
园艺学报 ›› 2022, Vol. 49 ›› Issue (2): 352-364.doi: 10.16420/j.issn.0513-353x.2020-1013
收稿日期:2021-07-01
修回日期:2021-12-23
出版日期:2022-02-25
发布日期:2022-02-28
通讯作者:
孙黎
E-mail:sunlishz@126.com
基金资助:
ZHOU Zhiming, YANG Jiabao, ZHANG Cheng, ZENG Linglu, MENG Wanqiu, SUN Li( )
)
Received:2021-07-01
Revised:2021-12-23
Online:2022-02-25
Published:2022-02-28
Contact:
SUN Li
E-mail:sunlishz@126.com
摘要:
利用生物信息学技术从向日葵(Helianthus annuus)基因组中鉴定了13个长链酰基辅酶A合成酶(long-chain acyl-CoA synthases,LACSs)家族基因,其分布在向日葵8条染色体上。系统发育树显示,HaLACS家族成员分为6个亚族,且同一亚族成员具有相似的基因结构和蛋白保守基序。基因复制分析发现HaLACS家族扩增的主要因素是片段复制。HaLACS的启动子区域富含逆境响应和植物激素调控相关元件。表达谱分析显示HaLACS在向日葵不同组织中的表达存在差异。比较2个向日葵品种(高/低亚油酸含量)中HaLACS在种子发育不同阶段的表达,HaLACS呈现差异表达。qRT-PCR结果表明,200 mmol · L-1 NaCl处理24 h后,7个HaLACS在茎中的表达量与同期对照相比显著上调;100 μmol · L-1 ABA 处理3 h后,8个HaLACS在根中的表达量显著上调;15% PEG 6000模拟干旱胁迫后,多数HaLACS在不同组织中呈现诱导表达。综上,向日葵LACS基因家族不仅参与了向日葵脂质的合成,而且在应对非生物胁迫时发挥重要作用。
中图分类号:
周至铭, 杨佳宝, 张程, 曾令露, 孟晚秋, 孙黎. 向日葵LACS家族鉴定及响应非生物胁迫表达分析[J]. 园艺学报, 2022, 49(2): 352-364.
ZHOU Zhiming, YANG Jiabao, ZHANG Cheng, ZENG Linglu, MENG Wanqiu, SUN Li. Genome-wide Identification and Expression Analyses of Long-chain Acyl-CoA Synthetases Under Abiotic Stresses in Helianthus annuus[J]. Acta Horticulturae Sinica, 2022, 49(2): 352-364.
| 基因Gene | 正向引物(5′-3′)Forward primer | 反向引物(5′-3′)Reverse primer |
|---|---|---|
| 18S rRNA | CTACCACATCCAAGGAAGGCAG | CGACAGAAGGGACGAGTAAACC |
| HaLACS1-1 | GAGCTCGAAAACGGAACGTG | ATTGCAAGCCTCCATCGCTA |
| HaLACS1-2 | TTGGACCATGCCGAAGTTGA | AAGGAACTCGTTCCACGCAT |
| HaLACS2 | AGAAACAGACAAACCGGGCA | ACGCCTTCTTTACAGTGCCT |
| HaLACS4-1 | GGTTATGAGCATGGAGGCGT | CCACGCCAAAATTTCCGACT |
| HaLACS4-2 | GGTTATGAGCATGGAGGCGT | GGTTATGAGCATGGAGGCGT |
| HaLACS4-3 | CCTCCGATTGACGGTTTGGA | CTCAACTCCACAAGCTCGGA |
| HaLACS4-4 | CGGTGAAAGACGGAACTCCA | CGACGCCACATGAACGGATA |
| HaLACS4-5 | GAAAACAATCGTGAGCTTTGGGA | AGTTCAACGATACTTTCGCGCT |
| HaLACS6 | CACTGCCGGAGAGTTCTTCA | TGCTTCCCCGTAAGTTGTCC |
| HaLACS7 | TCATCTTCTTCCCTCCACCGA | ATCTGGTGACAAGCTTCAGCG |
| HaLACS8 | GCAGATGGATGTTGGGGGAT | CGCCAACTTCAACTGGAACG |
| HaLACS9-1 | ACTCTCGCCCTTCGGAAATC | TTTCAAATGCCCTCCCGTCT |
| HaLACS9-2 | ATTGCATTGCAGGCGTGTTT | TTTCCAACTGCTGCTTCCGT |
表1 qRT-PCR引物序列
Table 1 Primers used for qRT-PCR
| 基因Gene | 正向引物(5′-3′)Forward primer | 反向引物(5′-3′)Reverse primer |
|---|---|---|
| 18S rRNA | CTACCACATCCAAGGAAGGCAG | CGACAGAAGGGACGAGTAAACC |
| HaLACS1-1 | GAGCTCGAAAACGGAACGTG | ATTGCAAGCCTCCATCGCTA |
| HaLACS1-2 | TTGGACCATGCCGAAGTTGA | AAGGAACTCGTTCCACGCAT |
| HaLACS2 | AGAAACAGACAAACCGGGCA | ACGCCTTCTTTACAGTGCCT |
| HaLACS4-1 | GGTTATGAGCATGGAGGCGT | CCACGCCAAAATTTCCGACT |
| HaLACS4-2 | GGTTATGAGCATGGAGGCGT | GGTTATGAGCATGGAGGCGT |
| HaLACS4-3 | CCTCCGATTGACGGTTTGGA | CTCAACTCCACAAGCTCGGA |
| HaLACS4-4 | CGGTGAAAGACGGAACTCCA | CGACGCCACATGAACGGATA |
| HaLACS4-5 | GAAAACAATCGTGAGCTTTGGGA | AGTTCAACGATACTTTCGCGCT |
| HaLACS6 | CACTGCCGGAGAGTTCTTCA | TGCTTCCCCGTAAGTTGTCC |
| HaLACS7 | TCATCTTCTTCCCTCCACCGA | ATCTGGTGACAAGCTTCAGCG |
| HaLACS8 | GCAGATGGATGTTGGGGGAT | CGCCAACTTCAACTGGAACG |
| HaLACS9-1 | ACTCTCGCCCTTCGGAAATC | TTTCAAATGCCCTCCCGTCT |
| HaLACS9-2 | ATTGCATTGCAGGCGTGTTT | TTTCCAACTGCTGCTTCCGT |
| 转录本ID Transcript ID | 基因名称 Gene name | CDS/bp | 大小/aa Size | 蛋白分子量/kD Molecular weight | 等电点 pI | 拟南芥同源基因 Arabidopsis homologous gene | |
|---|---|---|---|---|---|---|---|
| 登录号Accession No. | 名称Name | ||||||
| OTG27997 | HaLACS1-1 | 1 980 | 659 | 74.4 | 5.82 | AT2G47240 | AtLACS1 |
| OTF93791 | HaLACS1-2 | 1 986 | 661 | 74.8 | 7.23 | AT2G47240 | AtLACS1 |
| OTG12244 | HaLACS2 | 1 980 | 659 | 73.5 | 5.70 | AT1G49430 | AtLACS2 |
| OTG32156 | HaLACS4-1 | 1 986 | 661 | 73.5 | 6.07 | AT4G23850 | AtLACS4 |
| OTG33063 | HaLACS4-2 | 1 995 | 664 | 74.0 | 6.69 | AT4G23850 | AtLACS4 |
| OTG20245 | HaLACS4-3 | 1 980 | 659 | 73.5 | 6.50 | AT4G23850 | AtLACS4 |
| OTG02621 | HaLACS4-4 | 1 974 | 657 | 73.4 | 6.69 | AT4G23850 | AtLACS4 |
| OTG02629 | HaLACS4-5 | 1 983 | 660 | 73.8 | 8.49 | AT4G23850 | AtLACS4 |
| OTG11238 | HaLACS6 | 2 094 | 697 | 76.3 | 7.51 | AT3G05970 | AtLACS6 |
| OTG31091 | HaLACS7 | 2 097 | 698 | 77.0 | 6.58 | AT5G27600 | AtLACS7 |
| OTG29824 | HaLACS8 | 2 163 | 720 | 78.6 | 7.86 | AT2G04350 | AtLACS8 |
| OTG15459 | HaLACS9-1 | 2 091 | 696 | 76.2 | 6.10 | AT1G77590 | AtLACS9 |
| OTG07054 | HaLACS9-2 | 2 121 | 706 | 77.4 | 6.78 | AT1G77590 | AtLACS9 |
表2 向日葵LACS基因基本信息
Table 2 Detailed information of LACS genes in sunflower genome
| 转录本ID Transcript ID | 基因名称 Gene name | CDS/bp | 大小/aa Size | 蛋白分子量/kD Molecular weight | 等电点 pI | 拟南芥同源基因 Arabidopsis homologous gene | |
|---|---|---|---|---|---|---|---|
| 登录号Accession No. | 名称Name | ||||||
| OTG27997 | HaLACS1-1 | 1 980 | 659 | 74.4 | 5.82 | AT2G47240 | AtLACS1 |
| OTF93791 | HaLACS1-2 | 1 986 | 661 | 74.8 | 7.23 | AT2G47240 | AtLACS1 |
| OTG12244 | HaLACS2 | 1 980 | 659 | 73.5 | 5.70 | AT1G49430 | AtLACS2 |
| OTG32156 | HaLACS4-1 | 1 986 | 661 | 73.5 | 6.07 | AT4G23850 | AtLACS4 |
| OTG33063 | HaLACS4-2 | 1 995 | 664 | 74.0 | 6.69 | AT4G23850 | AtLACS4 |
| OTG20245 | HaLACS4-3 | 1 980 | 659 | 73.5 | 6.50 | AT4G23850 | AtLACS4 |
| OTG02621 | HaLACS4-4 | 1 974 | 657 | 73.4 | 6.69 | AT4G23850 | AtLACS4 |
| OTG02629 | HaLACS4-5 | 1 983 | 660 | 73.8 | 8.49 | AT4G23850 | AtLACS4 |
| OTG11238 | HaLACS6 | 2 094 | 697 | 76.3 | 7.51 | AT3G05970 | AtLACS6 |
| OTG31091 | HaLACS7 | 2 097 | 698 | 77.0 | 6.58 | AT5G27600 | AtLACS7 |
| OTG29824 | HaLACS8 | 2 163 | 720 | 78.6 | 7.86 | AT2G04350 | AtLACS8 |
| OTG15459 | HaLACS9-1 | 2 091 | 696 | 76.2 | 6.10 | AT1G77590 | AtLACS9 |
| OTG07054 | HaLACS9-2 | 2 121 | 706 | 77.4 | 6.78 | AT1G77590 | AtLACS9 |
| 重复基因对 Duplicated gene pairs | 非同义替 换率Ka | 同义替换率 Ks | Ka/Ks | 重复类型 Duplicated type | 选择压力 Selective pressure | 分化时间/ Mya Divergence time |
|---|---|---|---|---|---|---|
| HaLACS9-1/HaLACS9-2 | 0.0540 | 0.5316 | 0.1016 | 片段重复Segmental | 纯化Purifying | 17.72 |
| HaLACS4-1/HaLACS4-5 | 0.0739 | 0.6713 | 0.1101 | 片段重复Segmental | 纯化Purifying | 22.38 |
| HaLACS4-1/HaLACS4-4 | 0.0905 | 0.6463 | 0.1400 | 片段重复Segmental | 纯化Purifying | 21.54 |
| HaLACS4-1/HaLACS4-3 | 0.1003 | 1.0273 | 0.0976 | 片段重复Segmental | 纯化Purifying | 34.24 |
| HaLACS4-4/HaLACS4-5 | 0.0600 | 0.3516 | 0.1706 | 串联重复Tandem | 纯化Purifying | 11.72 |
表3 向日葵LACS家族成员基因复制事件和发散时间分析
Table 3 Gene duplication events and divergence time of the LACS genes in Helianthus annuus
| 重复基因对 Duplicated gene pairs | 非同义替 换率Ka | 同义替换率 Ks | Ka/Ks | 重复类型 Duplicated type | 选择压力 Selective pressure | 分化时间/ Mya Divergence time |
|---|---|---|---|---|---|---|
| HaLACS9-1/HaLACS9-2 | 0.0540 | 0.5316 | 0.1016 | 片段重复Segmental | 纯化Purifying | 17.72 |
| HaLACS4-1/HaLACS4-5 | 0.0739 | 0.6713 | 0.1101 | 片段重复Segmental | 纯化Purifying | 22.38 |
| HaLACS4-1/HaLACS4-4 | 0.0905 | 0.6463 | 0.1400 | 片段重复Segmental | 纯化Purifying | 21.54 |
| HaLACS4-1/HaLACS4-3 | 0.1003 | 1.0273 | 0.0976 | 片段重复Segmental | 纯化Purifying | 34.24 |
| HaLACS4-4/HaLACS4-5 | 0.0600 | 0.3516 | 0.1706 | 串联重复Tandem | 纯化Purifying | 11.72 |
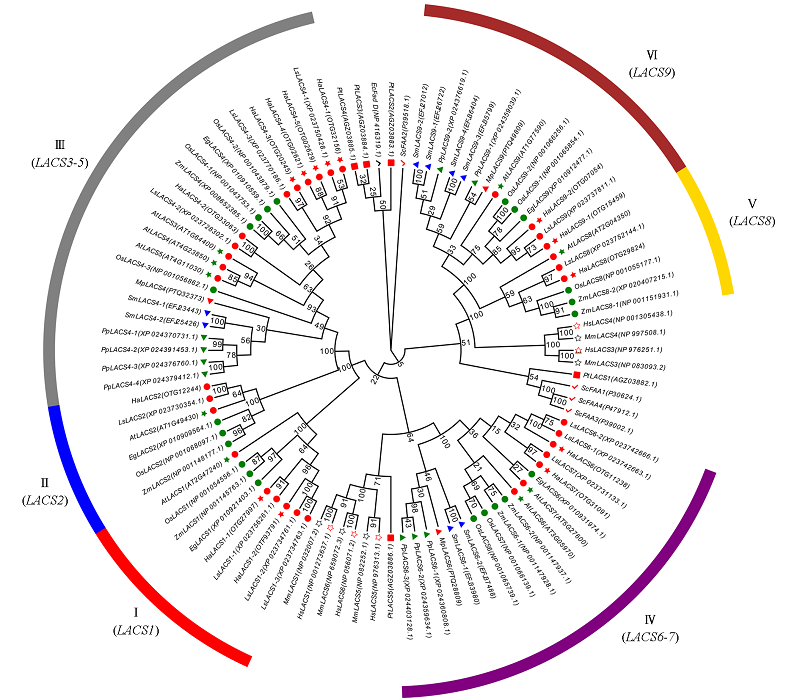
图2 向日葵和其他物种LACS基因家族系统进化分析 绿色圆圈代表单子叶植物,红色圆圈代表双子叶植物。 Ec:大肠杆菌(原核生物);Sc:酿酒酵母(真菌);Pt:三角褐指藻(藻类);Pp:小立碗藓(藓);Sm:江南卷柏(蕨);Mp:地钱(苔);At:拟南芥;Ha:向日葵;Ls:莴苣;Eg:油棕;Zm:玉米;Os:水稻;Hs:人;Mm:家鼠。
Fig. 2 Phylogenetic analysis of LACS gene family in sunflower and other species The green circles indicate monocotyledonous plants,the red circle indicates a dicot plant. Ec:Escherichia coli(prokaryote);Sc:Saccharomyces cerevisiae(fungi);Pt:Phaeodac tylumtricornutum(algae);Pp:Physcomitrella patens(moss);Sm:Selaginella moellendorffii(fern);Mp:Marchantia polymorpha(moss);At:Arabidopsis thaliana;Ha:Helianthus annuus;Ls:Lactuca sativa;Eg:Elaeis guineensis;Zm:Zea mays;Os:Oryza sativa;Hs:Homo sapiens;Mm:Mus musculus.

图4 向日葵HaLACS与其他同源蛋白的氨基酸多序列比对 Ec:大肠杆菌;At:拟南芥;Sc:酿酒酵母;Ha:向日葵。
Fig. 4 Multiple sequence alignment of the HaLACS amino acid sequences with other proteins Ec:Escherichia coli;At:Arabidopsis thaliana;Sc:Selaginella moellendorffii;Ha:Helianthus annuus.
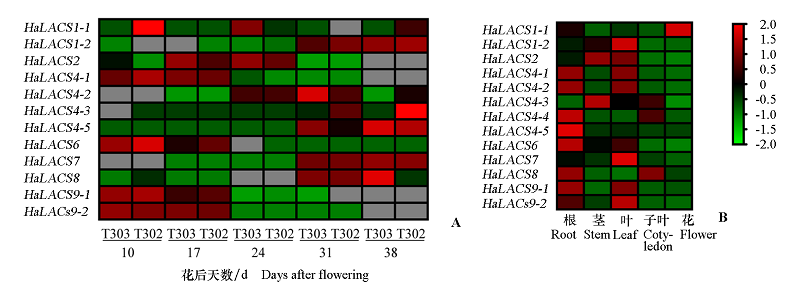
图7 HaLACS在2个向日葵品系不同发育阶段种子(A)和不同组织(B)的表达模式
Fig. 7 Expression levels in different seed development stages(A)of two sunflower cultivars and different tissues(B)
| [1] |
Aznar-Moreno J A, Calerón M V, Martínez-Force E, Garcés R, Mullen R, Gidda S K, Salas J J. 2014. Sunflower(Helianthus annuus L.)long-chain acyl-coenzyme A synthetases expressed at high levels in developing seeds. Physiologia Plantarum, 150 (3):363-373.
doi: 10.1111/ppl.2014.150.issue-3 URL |
| [2] |
Bernard A, Joubès J. 2013. Arabidopsis cuticular waxes:advances in synthesis,export and regulation. Progress in Lipid Research, 52 (1):110-129.
doi: 10.1016/j.plipres.2012.10.002 pmid: 23103356 |
| [3] |
Chen C J, Chen H, Zhang Y, Thomas H R, Frank M H, He Y H, Xia R. 2020. TBtools:an integrative toolkit developed for interactive analyses of big biological data. Molecular Plant, 13 (8):1194-1202.
doi: 10.1016/j.molp.2020.06.009 URL |
| [4] |
Dimopoulos N, Tindjau R, Wong D C J, Matzat T, Haslam T, Song C, Gambetta G A, Kunst L, Castellarin S D. 2020. Drought stress modulates cuticular wax composition of the grape berry. Journal of Experimental Botany, 71 (10):3126-3141.
doi: 10.1093/jxb/eraa046 pmid: 31985780 |
| [5] |
Ding L N, Gu S L, Zhu F G, Ma Z Y, Li J, Li M, Zheng W, Tan X L. 2020. Long-chain acyl-CoA synthetase 2 is involved in seed oil production in Brassica napus. BMC Plant Biology, 20 (1):21-34.
doi: 10.1186/s12870-020-2240-x pmid: 31931712 |
| [6] |
Etchegaray A, Ralf D, Paul C E, Geoffrey T, von Döhren H. 1998. Group specific antibodies against the putative AMP-binding domain signature SGTTGXPKG in peptide synthetases and related enzymes. Biochemistry and Molecular Biology International, 44 (2):235-243.
pmid: 9530507 |
| [7] |
Flagel L E, Wendel J F. 2009. Gene duplication and evolutionary novelty in plants. New Phytologist, 183 (3):557-564.
doi: 10.1111/j.1469-8137.2009.02923.x pmid: 19555435 |
| [8] |
Fulda M, Heinz E, Wolter F P. 1994. The fadD gene of Escherichia coli K 12 is located close to rnd at 39.6 min of the chromosomal-map and is a new member of the AMP-binding protein family. Molecular and General Genetics, 242 (3):241-249.
pmid: 8107670 |
| [9] |
Fulda M, Shockey J, Werber M, Wolter F P, Heinz E. 2002. Two long-chain acyl-CoA synthetases from Arabidopsis thaliana involved in peroxisomal fatty acid b-oxidation. The Plant Journal, 32 (1):93-103.
doi: 10.1046/j.1365-313X.2002.01405.x URL |
| [10] |
Iijima H, Fujino T, Minekura H, Suzuki H, Kang M J, Yamamoto T. 1996. Biochemical studies of two rat Acyl-CoA synthetases,ACS1 and ACS2. European Journal of Biochemistry, 242 (2):186-190.
doi: 10.1111/ejb.1996.242.issue-2 URL pmid: 8973631 |
| [11] |
Jessen D, Olbrich A, Knüfer J, Krüger A, Hoppert M, Polle A, Fulda M. 2011. Combined activity of LACS1 and LACS4 is required for proper pollen coat formation in Arabidopsis. The Plant Journal, 68 (4):715-726.
doi: 10.1111/j.1365-313X.2011.04722.x pmid: 21790813 |
| [12] |
Kosma D K, Bourdenx B, Bernard A, Parsons E P, Lv S Y, Joubès J, Jenks M A. 2009. The impact of water deficiency on leaf cuticle lipids of Arabidopsis. Plant Physiology, 151 (4):1918-1929.
doi: 10.1104/pp.109.141911 URL |
| [13] |
Li Xiao-cui, Kang Kai-cheng, Huang Xian-zhong, Fan Yong-bin, Song Miao-miao, Huang Yun-jie, Ding Jia-jia. 2020. Genome-wide identification,phylogenetic analysis and expression profiling of the MKK gene family in Arabidopsis pumila. Hereditas(Beijing), 42:403-421. (in Chinese)
doi: 10.16288/j.yczz.19-388 pmid: 32312709 |
|
李晓翠, 康凯程, 黄先忠, 范永斌, 宋苗苗, 黄韵杰, 丁佳佳. 2020. 小拟南芥MKK基因家族全基因组鉴定及进化和表达分析. 遗传, 42:403-421.
pmid: 32312709 |
|
| [14] |
Lü S Y, Song T, Kosma D K, Parsons E P, Rowland O, Jenks M A. 2009. Arabidopsis CER8 encodes LONG-CHAIN ACYL-COA SYNTHETASE 1 (LACS1) that has overlapping functions with LACS2 in plant wax and cutin synthesis. The Plant Journal, 59 (4):553-564.
doi: 10.1111/tpj.2009.59.issue-4 URL |
| [15] |
Mashek D G, Li L O, Coleman R A. 2007. Long-chain Acyl-CoA synthetases and fatty acid channeling. Future Lipidology, 2 (4):465-476.
doi: 10.2217/17460875.2.4.465 URL |
| [16] |
Schnurr J, Shockey J, Browse J. 2004. The Acyl-CoA synthetase encoded by LACS2 is essential for normal cuticle development in Arabidopsis. The Plant Cell, 16 (3):629-642.
doi: 10.1105/tpc.017608 URL |
| [17] |
Shockey J M, Fulda M S, Browse J A. 2002. Arabidopsis contains nine long-chain acyl-coenzyme a synthetase genes that participate in fatty acid and glycerolipid metabolism. Plant Physiology, 129 (4):1710-1722.
pmid: 12177484 |
| [18] |
Suh M C, Samuels A L, Jetter R, Kunst L, Pollard M, Ohlrogge J, Beisson F. 2005. Cuticular lipid composition,surface structure,and gene expression in Arabidopsis stem epidermis. Plant Physiology, 139 (4):1649-1665.
doi: 10.1104/pp.105.070805 URL |
| [19] |
Vatansever R, Koc I, Ozyigit I I, Sen U, Uras M E, Anjum N A, Pereira E, Filiz E. 2016. Genome-wide identification and expression analysis of sulfate transporter(SULTR)genes in potato(Solanum tuberosum L.). Planta, 244 (6):1167-1183.
pmid: 27473680 |
| [20] |
Wang X L, Li X B. 2009. The GhACS1 gene encodes an acyl-CoA synthetase which is essential for normal microsporogenesis in early anther development of cotton. The Plant Journal, 57 (3):473-486.
doi: 10.1111/tpj.2009.57.issue-3 URL |
| [21] |
Xiao Y, Li X T, Yao L H, Xu D X, Li Yang, Zhang X F, Li Z, Xiao Q L, Ni Y, Guo Y J. 2020. Chemical profiles of cuticular waxes on various organs of Sorghum bicolor and their antifungal activities. Plant Physiology and Biochemistry, 155:596-604.
doi: S0981-9428(20)30408-3 pmid: 32846395 |
| [22] |
Xiao Z C, Li N N, Wang S F, Sun J J, Zhang L Y, Zhang C, Yang H, Zhao H Y, Yang B, Wei L J, Du H, Qu C M, Lu K, Li J N. 2019. Genome-wide identification and comparative expression profile analysis of the long-chain Acyl-CoA synthetase(LACS)gene family in two different oil content cultivars of Brassica napus. Biochemical Genetics, 57 (6):781-800.
doi: 10.1007/s10528-019-09921-5 URL |
| [23] | Yi Ting, Zhang Zhishuo, Tang Bingqian, Xie Lingling, Zou Xuexiao. 2020. Identification and expression analysis of the KCS gene family in pepper. Acta Horticulturae Sinica, 47 (2):370-380. (in Chinese) |
| 易婷, 张志硕, 汤冰倩, 谢玲玲, 邹学校. 2020. 辣椒β-酮脂酰辅酶A合酶基因家族的鉴定与表达分析. 园艺学报, 47 (2):370-380. | |
| [24] |
Zhang C L, Mao K, Zhou L J, Wang G L, Zhang Y L, Li Y Y, Hao Y J. 2018. Genome-wide identification and characterization of apple long-chain Acyl-CoA synthetases and expression analysis under different stresses. Plant Physiology and Biochemistry, 132:320-332.
doi: 10.1016/j.plaphy.2018.09.004 URL |
| [25] |
Zhao L F, Haslam T M, Sonntag A, Molina I, Kunst L. 2019. Functional overlap of long-chain Acyl-CoA synthetases in Arabidopsis. Plant and Cell Physiology, 60 (5):1041-1054.
doi: 10.1093/pcp/pcz019 URL |
| [26] |
Zhao L F, Katavic V, Li F L, Haughn G W, Kunst L. 2010. Insertional mutant analysis reveals that long-chain acyl-CoA synthetase 1(LACS1),but not LACS8,functionally overlaps with LACS9 in Arabidopsis seed oil biosynthesis. The Plant Journal, 64 (6):1048-1058.
doi: 10.1111/tpj.2010.64.issue-6 URL |
| [27] | Zhu X D, Wang M Q, Li X P, Jiu S T, Wang C, Fang J G. 2017. Genome-wide analysis of the sucrose synthase gene family in grape(Vitis vinifera):structure,evolution,and expression profiles. Genes(Basel), 8 (4):111-135. |
| [1] | 王晓晨, 聂子页, 刘先菊, 段 伟, 范培格, 梁振昌, . 脱落酸对‘京香玉’葡萄果实单萜物质合成的影响[J]. 园艺学报, 2023, 50(2): 237-249. |
| [2] | 翟含含, 翟宇杰, 田义, 张叶, 杨丽, 温陟良, 陈海江. 桃SAUR家族基因分析及PpSAUR5功能鉴定[J]. 园艺学报, 2023, 50(1): 1-14. |
| [3] | 徐小萍, 曹清影, 蔡柔荻, 官庆栩, 张梓浩, 陈裕坤, 徐涵, 林玉玲, 赖钟雄. 龙眼miR408与DlLAC12克隆及其在球形胚发生和非生物胁迫下的表达分析[J]. 园艺学报, 2022, 49(9): 1866-1882. |
| [4] | 贾鑫, 曾臻, 陈月, 冯慧, 吕英民, 赵世伟. 月季‘月月粉’RcDREB2A的克隆与表达分析[J]. 园艺学报, 2022, 49(9): 1945-1956. |
| [5] | 张秋悦, 刘昌来, 于晓晶, 杨甲定, 封超年. 盐胁迫条件下杜梨叶片差异表达基因qRT-PCR内参基因筛选[J]. 园艺学报, 2022, 49(7): 1557-1570. |
| [6] | 马维峰, 李艳梅, 马宗桓, 陈佰鸿, 毛娟. 苹果POD家族基因的鉴定与MdPOD15的功能分析[J]. 园艺学报, 2022, 49(6): 1181-1199. |
| [7] | 李亚梅, 马福利, 张山奇, 黄锦秋, 陈梦婷, 周军永, 孙其宝, 孙俊. 酸枣愈伤组织转化体系构建及在ZjBRC1调控ZjYUCCA表达中的应用[J]. 园艺学报, 2022, 49(4): 749-757. |
| [8] | 张瑞, 张夏燚, 赵婷, 王双成, 张仲兴, 刘博, 张德, 王延秀. 基于转录组分析垂丝海棠响应盐碱胁迫的分子机制[J]. 园艺学报, 2022, 49(2): 237-251. |
| [9] | 乔军, 王利英, 刘婧, 李素文. 基于转录组测序的茄子萼下果色光敏相关基因表达分析[J]. 园艺学报, 2022, 49(11): 2347-2356. |
| [10] | 叶广继, 郑贞贞, 纳添仓, 王舰. 马铃薯资源糖苷生物碱含量评价及合成相关基因表达分析[J]. 园艺学报, 2022, 49(11): 2357-2366. |
| [11] | 侯天泽, 易双双, 张志群, 王健, 李崇晖. 秋石斛RT-qPCR内参基因的筛选与验证[J]. 园艺学报, 2022, 49(11): 2489-2501. |
| [12] | 周铁, 潘斌, 李菲菲, 马小川, 汤孟婧, 廉雪菲, 常媛媛, 陈岳文, 卢晓鹏. 膨大期干旱对温州蜜柑品质形成的影响及复水后树体水分吸收转运规律[J]. 园艺学报, 2022, 49(1): 11-22. |
| [13] | 谢思艺, 周承哲, 朱晨, 詹冬梅, 陈兰, 吴祖春, 赖钟雄, 郭玉琼. 茶树CsTIFY家族全基因组鉴定及非生物胁迫和激素处理中主要基因表达分析[J]. 园艺学报, 2022, 49(1): 100-116. |
| [14] | 贺琰, 孙艳丽, 赵芳芳, 代红军. 外源油菜素内酯处理对‘美乐’葡萄果实糖代谢的影响[J]. 园艺学报, 2022, 49(1): 117-128. |
| [15] | 李茂福, 杨媛, 王华, 范又维, 孙佩, 金万梅. 月季自交不亲和性S-RNase的鉴定与分析[J]. 园艺学报, 2022, 49(1): 157-165. |
| 阅读次数 | ||||||
|
全文 |
|
|||||
|
摘要 |
|
|||||
版权所有 © 2012 《园艺学报》编辑部 京ICP备10030308号-2 国际联网备案号 11010802023439
编辑部地址: 北京市海淀区中关村南大街12号中国农业科学院蔬菜花卉研究所 邮编: 100081
电话: 010-82109523 E-Mail: yuanyixuebao@126.com
技术支持:北京玛格泰克科技发展有限公司