
| LOC104603275△ | 亚复合体b4的光合B1亚基,叶绿体 Photosynthetic NDH subunit of subcomplex B 1, chloroplastic | 301.09 | 768.07 | 0.39 | 0.0223 | 下调Down |
| LOC104604690 | 未知 Unkown | 113.28 | 713.97 | 0.16 | 2.76E-09 | 下调Down |
| LOC104605527△ | 可溶性无机焦磷酸酶6,叶绿体 Soluble inorganic pyrophosphatase 6,chloroplastic | 36.45 | 234.95 | 0.16 | 6.66E-06 | 下调Down |
| LOC104605630 | 吲哚-3-乙酸-酰胺合成酶GH3.10 Indole-3-acetic acid-amido synthetase GH3.10 | 226.60 | 855.13 | 0.26 | 0.0063 | 下调Down |
| LOC104605653 | O-酰基转移酶WSD1 O-acyltransferase WSD1 | 6 766.33 | 22 004.70 | 0.31 | 0.0003 | 下调Down |
| LOC104605705△ | 类囊体形成蛋白1(THF1),类叶绿体 Protein thylakoid formation1,chloroplastic-like | 2 055.18 | 4 675.31 | 0.44 | 0.0290 | 下调Down |
| LOC104606133 | 干燥相关蛋白PCC13-62 Desiccation-related protein PCC13-62 | 151.66 | 844.91 | 0.18 | 3.84E-08 | 下调Down |
| LOC104606839 | 未知 Unkown | 58.90 | 370.51 | 0.16 | 3.85E-07 | 下调Down |
| LOC104606865 | AT-hook基序和定位蛋白 AHL29 AT-hook motif nuclear-localized protein 29 | 94.71 | 556.50 | 0.17 | 5.44E-05 | 下调Down |
| LOC104607352 | 果胶酯酶抑制剂7 PMEI7 Pectinesterase inhibitor 7 PMEI7 | 180.46 | 916.09 | 0.20 | 0.0390 | 下调Down |
| LOC104607711 | 富含赖氨酸的阿拉伯半乳糖蛋白18 Lysine-rich arabinogalactan protein 18 | 557.34 | 1 888.33 | 0.30 | 5.61E-05 | 下调Down |
| LOC104607973 | NRT1/ PTR蛋白家族5.8 Protein NRT1/ PTR FAMILY 5.8 | 345.98 | 92.02 | 3.76 | 0.0024 | 上调 Up |
| LOC104608668 | 排毒蛋白EXO EXO_ARATH Protein EXORDIUM | 798.37 | 2 167.39 | 0.37 | 0.0084 | 下调Down |
| LOC104608669 | 排毒蛋白EXO EXO_ARATH Protein EXORDIUM | 2 848.10 | 9590.01 | 0.30 | 0.0144 | 下调Down |
| LOC104610246 | 未知 Unkown | 256.78 | 869.88 | 0.30 | 0.0003 | 下调Down |
| LOC104610692 | 温度诱导的脂质运载蛋白1 Temperature-induced lipocalin-1 | 3 564.02 | 9 331.83 | 0.38 | 0.0024 | 下调Down |
| LOC104610780 | (S)-2-羟基酸氧化酶 GLO1 (S)-2-hydroxy-acid oxidase GLO1 | 883.52 | 2 867.34 | 0.31 | 0.0001 | 下调Down |
| LOC104611127 | 未知 Unkown | 48.52 | 177.72 | 0.27 | 0.0313 | 下调Down |
| LOC104612002 | 含BTB/POZ结构域和锚蛋白重复序列蛋白基因 COCH BTB/POZ domain and ankyrin repeat- containing protein COCH | 418.84 | 149.69 | 2.80 | 0.0178 | 上调 Up |
| LOC104612096 | 钠/钙交换器NCL Sodium/calcium exchanger NCL | 236.31 | 1 164.35 | 0.20 | 1.00E-05 | 下调Down |
| LOC104613105 | 转录因子bHLH30 Transcription factor bHLH30 | 154.16 | 28.88 | 5.34 | 0.0018 | 上调Up |
| LOC104613323 | 跨膜蛋白Transmembrane protein | 22.39 | 104.04 | 0.22 | 0.0325 | 下调Down |
| Novel00124* | 未知Unkown | 86.66 | 525.25 | 0.16 | 1.30E-08 | 下调Down |
| Novel00164* | 未知Unkown | 16.25 | 125.68 | 0.13 | 0.0481 | 下调Down |
| Novel00165* | 未知Unkown | 2.21 | 62.63 | 0.04 | 0.0001 | 下调Down |
| Novel00181* | 铁还原氧化酶8,线粒体 Ferric reduction oxidase 8,mitochondrial | 738.48 | 2 014.56 | 0.37 | 0.0023 | 下调Down |
| Novel00379* | 未知 Unkown | 19.31 | 114.75 | 0.17 | 0.0024 | 下调Down |
| Novel00435* | 类组氨酸激酶CKI1-like Histidine kinase CKI1-like | 902.48 | 2 282.27 | 0.40 | 0.0316 | 下调Down |
| Novel00455* | 热激蛋白HSP90.7 Heat shock protein HSP90.7 | 179.97 | 815.13 | 0.22 | 0.0061 | 下调Down |
Table 3 Information of differentially expressed genes between red and white petals
| 基因编号 Gene ID | 注释 Description | FPKM | FC | 校正的p值 padj | 变化 Change | |
|---|---|---|---|---|---|---|
| 红色Red | 白色White | |||||
| 104585760 | 吲哚-3-丙酮酸单氧酶YUCCA10 Indole-3-pyruvate monooxygenase YUCCA10 | 379.86 | 1 057.48 | 0.36 | 0.0061 | 下调Down |
| LOC104586332△ | 类囊体腔内15.0 kD蛋白2,类叶绿体 Thylakoid lumenal 15.0 kD protein 2, chloroplastic-like | 25.56 | 119.73 | 0.21 | 0.0146 | 下调Down |
| LOC104586359 | 包含 BTB/POZ 结构域蛋白 BTB/POZ domain-containing protein | 381.38 | 980.36 | 0.39 | 0.0204 | 下调Down |
| LOC104587048 | 酯酶 Esterase | 76.90 | 292.68 | 0.26 | 0.0023 | 下调Down |
| LOC104587342 | UDP-D-芹菜糖/UDP-D-木糖合酶1 UDP-D-apiose/UDP-D-xylose synthase 1 | 617.36 | 1 595.39 | 0.39 | 0.0118 | 下调Down |
| LOC104587387 | 未知 Unkown | 188.34 | 586.55 | 0.32 | 0.0023 | 下调Down |
| LOC104587394 | 包含 BTB/POZ 结构域蛋白 BTB/POZ domain-containing protein | 200.87 | 751.03 | 0.27 | 5.61E-05 | 下调Down |
| LOC104587400 | 未知 Unkown | 264.94 | 812.91 | 0.33 | 0.0481 | 下调Down |
| LOC104587793 | 未知 Unkown | 194.34 | 605.49 | 0.32 | 0.0023 | 下调Down |
| LOC104587865 | 未知 Unkown | 35.16 | 149.94 | 0.23 | 0.0146 | 下调Down |
| LOC104587866△ | 叶绿体运动蛋白CHUP1 Protein CHUP1,chloroplastic-like | 12.66 | 89.50 | 0.14 | 0.0053 | 下调Down |
| LOC104588342△ | 光系统I反应中心亚基 psaK,叶绿体 Photosystem I reaction center subunit psaK,chloroplastic | 188.65 | 553.88 | 0.34 | 0.0236 | 下调Down |
| LOC104588655△ | 组成型活性ROPs 2 的交互器,叶绿体 Interactor of constitutive active ROPs 2,chloroplastic | 62.64 | 422.51 | 0.15 | 2.18E-05 | 下调Down |
| LOC104589017 | GATA 锌指蛋白 GATA zinc finger protein | 974.90 | 2 752.10 | 0.35 | 0.0023 | 下调Down |
| LOC104590162△ | 氧进化增强蛋白3-1,叶绿体 Oxygen-evolving enhancer protein 3-1,chloroplastic | 1 231.52 | 2 756.21 | 0.45 | 0.0390 | 下调Down |
| LOC104590347 | ABC转运体G家族成员22 ABC transporter G family member 22 | 558.88 | 2 026.67 | 0.28 | 0.0263 | 下调Down |
| LOC104590803 | 轴向调节因子YABBY 2 Axial regulator YABBY 2 | 613.91 | 1 645.85 | 0.37 | 0.0054 | 下调Down |
| LOC104591558 | 豆肽链内切酶 Vignain | 1 308.09 | 6 381.86 | 0.20 | 0.0231 | 下调Down |
| LOC104591924△ | 甘油醛-3-磷酸脱氢酶A,叶绿体 Glyceraldehyde-3-phosphate dehydrogenase A,chloroplastic | 303.68 | 1 306.44 | 0.23 | 5.28E-05 | 下调Down |
| LOC104592073△ | 催泪因子合成酶,叶绿体 Lachrymatory-factor synthase | 140.18 | 437.15 | 0.32 | 0.0059425 | 下调Down |
| LOC104592306 | 赤霉素3-β-双加氧酶 1 Gibberellin 3-beta-dioxygenase 1 | 44.02 | 212.18 | 0.21 | 0.0009824 | 下调Down |
| LOC104592455 | 转录因子YABBY 5-like Axial regulator YABBY 5-like | 8.59 | 913.39 | 0.01 | 1.66E-05 | 下调Down |
| LOC104592502 | 14 kD富含脯氨酸的蛋白质DC2.15 14 kD proline-rich protein DC2.15 | 206.09 | 672.66 | 0.31 | 0.0056149 | 下调Down |
| LOC104593007△ | 光系统II核心复合蛋白psbY,叶绿体 Photosystem II core complex proteins psbY,chloroplastic | 179.71 | 547.87 | 0.33 | 0.039928 | 下调Down |
| LOC104593587 | α-L-阿拉伯呋喃糖酶 1 Alpha-L-arabinofuranosidase 1-like | 3 860.50 | 12 376.06 | 0.31 | 0.0016702 | 下调Down |
| LOC104594041 | S型阴离子通道SLAH1 S-type anion channel SLAH1 | 254.66 | 75.28 | 3.38 | 0.013928 | 上调Up |
| LOC104594703 | 谷胱甘肽 S-转移酶 L3 Glutathione S-transferase L3 | 18.45 | 125.60 | 0.15 | 0.0047725 | 下调Down |
| LOC104594809 | 过氧化物酶 21 Peroxidase 21 | 1 697.61 | 626.18 | 2.71 | 0.01402 | |
| LOC104594846 | α碳酸酐酶8 Alpha carbonic anhydrase 8 | 434.02 | 1 641.06 | 0.26 | 0.031317 | 下调Down |
| LOC104595273 | 赤霉素3-β-双加氧酶 1 Gibberellin 3-beta-dioxygenase 1 | 46.11 | 251.33 | 0.18 | 0.019816 | 下调Down |
| LOC104595276△ | 丙酮酸钠协同转运蛋白BASS2 sodium/pyruvate cotransporter BASS2 | 2 043.27 | 4 692.64 | 0.44 | 0.030206 | 下调Down |
| LOC104595371 | 包含EG45结构域蛋白 EG45-like domain containing protein | 1 011.13 | 367.72 | 2.75 | 0.032524 | 上调Up |
| LOC104595406 | 轴向调节因子YABBY 5-like Axial regulator YABBY 5-like | 33.82 | 1 727.01 | 0.02 | 6.39E-09 | 下调Down |
| LOC104596121 | 含重复WD 结构域的蛋白 RUP2 WD repeat-containing protein RUP2 | 97.23 | 387.20 | 0.25 | 0.0002 | 下调Down |
| LOC104596470 | 谷胱甘肽S-转移酶 GSTU17 Glutathione S-transferase U17 | 117.27 | 509.20 | 0.23 | 0.0390 | 下调Down |
| LOC104596509 | 亚油酸酯9S-脂氧合酶 5 Probable linoleate 9S-lipoxygenase 5 | 556.77 | 2 032.33 | 0.27 | 5.28E-05 | 下调Down |
| LOC104597338 | 未知 Unkown | 524.39 | 2 025.64 | 0.26 | 0.0111 | 下调Down |
| LOC104597400 | 蔗糖—肌醇半乳糖基转移酶2 Galactinol-sucrose galactosyltransferase 2 | 943.20 | 3 133.75 | 0.30 | 0.0020 | 下调Down |
| LOC104597497 | 细胞色素P450 93A3 Cytochrome P450 93A3 | 920.50 | 383.45 | 2.40 | 0.0483 | 上调Up |
| LOC104597628 | 种子胰凝乳蛋白酶抑制剂TI5-72 Seed trypsin/chymotrypsin inhibitor TI5-72 | 259.84 | 1 005.47 | 0.26 | 0.0061 | 下调Down |
| LOC104597911△ | 亚复合体b4的光合NDH亚基,叶绿体 Photosynthetic NDH subunit of subcomplex B 4, chloroplastic | 4.87 | 104.81 | 0.05 | 5.75E-06 | 下调Down |
| LOC104597930 | 含 NAC 结构域蛋白NAC100 NAC domain-containing protein 100 | 112.47 | 461.16 | 0.24 | 0.0072 | 下调Down |
| LOC104597931 | 转录因子 TCP5 Transcription factor TCP5 | 262.31 | 58.19 | 4.51 | 0.0004 | 上调Up |
| LOC104598813 | β-D-木糖酶 Beta-D-xylosidase | 1 221.24 | 5 693.74 | 0.21 | 0.0146 | 下调Down |
| LOC104598896 | 内切葡聚糖酶17 Endoglucanase 17 | 529.67 | 1 602.49 | 0.33 | 0.0139 | 下调Down |
| LOC104599017 | 支架蛋白 Cullin | 70.23 | 222.45 | 0.32 | 0.0271 | 下调Down |
| LOC104599122 | 木葡聚糖内糖基转移酶/水解酶 2 Xyloglucan endotransglucosylase/hydrolase 2 | 61.05 | 220.88 | 0.28 | 0.0111 | 下调Down |
| LOC104599774△ | 肽基脯氨酸顺反异构酶FKBP13,叶绿体 Peptidyl-prolyl cis-trans isomerase FKBP13, chloroplastic | 100.34 | 697.49 | 0.14 | 3.10E-10 | 下调Down |
| LOC104599896 | 脂质转移蛋白 Patellin | 29.09 | 160.76 | 0.18 | 0.0016 | 下调Down |
| LOC104600529 | 类异胡豆苷合成酶4基因SSL4 Protein STRICTOSIDINE SYNTHASE-LIKE 4-like | 1 001.83 | 2 658.36 | 0.38 | 0.0054 | 下调Down |
| LOC104600691 | β-果糠糖苷酶,不溶同工酶1 Beta-fructofuranosidase,insoluble isoenzyme 1 | 218.91 | 48.72 | 4.49 | 0.0009 | 上调Up |
| LOC104600849 | 转录因子 EMB1444 Transcription factor EMB1444 | 25.06 | 108.77 | 0.23 | 0.0393 | 下调Down |
| LOC104600890 | 未知 Unkown | 11.48 | 70.98 | 0.16 | 0.0474 | 下调Down |
| LOC104601317 | 普遍应激蛋白 PHOS34 Universal stress protein PHOS34-like | 4.66 | 46.12 | 0.10 | 0.0483 | 下调Down |
| LOC104601584 | 钠转运体 HKT1 Sodium transporter HKT1 | 349.98 | 1 418.70 | 0.25 | 6.66E-06 | 下调Down |
| LOC104601751 | 类枯草菌素蛋白酶SBT1.9 Subtilisin-like protease SBT1.9 | 82.89 | 385.71 | 0.21 | 0.0438 | 下调Down |
| LOC104601926 | 生长素外排载体成分1 Auxin efflux carrier component 1 | 240.85 | 721.72 | 0.33 | 0.0054 | 下调Down |
| LOC104602271 | 21 kD 蛋白 21 kD protein | 1 519.82 | 4 115.71 | 0.37 | 0.0024 | 下调Down |
| LOC104602296 | β-果糠糖苷酶,可溶性同工酶I Beta-fructofuranosidase,soluble isoenzyme I | 3 037.56 | 10 350.37 | 0.29 | 3.83E-05 | 下调Down |
| LOC104602308 | 14 kD富含脯氨酸的蛋白质DC2.15 14 kD proline-rich protein DC2.15 | 14.11 | 86.45 | 0.16 | 0.0231 | 下调Down |
| LOC104602311 | 14 kD富含脯氨酸的蛋白质DC2.15 14 kD proline-rich protein DC2.15 | 2 417.69 | 8 525.97 | 0.28 | 5.44E-05 | 下调Down |
| LOC104603231 | 天冬氨酸蛋白酶家族蛋白 Aspartyl protease family protein | 3.11 | 130.60 | 0.02 | 0.0023 | 下调Down |
| LOC104603275△ | 亚复合体b4的光合B1亚基,叶绿体 Photosynthetic NDH subunit of subcomplex B 1, chloroplastic | 301.09 | 768.07 | 0.39 | 0.0223 | 下调Down |
| LOC104604690 | 未知 Unkown | 113.28 | 713.97 | 0.16 | 2.76E-09 | 下调Down |
| LOC104605527△ | 可溶性无机焦磷酸酶6,叶绿体 Soluble inorganic pyrophosphatase 6,chloroplastic | 36.45 | 234.95 | 0.16 | 6.66E-06 | 下调Down |
| LOC104605630 | 吲哚-3-乙酸-酰胺合成酶GH3.10 Indole-3-acetic acid-amido synthetase GH3.10 | 226.60 | 855.13 | 0.26 | 0.0063 | 下调Down |
| LOC104605653 | O-酰基转移酶WSD1 O-acyltransferase WSD1 | 6 766.33 | 22 004.70 | 0.31 | 0.0003 | 下调Down |
| LOC104605705△ | 类囊体形成蛋白1(THF1),类叶绿体 Protein thylakoid formation1,chloroplastic-like | 2 055.18 | 4 675.31 | 0.44 | 0.0290 | 下调Down |
| LOC104606133 | 干燥相关蛋白PCC13-62 Desiccation-related protein PCC13-62 | 151.66 | 844.91 | 0.18 | 3.84E-08 | 下调Down |
| LOC104606839 | 未知 Unkown | 58.90 | 370.51 | 0.16 | 3.85E-07 | 下调Down |
| LOC104606865 | AT-hook基序和定位蛋白 AHL29 AT-hook motif nuclear-localized protein 29 | 94.71 | 556.50 | 0.17 | 5.44E-05 | 下调Down |
| LOC104607352 | 果胶酯酶抑制剂7 PMEI7 Pectinesterase inhibitor 7 PMEI7 | 180.46 | 916.09 | 0.20 | 0.0390 | 下调Down |
| LOC104607711 | 富含赖氨酸的阿拉伯半乳糖蛋白18 Lysine-rich arabinogalactan protein 18 | 557.34 | 1 888.33 | 0.30 | 5.61E-05 | 下调Down |
| LOC104607973 | NRT1/ PTR蛋白家族5.8 Protein NRT1/ PTR FAMILY 5.8 | 345.98 | 92.02 | 3.76 | 0.0024 | 上调 Up |
| LOC104608668 | 排毒蛋白EXO EXO_ARATH Protein EXORDIUM | 798.37 | 2 167.39 | 0.37 | 0.0084 | 下调Down |
| LOC104608669 | 排毒蛋白EXO EXO_ARATH Protein EXORDIUM | 2 848.10 | 9590.01 | 0.30 | 0.0144 | 下调Down |
| LOC104610246 | 未知 Unkown | 256.78 | 869.88 | 0.30 | 0.0003 | 下调Down |
| LOC104610692 | 温度诱导的脂质运载蛋白1 Temperature-induced lipocalin-1 | 3 564.02 | 9 331.83 | 0.38 | 0.0024 | 下调Down |
| LOC104610780 | (S)-2-羟基酸氧化酶 GLO1 (S)-2-hydroxy-acid oxidase GLO1 | 883.52 | 2 867.34 | 0.31 | 0.0001 | 下调Down |
| LOC104611127 | 未知 Unkown | 48.52 | 177.72 | 0.27 | 0.0313 | 下调Down |
| LOC104612002 | 含BTB/POZ结构域和锚蛋白重复序列蛋白基因 COCH BTB/POZ domain and ankyrin repeat- containing protein COCH | 418.84 | 149.69 | 2.80 | 0.0178 | 上调 Up |
| LOC104612096 | 钠/钙交换器NCL Sodium/calcium exchanger NCL | 236.31 | 1 164.35 | 0.20 | 1.00E-05 | 下调Down |
| LOC104613105 | 转录因子bHLH30 Transcription factor bHLH30 | 154.16 | 28.88 | 5.34 | 0.0018 | 上调Up |
| LOC104613323 | 跨膜蛋白Transmembrane protein | 22.39 | 104.04 | 0.22 | 0.0325 | 下调Down |
| Novel00124* | 未知Unkown | 86.66 | 525.25 | 0.16 | 1.30E-08 | 下调Down |
| Novel00164* | 未知Unkown | 16.25 | 125.68 | 0.13 | 0.0481 | 下调Down |
| Novel00165* | 未知Unkown | 2.21 | 62.63 | 0.04 | 0.0001 | 下调Down |
| Novel00181* | 铁还原氧化酶8,线粒体 Ferric reduction oxidase 8,mitochondrial | 738.48 | 2 014.56 | 0.37 | 0.0023 | 下调Down |
| Novel00379* | 未知 Unkown | 19.31 | 114.75 | 0.17 | 0.0024 | 下调Down |
| Novel00435* | 类组氨酸激酶CKI1-like Histidine kinase CKI1-like | 902.48 | 2 282.27 | 0.40 | 0.0316 | 下调Down |
| Novel00455* | 热激蛋白HSP90.7 Heat shock protein HSP90.7 | 179.97 | 815.13 | 0.22 | 0.0061 | 下调Down |
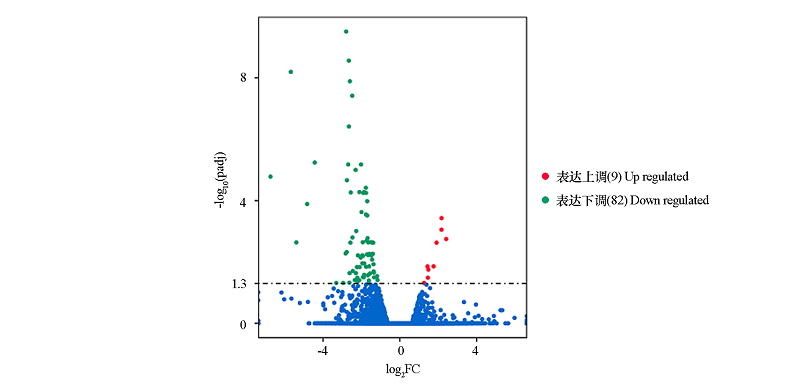
Fig. 2 The screening results of differentially expressed genes between red and white petals of bicolor lotus‘Dasajin’by volcano plots FC:Fold change,red/white.
| [1] |
An R, Liu X, Wang R, Wu H, Liang S, Shao J, Qi Y, An L, Yu F. 2014. The over-expression of two transcription factors,ABS5/bHLH30 and ABS7/MYB101,leads to upwardly curly leaves. PLoS ONE, 9 (9):e107637.
doi: 10.1371/journal.pone.0107637 URL |
| [2] | Bi Mengmeng, Cao Yuwei, Song Meng, Tang Yuchao, He Guoren, Yang Yue, Yang Panpan, Xu Leifeng, Ming Jun. 2021. Advances in flower color research of Lilium. Acta Horticulturae Sinica, 48 (10):2073-2086. (in Chinese) |
| 毕蒙蒙, 曹雨薇, 宋蒙, 唐玉超, 何国仁, 杨悦, 杨盼盼, 徐雷锋, 明军. 2021. 百合花色研究进展. 园艺学报, 48 (10):2073-2086. | |
| [3] |
Boter M, Golz J F, Giménez-Ibañez S, Fernandez-Barbero G, Franco-Zorrilla J M, Solano R. 2015. FILAMENTOUS FLOWER is a direct target of JAZ3 and modulates responses to jasmonate. Plant Cell, 27 (11):3160-3174.
doi: 10.1105/tpc.15.00220 URL |
| [4] |
Chatel G, Montiel G, Pré M, Memelink J, Thiersault M, Saint-Pierre B, Doireau P, Gantet P. 2003. CrMYC1,a Catharanthus roseus elicitor- and jasmonate-responsive bHLH transcription factor that binds the G-box element of the strictosidine synthase gene promoter. J Exp Bot, 54 (392):2587-2588.
doi: 10.1093/jxb/erg275 URL |
| [5] |
Cheng J, Liao L, Zhou H, Gu C, Wang L, Han Y. 2015. A small indel mutation in an anthocyanin transporter causes variegated colouration of peach flowers. J Exp Bot, 66 (22):7227-7239.
doi: 10.1093/jxb/erv419 pmid: 26357885 |
| [6] |
Deng J, Chen S, Yi X, Wang K, Liu Y, Li S, Yang P. 2013. Systematic qualitative and quantitative assessment of anthocyanins,flavones and flavonols in the petals of 108 lotus(Nelumbo nucifera)cultivars. Food Chem, 139:307-312.
doi: 10.1016/j.foodchem.2013.02.010 URL |
| [7] |
Deng J, Fu Z, Chen S, Damaris R N, Wang K, Li T, Yang P. 2015. Proteomic and epigenetic analyses of lotus(Nelumbo nucifera)petals between red and white cultivars. Plant Cell Physiol, 56 (8):1546-1555.
doi: 10.1093/pcp/pcv077 URL |
| [8] |
Ding B, Patterson E L, Holalu S, Li J J, Johnson G, Stanley L, Greenlee A, Peng F, Blinov M, Blackman B. 2020. Two MYB Proteins in a self-organizing activator-inhibitor system produce spotted pigmentation patterns. Curr Biol, 30 (5):1-13.
doi: 10.1016/j.cub.2019.10.048 URL |
| [9] |
Fu Y R, Liu F L, Li S, Tian D K, Dong L, Chen Y C, Su Y. 2021. Genetic diversity of wild Asian lotus(Nelumbo nucifera)from Northern China. Horticultural Plant Journal, 7 (5):488-500.
doi: 10.1016/j.hpj.2021.04.005 URL |
| [10] |
Gan Y, Li H, Xie Y, Wu W, Li M, Wang X, Huang J. 2014. THF1 mutations lead to increased basal and wound-induced levels of oxylipins that stimulate anthocyanin biosynthesis via COI1 signaling in Arabidopsis. J Integr Plant Biol, 56 (9):916-927.
doi: 10.1111/jipb.12177 |
| [11] |
Liu X J, Chuang Y N, Chiou C Y, Chin D C, Shen F Q, Yeh K W. 2012. Methylation effect on chalcone synthase gene expression determines anthocyanin pigmentation in floral tissues of two Oncidium orchid cultivars. Planta, 236 (2):401-409.
doi: 10.1007/s00425-012-1616-z URL |
| [12] |
Mehrtens F, Kranz H, Bednarek P, Weisshaar B. 2005. The Arabidopsis transcription factor MYB 12 is a flavonol-specific regulator of phenylpropanoid biosynthesis. Plant Physiol, 138 (2):1083-1096.
pmid: 15923334 |
| [13] |
Ming R, VanBuren R, Liu Y, Yang M, Han Y, Li LT, Zhang Q, Kim M J, Schatz M C, Campbell M, Li J, Bowers J E, Tang H, Lyons E, Ferguson A A, Narzisi G, Nelson D R, Blaby-Haas C E, Gschwend A R, Jiao Y, Der J P, Zeng F, Han J, Min X J, Hudson K A, Singh R, Grennan A K, Karpowicz S J, Watling J R, Ito K, Robinson S A, Hudson M E, Yu Q, Mockler T C, Carroll A, Zheng Y, Sunkar R, Jia R, Chen N, Arro J, Wai C M, Wafula E, Spence A, Han Y, Xu L, Zhang J, Peery R, Haus M J, Xiong W, Walsh J A, Wu J, Wang M L, Zhu Y J, Paull R E, Britt A B, Du C, Downie S R, Schuler M A, Michael T P, Long S P, Ort D R, Schopf J W, Gang D R, Jiang N, Yandell M, de Pamphilis C W, Merchant S S, Paterson A H, Buchanan B B, Li S, Shen-Miller J. 2013. Genome of the long-living sacred lotus(Nelumbo nucifera Gaertn.). Genome Biol, 14 (5):R41.
doi: 10.1186/gb-2013-14-5-r41 URL |
| [14] |
Morita Y, Hoshino A. 2018. Recent advances in flower color variation and patterning of Japanese morning glory and petunia. Breed Sci, 68 (1):128-138.
doi: 10.1270/jsbbs.17107 URL |
| [15] |
Ohno S, Hosokawa M, Kojima M, Kitamura Y, Hoshino A, Tatsuzawa F, Doi M, Yazawa S. 2011. Simultaneous post-transcriptional gene silencing of two different chalcone synthase genes resulting in pure white flowers in the octoploid dahlia. Planta, 234 (5):945-958.
doi: 10.1007/s00425-011-1456-2 pmid: 21688014 |
| [16] |
Shamimuzzaman M, Vodkin L. 2013. Genome-wide identification of binding sites for NAC and YABBY transcription factors and co-regulated genes during soybean seedling development by ChIP-Seq and RNA-Seq. BMC Genomics, 14:477.
doi: 10.1186/1471-2164-14-477 pmid: 23865409 |
| [17] |
Saito R, Kuchitsu K, Ozeki Y, Nakayama M. 2007. Spatiotemporal metabolic regulation of anthocyanin and related compounds during the development of marginal picotee petals in Petunia hybrida(Solanaceae). J Plant Res, 120 (4):563-568.
doi: 10.1007/s10265-007-0086-z URL |
| [18] |
Tateishi N, Ozaki Y, Okubo H. 2010. White marginal picotee formation in the petals of Camellia japonica‘Tamanoura’. J Jpn Soc Hortic Sci, 79 (2):207-214.
doi: 10.2503/jjshs1.79.207 URL |
| [19] |
van Es S W, Silveira S R, Rocha D I, Bimbo A, Martinelli A P, Dornelas M C, Angenent G C, Immink R G H. 2018. Novel functions of the Arabidopsis transcription factor TCP 5 in petal development and ethylene biosynthesis. Plant J, 94 (5):867-879.
doi: 10.1111/tpj.2018.94.issue-5 URL |
| [20] | Wu Qian, Zhang Huijin, Wang Xiaohan, Zhao Wen, Zhou Xian, Wang Liangsheng. 2021. Research progress on flower color of waterlily(Nymphaea). Acta Horticulturae Sinica, 48 (10):2087-2099. (in Chinese) |
| 吴倩, 张会金, 王晓晗, 赵文, 周娴, 王亮生. 2021. 睡莲花色研究进展. 园艺学报, 48 (10):2087-2099. | |
| [21] | Xu Lei-feng. 2017. Transcriptome analusis of bicolor tepal development in lilies and functional analysis of LhUFGT and LhSGR[Ph. D. Dissertation]. Beijing: Chinese Academy of Agricultural Sciences. (in Chinese) |
| 徐雷锋. 2017. 百合双色花形成的转录组分析及基因LhUFGT和LhSGR的功能研究[博士论文]. 北京: 中国农业科学院. | |
| [22] | Xu Yi, Qiao Zheng-lin, Zhao Lin, Zhao Wan-yue, Chen Long-qing, Hu Hui-zhen. 2021. Genome-wide and transcriptome analysis of YABBy family gene in lotus(Nelumbo nucifera Gaertn.). Mol Plant Breed,http://kns.cnki.net/kcms/detail/46.1068.S.20210326.1013.006.html. (in Chinese) |
| 徐逸, 谯正林, 赵琳, 赵婉玥, 陈龙清, 胡慧贞. 2021. 荷花YABBY家族的全基因组和转录组分析. 分子植物育种,http://kns.cnki.net/kcms/detail/46.1068.S.20210326.1013.006.html. | |
| [23] |
Zhao Y, Liu C, Ge D, Yan M, Ren Y, Huang X, Yuan Z. 2020. Genome-wide identification and expression of YABBY genes family during flower development in Punica granatum L. Gene, 752:144784.
doi: 10.1016/j.gene.2020.144784 URL |
| [1] | JIANG Yu, TU Xunliang, and HE Junrong. Analysis of Differential Expression Genes in Leaves of Leaf Color Mutant of Chinese Orchid [J]. Acta Horticulturae Sinica, 2023, 50(2): 371-381. |
| [2] | LIU Yiping, NI Menghui, WU Fangfang, LIU Hongli, HE Dan, KONG Dezheng. Association Analysis of Organ Traits with SSR Markers in Lotus(Nelumbo nucifera) [J]. Acta Horticulturae Sinica, 2023, 50(1): 103-115. |
| [3] | ZHAO Xueyan, WANG Qi, WANG Li, WANG Fangyuan, WANG Qing, LI Yan. Comparative Transcriptome Analysis of Differential Expression in Different Tissues of Corydalis yanhusuo [J]. Acta Horticulturae Sinica, 2023, 50(1): 177-187. |
| [4] | LI Qingqing, ZHAO Guangsheng, JI Jianbin, and CUI Jianping. A New Ornamental Lotus Cultivar‘Yanzhao Mudan’ [J]. Acta Horticulturae Sinica, 2022, 49(S2): 211-212. |
| [5] | JI Jianbin, LI Qingqing, ZHAO Gangsheng, and CUI Jianping. A New Ornamental Lotus Cultivar‘Longfu zhi Guang’ [J]. Acta Horticulturae Sinica, 2022, 49(S2): 213-214. |
| [6] | WANG Jingjing, YU Linjie, LIU Bin, YANG Lijuan, YANG Xinyu, and TANG Fei, . A New Ornamental Lotus Cultivar‘Yutangchun’ [J]. Acta Horticulturae Sinica, 2022, 49(S2): 215-216. |
| [7] | ZOU Xue, DING Fan, LIU Lifang, YU Hankaizong, CHEN Nianwei, and RAO Liping. A New Purple Potato Cultivar‘Mianziyu 1’ [J]. Acta Horticulturae Sinica, 2022, 49(S1): 93-94. |
| [8] | ZHANG Zhiyuan, JIANG Haicui, ZHANG Lingling, WAN Jindan, HU Junting, and GU Qi, . A New Ornamental Lotus Cultivar‘Zhongshan Nuanyang’ [J]. Acta Horticulturae Sinica, 2022, 49(S1): 159-160. |
| [9] | WANG Sha, ZHANG Xinhui, ZHAO Yujie, LI Bianbian, ZHAO Xueqing, SHEN Yu, DONG Jianmei, YUAN Zhaohe. Cloning and Functional Analysis of PgMYB111 Related to Anthocyanin Synthesis in Pomegranate [J]. Acta Horticulturae Sinica, 2022, 49(9): 1883-1894. |
| [10] | HUANG Ling, HU Xianmei, LIANG Zehui, WANG Yanping, CHAN Zhulong, XIANG Lin. Cloning and Function Identification of Anthocyanidin Synthase Gene TgANS in Tulipa gesneriana [J]. Acta Horticulturae Sinica, 2022, 49(9): 1935-1944. |
| [11] | LI Maofu, YANG Yuan, WANG Hua, FAN Youwei, SUN Pei, JIN Wanmei. Analysis the Function of R2R3 MYB Transcription Factor RhMYB113c on Regulating Anthocyanin Synthesis in Rosa hybrida [J]. Acta Horticulturae Sinica, 2022, 49(9): 1957-1966. |
| [12] | QIAN Jieyu, JIANG Lingli, ZHENG Gang, CHEN Jiahong, LAI Wuhao, XU Menghan, FU Jianxin, ZHANG Chao. Identification and Expression Analysis of MYB Transcription Factors Regulating the Anthocyanin Biosynthesis in Zinnia elegans and Function Research of ZeMYB9 [J]. Acta Horticulturae Sinica, 2022, 49(7): 1505-1518. |
| [13] | ZHANG Lugang, LU Qianqian, HE Qiong, XUE Yihua, MA Xiaomin, MA Shuai, NIE Shanshan, YANG Wenjing. Creation of Novel Germplasm of Purple-orange Heading Chinese Cabbage [J]. Acta Horticulturae Sinica, 2022, 49(7): 1582-1588. |
| [14] | CHEN Daozong, LIU Yi, SHEN Wenjie, ZHU Bo, TAN Chen. Identification and Analysis of PAP1/2 Homologous Genes in Brassica rapa,B. oleracea and B. napus [J]. Acta Horticulturae Sinica, 2022, 49(6): 1301-1312. |
| [15] | LI Xiaoming, YU Junchi, WANG Chunxia. Comparison of Growth and Secondary Metabolites of Purple and White Flower Dracocephalum moldavica Under Field,Greenhouse and Greenhouse Shading Conditions [J]. Acta Horticulturae Sinica, 2022, 49(6): 1363-1370. |
| Viewed | ||||||
|
Full text |
|
|||||
|
Abstract |
|
|||||
Copyright © 2012 Acta Horticulturae Sinica 京ICP备10030308号-2 国际联网备案号 11010802023439
Tel: 010-82109523 E-Mail: yuanyixuebao@126.com
Support by: Beijing Magtech Co.Ltd