
Acta Horticulturae Sinica ›› 2022, Vol. 49 ›› Issue (4): 827-840.doi: 10.16420/j.issn.0513-353x.2020-1069
• Research Papers • Previous Articles Next Articles
WANG Ying, AI Penghui, LI Shuailei, KANG Dongru, LI Zhongai, WANG Zicheng**( )
)
Received:2021-07-16
Revised:2021-11-15
Online:2022-04-25
Published:2022-04-24
Contact:
WANG Zicheng
E-mail:wzc@henu.edu.cn
CLC Number:
WANG Ying, AI Penghui, LI Shuailei, KANG Dongru, LI Zhongai, WANG Zicheng. Identification and Expression Analysis of Genes Related to DNA Methylation in Chrysanthemum × morifolium and C. nankingense[J]. Acta Horticulturae Sinica, 2022, 49(4): 827-840.
Add to citation manager EndNote|Ris|BibTeX
URL: https://www.ahs.ac.cn/EN/10.16420/j.issn.0513-353x.2020-1069
| 基因 Gene | 上游引物序列(5′-3′) Forward primer sequence | 下游引物序列(5′-3′) Reverse primer sequence |
|---|---|---|
| Cm-methylase1 | AACTGGTCACTATACAACTG | ATCATCTGCCGTCATTAC |
| Cm-methylase2 | GACGCTCTCCAAGATTAG | ACCACCATCATTAGATTCAA |
| Cm-methylase10 | TGACTCCGATAGGCTACTT | GGTATCCGAATCAATATCAACATT |
| Cm-demethylase3 | TGGAGAATGATGGAAGTG | GCCTGTTACCTTGGATAA |
| Cn-methylase1 | TGTCAAAATCTGCTGGAGG | TTGTCATTATTGTTGGGGG |
| Cn-methylase2 | AAAAAAGAAACGAAAGGGTC | ATAAAGAAAGCGAGAAATGG |
| Cn-methylase3 | TACAATTCATGGTGCCTTGC | ATTTTGCGCTTCCATATGCT |
| Cn-methylase4 | GCTTCTTCCACTTCCTCCCC | CTTAAACCCCCACCTCTCGT |
| Cn-methylase5 | TTGCCTCAGTTTCCTTTACC | AGCACCTCCATACTCCATTT |
| Cn-methylase6 | AATCACTTTGCTATCTCTCT | TTACTTACTTGTGTCTCTGG |
| Cn-methylase7 | TCTTTTGCTCTTGCTCACAC | CACACACAGCCCACACATTT |
| Cn-methylase8 | CCAAGCTAATGTGTGGCTTC | GCTGTCCATTAAACTTTGGG |
| Cn-methylase9 | TTTTGGAAGTGAATACAGGG | ATGGTTGACATAGTGGAGAA |
| Cn-methylase10 | ATGACAGTGAAGGACAGCGA | GAAATGAGGGGTATGAAAGG |
| Cn-demethylase1 | CAAACCTAAGAAACCGCCAA | CAACATCTCCAAAGCCACAA |
| Cn-demethylase2 | CTGAAGAAACTGGTAGACAA | CAAAATCAAGAACCTAATGG |
| Cn-demethylase3 | ACAGATAAGGACCACGAATG | GGACTGAATAACCAGCAAAC |
| Cn-demethylase4 | CCACCAGAAGGAAAATGTCG | CTCACCAGGATTGTCCCACG |
| Cn-demethylase5 | AAACCGAAGCCAATACTGAG | TTCTAAGAGTGGGTGGGAGT |
| Actin | AGCTTGCATATGTTGCTCTTGA | TTACCGTAAAGGTCCTTCCTGA |
Table 1 Appendix a sequences for fluorescent quantitative
| 基因 Gene | 上游引物序列(5′-3′) Forward primer sequence | 下游引物序列(5′-3′) Reverse primer sequence |
|---|---|---|
| Cm-methylase1 | AACTGGTCACTATACAACTG | ATCATCTGCCGTCATTAC |
| Cm-methylase2 | GACGCTCTCCAAGATTAG | ACCACCATCATTAGATTCAA |
| Cm-methylase10 | TGACTCCGATAGGCTACTT | GGTATCCGAATCAATATCAACATT |
| Cm-demethylase3 | TGGAGAATGATGGAAGTG | GCCTGTTACCTTGGATAA |
| Cn-methylase1 | TGTCAAAATCTGCTGGAGG | TTGTCATTATTGTTGGGGG |
| Cn-methylase2 | AAAAAAGAAACGAAAGGGTC | ATAAAGAAAGCGAGAAATGG |
| Cn-methylase3 | TACAATTCATGGTGCCTTGC | ATTTTGCGCTTCCATATGCT |
| Cn-methylase4 | GCTTCTTCCACTTCCTCCCC | CTTAAACCCCCACCTCTCGT |
| Cn-methylase5 | TTGCCTCAGTTTCCTTTACC | AGCACCTCCATACTCCATTT |
| Cn-methylase6 | AATCACTTTGCTATCTCTCT | TTACTTACTTGTGTCTCTGG |
| Cn-methylase7 | TCTTTTGCTCTTGCTCACAC | CACACACAGCCCACACATTT |
| Cn-methylase8 | CCAAGCTAATGTGTGGCTTC | GCTGTCCATTAAACTTTGGG |
| Cn-methylase9 | TTTTGGAAGTGAATACAGGG | ATGGTTGACATAGTGGAGAA |
| Cn-methylase10 | ATGACAGTGAAGGACAGCGA | GAAATGAGGGGTATGAAAGG |
| Cn-demethylase1 | CAAACCTAAGAAACCGCCAA | CAACATCTCCAAAGCCACAA |
| Cn-demethylase2 | CTGAAGAAACTGGTAGACAA | CAAAATCAAGAACCTAATGG |
| Cn-demethylase3 | ACAGATAAGGACCACGAATG | GGACTGAATAACCAGCAAAC |
| Cn-demethylase4 | CCACCAGAAGGAAAATGTCG | CTCACCAGGATTGTCCCACG |
| Cn-demethylase5 | AAACCGAAGCCAATACTGAG | TTCTAAGAGTGGGTGGGAGT |
| Actin | AGCTTGCATATGTTGCTCTTGA | TTACCGTAAAGGTCCTTCCTGA |
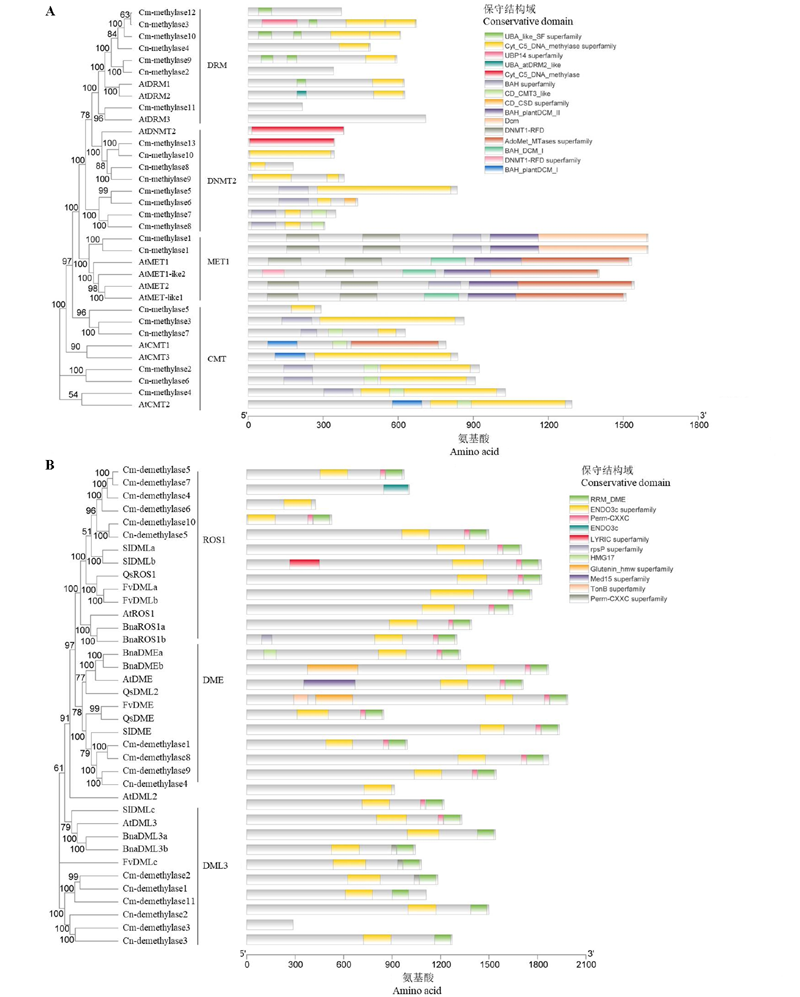
Fig. 1 Phylogenetic tree and conserved domains of DNA methylases(A)and demethylases(B)of Chrysanthemum ‘Jinbeidahong’(Cm)and Chrysanthemum nankingense(Cn) Using the Neighbour-joining method in MEGA 7 software. The value of bootstrap is 1 000. Numbers indicate bootstrap values. At:Arabidopsis thaliana;Sl:Solanum lycopersicum;Qs:Quercus suber;Fv:Fragaria vesca;Bna:Brassica napus.
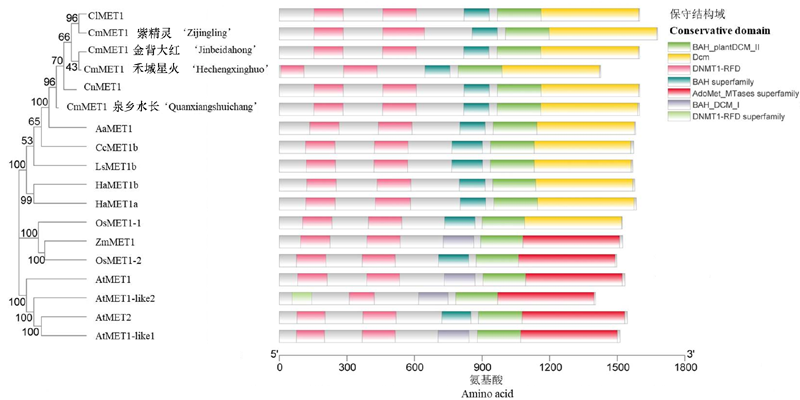
Fig. 2 Phylogenetic tree and conserved domains of MET1 family in different species Using the Neighbour-joining method in MEGA 7 software. The value of bootstrap is 1 000. Numbers indicate bootstrap values. Cl:C. lavandulifolium;Cn:C. nankingense;Aa:Artemisia annua;Cc:Cynara cardunculus;Ls:Lactuca saliva;Ha:Helianthus annuus;Os:Oryza sativa;Zm:Zea mays;At:Arabidopsis thaliana.
| 酶 Enzyme | 基因 Gene name | CDS长度/bp CDS length | 蛋白质 Protein | 类型 Type | ||
|---|---|---|---|---|---|---|
| 长度/aa Length | 分子量/Da Mw | 等电点 pI | ||||
| 甲基转移酶 | Cm-methylase1 | 4 800 | 1 599 | 178 739.29 | 5.75 | MET1 |
| Methyltransferase | Cm-methylase2 | 2 788 | 925 | 103 843.57 | 6.14 | CMT |
| Cm-methylase3 | 2 595 | 864 | 97 299.75 | 4.91 | CMT | |
| Cm-methylase4 | 3 093 | 1 030 | 116 027.35 | 5.92 | CMT | |
| Cm-methylase5 | 2 511 | 836 | 94 321.79 | 5.95 | DNMT2 | |
| Cm-methylase6 | 1 317 | 438 | 49 870.56 | 5.24 | DNMT2 | |
| Cm-methylase7 | 1 053 | 350 | 39 764.17 | 5.13 | DNMT2 | |
| Cm-methylase8 | 921 | 306 | 34 963.24 | 4.91 | DNMT2 | |
| Cm-methylase9 | 1 788 | 595 | 66 491.85 | 4.89 | DRM | |
| Cm-methylase10 | 1 833 | 610 | 68 071.51 | 4.96 | DRM | |
| Cm-methylase11 | 654 | 217 | 24 671.57 | 8.97 | DRM | |
| Cm-methylase12 | 1 122 | 373 | 40 971.16 | 4.34 | DRM | |
| Cm-methylase13 | 1 041 | 346 | 39 705.42 | 5.83 | DNMT2 | |
| 去甲基化酶 | Cm-demethylase1 | 4 638 | 1 545 | 173 065.09 | 8.26 | DME |
| Demethylase | Cm-demethylase2 | 4 503 | 1 500 | 170 278.85 | 6.35 | DML3 |
| Cm-demethylase3 | 3 822 | 1 273 | 144 744.28 | 8.93 | DML3 | |
| Cm-demethylase4 | 1 281 | 426 | 47 988.14 | 5.92 | ROS1 | |
| Cm-demethylase5 | 2 928 | 975 | 110 623.30 | 5.53 | ROS1 | |
| Cm-demethylase6 | 1 584 | 527 | 60 170.57 | 5.78 | ROS1 | |
| Cm-demethylase7 | 3 027 | 1 008 | 115 750.42 | 5.78 | ROS1 | |
| Cm-demethylase8 | 2 751 | 916 | 103 280.83 | 8.51 | DME | |
| Cm-demethylase9 | 3 672 | 1 223 | 137 284.38 | 6.74 | DME | |
| Cm-demethylase10 | 4 503 | 1 500 | 169 026.50 | 6.05 | ROS1 | |
| Cm-demethylase11 | 864 | 287 | 32 569.63 | 4.37 | DML3 | |
Table 2 Structural analysis of DNA methyltransferases and demethylase genes of Chrysanthemum‘Jinbeidahong’
| 酶 Enzyme | 基因 Gene name | CDS长度/bp CDS length | 蛋白质 Protein | 类型 Type | ||
|---|---|---|---|---|---|---|
| 长度/aa Length | 分子量/Da Mw | 等电点 pI | ||||
| 甲基转移酶 | Cm-methylase1 | 4 800 | 1 599 | 178 739.29 | 5.75 | MET1 |
| Methyltransferase | Cm-methylase2 | 2 788 | 925 | 103 843.57 | 6.14 | CMT |
| Cm-methylase3 | 2 595 | 864 | 97 299.75 | 4.91 | CMT | |
| Cm-methylase4 | 3 093 | 1 030 | 116 027.35 | 5.92 | CMT | |
| Cm-methylase5 | 2 511 | 836 | 94 321.79 | 5.95 | DNMT2 | |
| Cm-methylase6 | 1 317 | 438 | 49 870.56 | 5.24 | DNMT2 | |
| Cm-methylase7 | 1 053 | 350 | 39 764.17 | 5.13 | DNMT2 | |
| Cm-methylase8 | 921 | 306 | 34 963.24 | 4.91 | DNMT2 | |
| Cm-methylase9 | 1 788 | 595 | 66 491.85 | 4.89 | DRM | |
| Cm-methylase10 | 1 833 | 610 | 68 071.51 | 4.96 | DRM | |
| Cm-methylase11 | 654 | 217 | 24 671.57 | 8.97 | DRM | |
| Cm-methylase12 | 1 122 | 373 | 40 971.16 | 4.34 | DRM | |
| Cm-methylase13 | 1 041 | 346 | 39 705.42 | 5.83 | DNMT2 | |
| 去甲基化酶 | Cm-demethylase1 | 4 638 | 1 545 | 173 065.09 | 8.26 | DME |
| Demethylase | Cm-demethylase2 | 4 503 | 1 500 | 170 278.85 | 6.35 | DML3 |
| Cm-demethylase3 | 3 822 | 1 273 | 144 744.28 | 8.93 | DML3 | |
| Cm-demethylase4 | 1 281 | 426 | 47 988.14 | 5.92 | ROS1 | |
| Cm-demethylase5 | 2 928 | 975 | 110 623.30 | 5.53 | ROS1 | |
| Cm-demethylase6 | 1 584 | 527 | 60 170.57 | 5.78 | ROS1 | |
| Cm-demethylase7 | 3 027 | 1 008 | 115 750.42 | 5.78 | ROS1 | |
| Cm-demethylase8 | 2 751 | 916 | 103 280.83 | 8.51 | DME | |
| Cm-demethylase9 | 3 672 | 1 223 | 137 284.38 | 6.74 | DME | |
| Cm-demethylase10 | 4 503 | 1 500 | 169 026.50 | 6.05 | ROS1 | |
| Cm-demethylase11 | 864 | 287 | 32 569.63 | 4.37 | DML3 | |
| 酶 Enzyme | 基因 Gene Name | CDS长度/bp CDS length | 外显子数 Number of exons | 蛋白质Protein | 类型 Type | ||
|---|---|---|---|---|---|---|---|
| 长度/aa Length | 分子量/Da Mw | 等电点 pI | |||||
| 甲基转移酶 | Cn-methylase1 | 4 803 | 3 | 1 600 | 178 962.60 | 5.78 | MET1 |
| Methyltransferase | Cn-methylase2 | 1 029 | 4 | 342 | 39 472.94 | 9.68 | DRM |
| Cn-methylase3 | 2 019 | 12 | 672 | 75 038.30 | 5.01 | DRM | |
| Cn-methylase4 | 1 470 | 9 | 489 | 54 992.14 | 5.19 | DRM | |
| Cn-methylase5 | 879 | 10 | 292 | 32 439.25 | 7.63 | CMT | |
| Cn-methylase6 | 2 730 | 20 | 909 | 101 888.25 | 6.20 | CMT | |
| Cn-methylase7 | 1 887 | 18 | 628 | 71 372.50 | 4.99 | CMT | |
| Cn-methylase8 | 543 | 4 | 181 | 20 947.95 | 7.57 | DNMT2 | |
| Cn-methylase9 | 1 155 | 9 | 384 | 44 105.80 | 6.36 | DNMT2 | |
| Cn-methylase10 | 1 038 | 9 | 345 | 39 700.18 | 5.27 | DNMT2 | |
| 去甲基化酶 | Cn-demethylase1 | 4 254 | 18 | 1 417 | 160 835.45 | 7.46 | DML3 |
| Demethylase | Cn-demethylase2 | 2 805 | 14 | 934 | 106 836.11 | 8.79 | DML3 |
| Cn-demethylase3 | 2 613 | 15 | 870 | 99 036.82 | 9.20 | DML3 | |
| Cn-demethylase4 | 5 244 | 20 | 1 747 | 194 573.45 | 6.65 | DME | |
| Cn-demethylase5 | 4 707 | 18 | 1 568 | 175 882.89 | 5.96 | ROS1 | |
Table 3 Structural analysis of DNA methyltransferases and demethylase genes of Chrysanthemum nankingense
| 酶 Enzyme | 基因 Gene Name | CDS长度/bp CDS length | 外显子数 Number of exons | 蛋白质Protein | 类型 Type | ||
|---|---|---|---|---|---|---|---|
| 长度/aa Length | 分子量/Da Mw | 等电点 pI | |||||
| 甲基转移酶 | Cn-methylase1 | 4 803 | 3 | 1 600 | 178 962.60 | 5.78 | MET1 |
| Methyltransferase | Cn-methylase2 | 1 029 | 4 | 342 | 39 472.94 | 9.68 | DRM |
| Cn-methylase3 | 2 019 | 12 | 672 | 75 038.30 | 5.01 | DRM | |
| Cn-methylase4 | 1 470 | 9 | 489 | 54 992.14 | 5.19 | DRM | |
| Cn-methylase5 | 879 | 10 | 292 | 32 439.25 | 7.63 | CMT | |
| Cn-methylase6 | 2 730 | 20 | 909 | 101 888.25 | 6.20 | CMT | |
| Cn-methylase7 | 1 887 | 18 | 628 | 71 372.50 | 4.99 | CMT | |
| Cn-methylase8 | 543 | 4 | 181 | 20 947.95 | 7.57 | DNMT2 | |
| Cn-methylase9 | 1 155 | 9 | 384 | 44 105.80 | 6.36 | DNMT2 | |
| Cn-methylase10 | 1 038 | 9 | 345 | 39 700.18 | 5.27 | DNMT2 | |
| 去甲基化酶 | Cn-demethylase1 | 4 254 | 18 | 1 417 | 160 835.45 | 7.46 | DML3 |
| Demethylase | Cn-demethylase2 | 2 805 | 14 | 934 | 106 836.11 | 8.79 | DML3 |
| Cn-demethylase3 | 2 613 | 15 | 870 | 99 036.82 | 9.20 | DML3 | |
| Cn-demethylase4 | 5 244 | 20 | 1 747 | 194 573.45 | 6.65 | DME | |
| Cn-demethylase5 | 4 707 | 18 | 1 568 | 175 882.89 | 5.96 | ROS1 | |
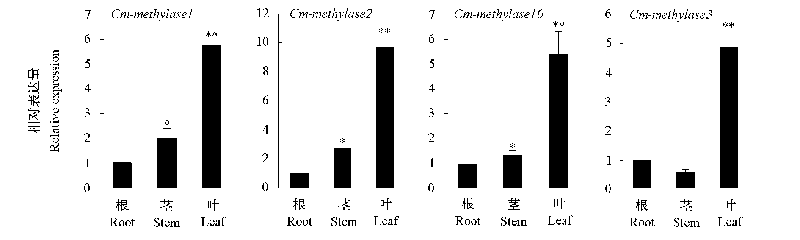
Fig. 3 Relative expression of DNA methylases and demethylases gene in root,stem and leaf of Chrysanthemum‘Jinbeidahong’seedling stage Expression leves in root were arbitrarily set to 1. Use * to indicate that the difference is significant(P < 0.05),** to indicate that the difference is significant extremely(P < 0.01). The same below
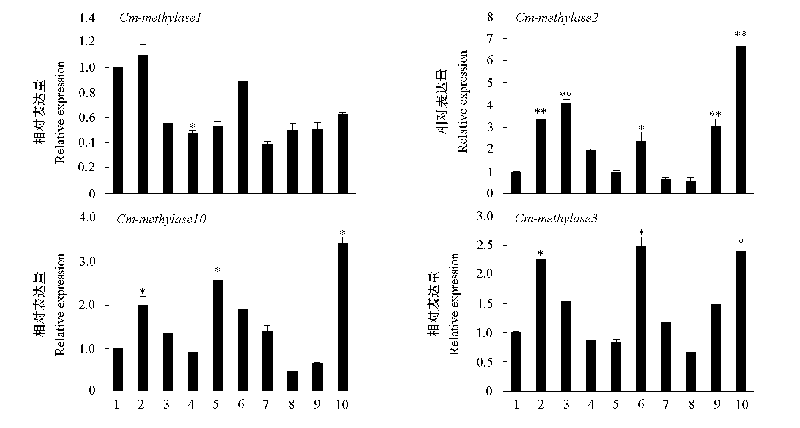
Fig. 5 Relative expression of DNA methylases and demethylases in different chrysanthemum cultivars 1:‘Jinbeidahong’;2:‘Qiansiwanlü’;3:‘Yinlongfenshui’;4:‘Guoqinghong’;5:‘Nannong Hongxiu’;6:‘Zijingling’;7:‘Hangbaiju’;8:‘Zidie Fanfei’;9:‘Tiannvdeshenmi’;10:‘Yutaiyihao’. Expression leves of‘Jinbeidahong’were arbitrarily set to 1, use * to indicate that the difference is significant(P < 0.05),** to indicate that the difference is significant extremely(P < 0.01).
| [1] |
Aparicio G, Gotz S, Conesa A, Segrelles D, Blanquer I, Garcia J M, Hernandez V, Robles M, Talon M. 2006. Blast2GO goes grid:developing a grid-enabled prototype for functional genomics analysis. Studies in Health Technology and Informatics, 120:194-204.
pmid: 16823138 |
| [2] | Ashapkin V V, Kutueva L I, Vanyushin B F. 2016. Plant DNA methyltransferase genes:multiplictiy,expression,methylation patterns. Biochemistry, 81 (2):141-151. |
| [3] |
Chan W L, Henderson I R, Jacobsen S E. 2005. Gardening the genome:DNA methylation in Arabidopsis thaliana. Nature Reviews Genetics, 6 (5):351-360.
doi: 10.1038/nrg1601 URL |
| [4] | Chang Lin-lin. 2009. Isolation and expression analysis of DNA methyltransferase genes in strawberry and apple[Ph. D. Dissertation]. Shenyang: Shenyang Agricultural University. (in Chinese) |
| 常琳琳. 2009. 草莓和苹果DNA甲基转移酶基因分离与表达分析[博士论文]. 沈阳: 沈阳农业大学. | |
| [5] | Dai Si-lan, Wang Wen-kui, Huang Jia-ping. 2002. Advances of researches on phylogeny of Dendranthema and origin of chrysanthemum. Journal of Beijing Forestry University, 24 (6):230-234. (in Chinese) |
| 戴思兰, 王文奎, 黄家平. 2002. 菊属系统学及菊花起源的研究进展. 北京林业大学学报, 24 (6):230-234. | |
| [6] |
Dong W, Li M M, Li Z A, Li S L, Zhu Y, Hong X, Wang Z C. 2020. Transcriptome analysis of the molecular mechanism of Chrysanthemum flower color change under short-day photoperiods. Plant Physiology and Biochemistry, 146:315-328.
doi: S0981-9428(19)30492-9 pmid: 31785518 |
| [7] |
Fan S H, Liu H F, Liu J, Hua W, Xu S M, Li J. 2020. Systematic analysis of the DNA methylase and demethylase gene families in rapeseed(Brassica napus L.)and their expression variations after salt and heat stresses. International Journal of Molecular Sciences, 21 (3):953.
doi: 10.3390/ijms21030953 URL |
| [8] | Fu Q S, Song A X, Hu H Y. 2012. Structural aspects of ubiquitin binding specificities. Current Protein & Peptide Science, 13 (5):482-489. |
| [9] |
Furner I J, Matzke M. 2011. Methylation and demethylation of the Arabidopsis genome. Current Opinion in Plant Biology, 14 (2):137-141.
doi: 10.1016/j.pbi.2010.11.004 pmid: 21159546 |
| [10] |
Gong Z Z, Morales-Ruiz T, Ariza R R, Roldan-Arjona T, David L, Zhu J K. 2002. ROS1,a repressor of transcriptional gene silencing in Arabidopsis,encodes a DNA glycosylaselyase. Cell, 111 (6):803-814.
doi: 10.1016/S0092-8674(02)01133-9 URL |
| [11] |
Gu T T, Ren S, Wang Y H, Han Y H, Li Y. 2016. Characterization of DNA methyltransferase and demethylase genes in Fragaria vesca. Molecular Genetics and Genomics, 291 (3):1333-1345.
doi: 10.1007/s00438-016-1187-y URL |
| [12] |
Kang D R, Dai S L, Gao K, Zhang F, Luo H. 2019a. Morphological variation of Chrysanthemum lavandulifolium induced by 5-azaC treatment. Scientia Horticulturae, 257:108645.
doi: 10.1016/j.scienta.2019.108645 URL |
| [13] |
Kang D R, Dai S L, Gao K, Zhang F, Luo H. 2019b. Morphological variation of five cut chrysanthemum cultivars induced by 5-azaC treatment. HortScience, 54 (7):1208-1216.
doi: 10.21273/HORTSCI14012-18 URL |
| [14] | Li J, Li C L, Lu S F. 2018. Identification and characterization of the cytosine-5 DNA methyltransferase gene family in Salvia miltiorrhiza. Peer J, 6:4461. |
| [15] |
Li S L, Li M M, Li Z A, Zhu Y, Ding H X, Fan X X, Li F, Wang Z C. 2019. Effects of the silencing of CmMET1 by RNA interference in chrysanthemum(Chrysanthemum morifolium). Plant Biotechnology Reports, 13 (1):63-72.
doi: 10.1007/s11816-019-00516-5 URL |
| [16] | Liu Yan-hua, Li Zhong-ai, Li Jie, Liu Xiao, Wang Zi-cheng. 2017. Effect of curcumin on DNA methylation and growth of chrysanthemum. Henan Agricultural Sciences, 46 (5):100-105. (in Chinese) |
| 刘艳华, 李忠爱, 李杰, 刘晓, 王子成. 2017. 姜黄素对菊花DNA甲基化及其生长发育的影响. 河南农业科学, 46 (5):100-105. | |
| [17] |
Matzke M A, Mette M F, Matzke A J M. 2000. Transgene silencing by the host genome defense:implications for the evolution of epigenetic control mechanisms in plants and vertebrates. Plant Molecular Biology, 43 (2-3):401-415.
pmid: 10999419 |
| [18] | Mok Y G, Uzawa R, Lee J, Werner G M, Eichman B F, Fischer R L, Huh J H. 2010. Domain structure of the DEMETER 5-methylcytosine DNA glycosylase. Proceedings of the National Academy of Sciences of the United States of America, 107 (45):19225-19230. |
| [19] | Morales-Ruiz T, Ortega-Galisteo A P, Ponferrada-Marin M I, Martinez-Macias M I, Ariza R R, Roldan-Arjona T. 2006. DEMETER and pepressor of silencing 1 encode 5-methylcytosine DNA glycosylases. Proceedings of the National Academy of Sciences of the United States of America, 103 (18):6853-6858. |
| [20] |
Nielsen P R, Nietlispach D, Mott H R, Callaghan J, Bannister A, Kouzarides T, Murzin A G, Murzina N V, Laue E D. 2002. Structure of the HP 1 chromodomain bound to histone H3 methylated at lysine 9. Nature, 416 (6876):103-107.
doi: 10.1038/nature722 URL |
| [21] |
Oliver A W, Jones S A, Roe S M, Matthews S, Goodwin G H, Pearl L H. 2005. Crystal structure of the proximal BAH domain of the polybromo protein. Biochemical Journal, 389 (3):657-664.
doi: 10.1042/BJ20050310 URL |
| [22] |
Ortega-Galisteo A P, Morales-Ruiz T, Ariza R R, Roldan-Arjona, 2008. Arabidopsis DEMETER-LIKE proteins DML2 and DML 3 are required for appropriate distribution of DNA methylation marks. Plant Molecular Biology, 67 (6):671-681.
doi: 10.1007/s11103-008-9346-0 pmid: 18493721 |
| [23] |
Qi X Y, Wang H B, Song A P, Jiang J F, Chen S M, Chen F D. 2018. Genomic and transcriptomic alterations following intergeneric hybridization and polyploidization in the Chrysanthemum nankingense × Tanacetum vulgare hybrid and allopolyploid (Asteraceae). Horticulture Research, 5:5.
doi: 10.1038/s41438-017-0003-0 URL |
| [24] |
Qian Y X, Xi Y L, Cheng B J, Zhu S W. 2014. Genome-wide identification and expression profiling of DNA methyltransferase gene family in maize. Plant Cell Reports, 33 (10):1661-1672.
doi: 10.1007/s00299-014-1645-0 URL |
| [25] |
Silva H G, Sobral R S, Magalhes A P, Morais-Cecilio L, Costa M M R. 2020. Genome-wide identification of epigenetic regulators in Quercus suber L. International Journal of Molecular Sciences, 21 (11):3783.
doi: 10.3390/ijms21113783 URL |
| [26] |
Song C, Liu Y F, Song A P, Dong G Q, Zhao H B, Sun W, Ramakrishnan S, Wang Y, Wang S B, Li T Z, Niu Y, Jiang J F, Dong B, Xia Y, Chen S M, Hu Z G, Chen F D, Chen S L. 2018. The Chrysanthemum nankingense genome provides insights into the evolution and diversification of chrysanthemum flowers and medicinal traits. Molecular Plant, 11 (12):1482-1491.
doi: S1674-2052(18)30308-3 pmid: 30342096 |
| [27] | Wang H B, Qi X Y, Chen S M, Fang W M, Guan Z Y, Teng N J, Liao Y, Jiang J F, Chen F D. 2015. Limited DNA methylation variation and the transcription of MET1 and DDM1 in the genus Chrysanthemum(Asteraceae):following the track of polyploidy. Frontiers in Plant Science, 6:68. |
| [28] | Xu Sheng, Zhang Xin, Liu Decai, Gu Tingting. 2020. Identification and functional analysis of N6-Adenine methylation(6mA)methyltransferase genes in Fragaria vesca. Acta Horticulturae Sinica, 47 (11):2194-2206. (in Chinese) |
| 徐昇, 张鑫, 刘德才, 顾婷婷. 2020. 森林草莓6mA甲基转移酶基因鉴定及功能分析. 园艺学报, 47 (11):2194-2206. | |
| [29] |
Yamauchi T, Johzuka-Hisatomi Y, Terada R, Nakamura I, Lida S. 2014. The MET1b gene encoding a maintenance DNA methyltransferase is indispensable for normal development in rice. Plant Molecular Biology, 85 (3):219-232.
doi: 10.1007/s11103-014-0178-9 pmid: 24535433 |
| [30] |
Yu Yuan-yuan, Guo Yun-hui, Wen Li-zhu, Sun Cui-hui, Fan Hong-mei, Sun Xian-zhi, Wang Wen-li, Sun Xia, Zheng Cheng-shu. 2018. Effects of DNA methylation level on the nitrate uptake of roots in Chrysanthemum morifolium. Plant Physiology Journal, 54 (5):886-894. (in Chinese)
doi: 10.1104/pp.54.6.886 URL |
| 于媛媛, 郭芸珲, 温立柱, 孙翠慧, 樊红梅, 孙宪芝, 王文莉, 孙霞, 郑成淑. 2018. 菊花基因组DNA甲基化水平对根系硝态氮吸收的影响. 植物生理学报, 54 (5):886-894. | |
| [31] |
Zhang C M, Hao Y J. 2020. Advances in genomic,transcriptomic,and metabolomic analyses of fruit quality in fruit crops. Horticultural Plant Journal, 6 (6):361-371.
doi: 10.1016/j.hpj.2020.11.001 URL |
| [32] |
Zhang F, Deng C Y, Dai S L. 2020. MSAP analysis reveals the DNA methylation dynamics during capitulum development in Chrysanthemum lavandulifolium. Canadian Journal of Plant Science, 101 (3):1-31.
doi: 10.1139/cjps-2019-0318 URL |
| [33] |
Zhang H M, Lang Z B, Zhu J K. 2018. Dynamics and function of DNA methylation in plants. Nature Reviews Molecular Cell Biology, 19 (8):489-506.
doi: 10.1038/s41580-018-0016-z URL |
| [34] | Zhou Chenping, Yang Min, Guo Jinju, Kuang Ruibin, Yang Hu, Huang Bingxiong, Wei Yuerong. 2022. Dynamic changes in DNA methylome and transcriptome patterns during papaya fruit ripening. Acta Horticulturae Sinica, 49 (3):519-532. (in Chinese) |
| 周陈平, 杨敏, 郭金菊, 邝瑞彬, 杨护, 黄炳雄, 魏岳荣. 2022. 番木瓜成熟过程中全基因组DNA甲基化和转录组变化分析. 园艺学报, 49 (3):519-532. |
| [1] | WANG Xiaochen, NIE Ziye, LIU Xianju, DUAN Wei, FAN Peige, and LIANG Zhenchang, . Effects of Abscisic Acid on Monoterpene Synthesis in‘Jingxiangyu’Grape Berries [J]. Acta Horticulturae Sinica, 2023, 50(2): 237-249. |
| [2] | ZHAI Hanhan, ZHAI Yujie, TIAN Yi, ZHANG Ye, YANG Li, WEN Zhiliang, CHEN Haijiang. Genome-wide Identification of Peach SAUR Gene Family and Characterization of PpSAUR5 Gene [J]. Acta Horticulturae Sinica, 2023, 50(1): 1-14. |
| [3] | SHAO Fengqing, LUO Xiurong, WANG Qi, ZHANG Xianzhi, WANG Wencai. Advances in Research of DNA Methylation Regulation During Fruit Ripening [J]. Acta Horticulturae Sinica, 2023, 50(1): 197-208. |
| [4] | YE Zhiqin and YANG Juan. A New Chrysanthemum Cultivar‘Zibinyun’ [J]. Acta Horticulturae Sinica, 2022, 49(S2): 173-174. |
| [5] | ZHAO Yanli, LI Zhanhong, CAO Qin, DAI Miaofei, GAO Mengmeng, SUN Zhenzhu, LI Huikuan, LI Yonghua, BIAN Shuxun, and HUANG Gan. A New Chrysanthemum Cultivar‘Bianjing Qingdianhuang’ [J]. Acta Horticulturae Sinica, 2022, 49(S2): 175-176. |
| [6] | CAO Qin, ZHAO Yanli, LI Zhanhong, GAO Mengmeng, DAI Miaofei, LI Yonghua, LI Huikuan, SUN Zhenzhu, BIAN Shuxun, and LI Fei. A New Muitiflora Chrysanthemum Cultivar‘Bianjing Zijingling’ [J]. Acta Horticulturae Sinica, 2022, 49(S2): 177-178. |
| [7] | CHU Zhuannan, LI Weiwen, DONG Ling, PENG Xingxing, CUI Guangsheng, TANG Maogui, ZHANG Qinghua, SHEN Wensheng, ZHAO Wei, XIONG Rui, HAN Piao, WU Jiqiu, LI Yulong, YANG Yueying, and ZHANG Qiling. A New Chrysanthemum morifolium Cultivar‘Gongju 1’ [J]. Acta Horticulturae Sinica, 2022, 49(S2): 179-180. |
| [8] | YANG Xiuwei, SU Jiangshuo, ZHANG Fei, GUAN Zhiyong, FANG Weimin, CHEN Fadi. Quantitative Evaluation and Genetic Variation of Flower Opening Angles in Spray Cut Chrysanthemums [J]. Acta Horticulturae Sinica, 2022, 49(8): 1723-1734. |
| [9] | ZHANG Qiuyue, LIU Changlai, YU Xiaojing, YANG Jiading, FENG Chaonian. Screening of Reference Genes for Differentially Expressed Genes in Pyrus betulaefolia Plant Under Salt Stress by qRT-PCR [J]. Acta Horticulturae Sinica, 2022, 49(7): 1557-1570. |
| [10] | LI Yamei, MA Fuli, ZHANG Shanqi, HUANG Jinqiu, CHEN Mengting, ZHOU Junyong, SUN Qibao, SUN Jun. Optimization of Jujube Callus Transformation System and Application of ZjBRC1 in Regulating ZjYUCCA Expression [J]. Acta Horticulturae Sinica, 2022, 49(4): 749-757. |
| [11] | LIU Xiaowei, XIA Bin, LI Ziwei, YANG Yujia, CHEN Bin, ZHOU Yunwei, HE Miao. Overexpression of Chrysanthemum indicum miR396a Gene in Arabidopsis Enhances Its Salt Tolerance [J]. Acta Horticulturae Sinica, 2022, 49(4): 816-826. |
| [12] | ZHOU Chenping, YANG Min, GUO Jinju, KUANG Ruibin, YANG Hu, HUANG Bingxiong, WEI Yuerong. Dynamic Changes in DNA Methylome and Transcriptome Patterns During Papaya Fruit Ripening [J]. Acta Horticulturae Sinica, 2022, 49(3): 519-532. |
| [13] | NIE Wenfeng, WANG Jinyu, GAO Chunjuan, CHEN Xuehao. A Review on Epigenetic Modifications in Regulating Fruit Development of Horticultural Crops [J]. Acta Horticulturae Sinica, 2022, 49(3): 671-686. |
| [14] | ZHANG Rui, ZHANG Xiayi, ZHAO Ting, WANG Shuangcheng, ZHANG Zhongxing, LIU Bo, ZHANG De, WANG Yanxiu. Transcriptome Analysis of the Molecular Mechanism of Saline-alkali Stress Response in Malus halliana Leaves [J]. Acta Horticulturae Sinica, 2022, 49(2): 237-251. |
| [15] | ZHOU Zhiming, YANG Jiabao, ZHANG Cheng, ZENG Linglu, MENG Wanqiu, SUN Li. Genome-wide Identification and Expression Analyses of Long-chain Acyl-CoA Synthetases Under Abiotic Stresses in Helianthus annuus [J]. Acta Horticulturae Sinica, 2022, 49(2): 352-364. |
| Viewed | ||||||
|
Full text |
|
|||||
|
Abstract |
|
|||||
Copyright © 2012 Acta Horticulturae Sinica 京ICP备10030308号-2 国际联网备案号 11010802023439
Tel: 010-82109523 E-Mail: yuanyixuebao@126.com
Support by: Beijing Magtech Co.Ltd