
园艺学报 ›› 2022, Vol. 49 ›› Issue (6): 1181-1199.doi: 10.16420/j.issn.0513-353x.2021-0275
• 研究论文 • 下一篇
收稿日期:2022-01-13
修回日期:2022-03-07
出版日期:2022-06-25
发布日期:2022-07-04
通讯作者:
毛娟
E-mail:maojuan@gsau.edu.cn
基金资助:
MA Weifeng, LI Yanmei, MA Zonghuan, CHEN Baihong, MAO Juan( )
)
Received:2022-01-13
Revised:2022-03-07
Online:2022-06-25
Published:2022-07-04
Contact:
MAO Juan
E-mail:maojuan@gsau.edu.cn
摘要:
为探究苹果过氧化物酶(POD)基因家族成员在苹果逆境下的作用,利用已报道的拟南芥POD基因,通过多序列比对鉴定出51个苹果POD(MdPOD)家族成员,并分析其在逆境条件下的表达规律。结果表明51个家族成员分布于13条染色体上,其编码95 ~ 1 443个氨基酸,蛋白分子量为10.18 ~ 161.02 kD,等电点为4.36 ~ 9.90。分析表明该基因家族可分为6个亚族,外显子数介于1 ~ 40。选择压分析表明20个基因在进化过程中受到纯化选择作用。亚细胞定位发现MdPOD15主要存在于细胞膜、细胞质、细胞核和叶绿体中。基因芯片表达图谱显示大部分MdPOD在花、叶和果实中表达量较高。实时荧光定量PCR 表明,ABA处理2 h大部分基因的表达量上调,之后出现下降,PEG处理12 h后51个MdPOD基因表达量达到峰值,而用NaCl处理时MdPOD的表达量到24 h时达到峰值。烟草叶片瞬时表达MdPOD15发现,其在细胞膜、细胞质和细胞核中表达,且苹果愈伤组织中MdPOD15的超表达可缓和非生物胁迫对细胞的损伤程度,由此可知MdPOD在不同非生物胁迫下的不同时间段可能发挥不同的作用。
中图分类号:
马维峰, 李艳梅, 马宗桓, 陈佰鸿, 毛娟. 苹果POD家族基因的鉴定与MdPOD15的功能分析[J]. 园艺学报, 2022, 49(6): 1181-1199.
MA Weifeng, LI Yanmei, MA Zonghuan, CHEN Baihong, MAO Juan. Identification of Apple POD Gene Family and Functional Analysis of MdPOD15 Gene[J]. Acta Horticulturae Sinica, 2022, 49(6): 1181-1199.
| 基因 Gene | 基因ID Gene ID | 上游引物(5′-3′) Forward primer | 下游引物(5′-3′) Reverse primer |
|---|---|---|---|
| MdPOD01 | MDP0000207215 | CCTGCTGCTCTGCTTATCGTCTTC | TCCTTCTGTTGCCAACCAAGTTCC |
| MdPOD02 | MDP0000449389 | ACCAAATGCTCTAAGCCGAACAGG | CCGATCCAAACACGCCCTTACC |
| MdPOD03 | MDP0000451182 | CGCATGTCCACACACTGTTTCTTG | CTTCCGTCTTTCCTCCCCTTTAGC |
| MdPOD04 | MDP0000226365 | GGGAAGGAGAGACAGTAGGACAGC | TGTATGGGCACCTGATAACGCAAC |
| MdPOD05 | MDP0000272643 | AGGGAGGCAACAAAGCTGAGAAAG | ATGAAACGACACCAGGGCAAGAAG |
| MdPOD06 | MDP0000234905 | GACAAGGCGTTGGTGGAGGATC | CGGAGCCAAGAAGGTTGAGTGAAG |
| MdPOD07 | MDP0000151864 | CGTGATGGACTTTGTGAGCCCTAC | CACCAACTGCTTCGACACCTGAC |
| MdPOD08 | MDP0000488361 | CAACCCACCACCACCTTCATCATC | GTCCACAGTTCACGCCCACAC |
| MdPOD09 | MDP0000021998 | GCCCCGCCATTGATATTTCTCCAG | CCTTGTCAGACAGCCCCATCATTC |
| MdPOD10 | MDP0000611163 | CGGAGCGGAGATACGTTGAATGAC | AGCACCTGATCGGTCTGGAGAAG |
| MdPOD11 | MDP0000229817 | TCAGCAGCTTTTCAGTGGTGGATC | GTCGTTCCAGTAAGCGGGCTAAG |
| MdPOD12 | MDP0000241173 | GCCTCATCGCCGAGAAGAACTG | TGCCATTCCCACACAAACAGAGAG |
| MdPOD13 | MDP0000261341 | GGCCTTCTTCCTGATGCTACCAAG | GATCGCTCCTTGTGGCACCTTC |
| MdPOD14 | MDP0000678562 | TGAAGAGGCTAGGAACACCGTAGG | TTTGGGACGCTGCATGGAACG |
| MdPOD15 | MDP0000209189 | CCAAAGGGCCTATTGCACTCTGAC | TCGGTTCTGAAGGTTGCTGGATTG |
| MdPOD16 | MDP0000727958 | CGTACTAAGCGGTGGACCTGATTG | AGAGATCGGGTTTGTGCTGGAATG |
| MdPOD17 | MDP0000283650 | ACACATTCCGCTCACGCATCTAC | TGTCGTCATTTCCGCTTCTTGGG |
| MdPOD18 | MDP0000194474 | AGTAATCAGCACGGCGGCAATG | TTGAGAGTGGCAGGAGGAAGTGG |
| MdPOD19 | MDP0000316890 | GCTTGCGGGTGTTATTGCAGTTG | ACCTCGTTTAGCATCTGGAAGTCG |
| MdPOD20 | MDP0000292921 | AGAGACATTCCAGCACCACTACCC | TGACGAACGAACACTTAGCGAACC |
| MdPOD21 | MDP0000151342 | GTCATACACTGGGAAGGGCACATC | CTCGGACTCCTCGTTCAACAGTTC |
| MdPOD22 | MDP0000214851 | GCTGATTCCGACGAAGGAGATACC | ACTCCCACTTCTCTCCCTACCTTG |
| MdPOD23 | MDP0000143123 | GCATGATTGGCCTTCCCTCAGAC | GGTCCTCGGCATACTTTGTGATCC |
| MdPOD24 | MDP0000903820 | GCCGCTGATGTTGGTGGACTC | CAGGACACTGGGTTGACTGCATC |
| MdPOD25 | MDP0000248823 | CTCTCCCAATCCTCCTCCTCACTC | GGGTCGGAAGCCTGGCATTTG |
| MdPOD26 | MDP0000171732 | GCGAACGAGAGGAGGGCTTTTG | TGGTTGCGGCAGTGAGAATGTC |
| MdPOD27 | MDP0000509183 | TCAGCCACTGGAATCACGAAGTTG | CAGCATCGTAGTCGCCACCATG |
| MdPOD28 | MDP0000199034 | TGCCACTCCATCGCTGCTAGG | GCCGTCTTGTACTCTTCGCTCAC |
| MdPOD29 | MDP0000136412 | ACAGGACGCAGGGATGGGATG | GCTCCAAGAAGGGCAACAAGATCC |
| MdPOD30 | MDP0000598572 | CTCCACCCACTTTGAATGCCTCTG | ACGAGCACCTGGCTACACCTAG |
| MdPOD31 | MDP0000126107 | CGGATCATCTGAGGGCTGTCTTTG | GAACGCTCCTTGTGGCACCTAC |
| MdPOD32 | MDP0000301828 | TGATTCGAGGATTGGTGCAAGCC | CGAGCGGAGTTGGTGTTTGGAG |
| MdPOD33 | MDP0000210077 | GCTGGCTGGAGTTGTTGCTGTAG | CGACCTTCTGGTGGTGGTTCTTG |
| MdPOD34 | MDP0000399965 | GTGTTGGTGCTCTAGGTTCGGATC | AATCCAGAACGCTCCTTGTGACAC |
| MdPOD35 | MDP0000319048 | CGTTGCTGCTCGGGACTCTATTG | GCTTGCTGTGGTGGAGTCTCTTC |
| MdPOD36 | MDP0000179781 | TCGGCAGAAGAGACTCCACCAC | GCAACTAGGTCCTTGGCAGTGAAG |
| MdPOD37 | MDP0000672440 | TTCCACCACAGCAAGCCAAACC | CCTAAGTCGCCAAATCGCTCTCTC |
| MdPOD38 | MDP0000264519 | TGGCAGTGTGGCAGTAAGTGTAAG | GCCTAAGTCGCCAAATCGCTCTC |
| MdPOD39 | MDP0000305310 | GATTCACATCAAAGACGGCAAGGC | ACAACTGGCACGCACGGATTAC |
| MdPOD40 | MDP0000208152 | TGCGATGGATCAATTCTGCTGGAC | GACGGCGGGACAAGCATTCTC |
| MdPOD41 | MDP0000124622 | CGGCATTTGTGTTGAAGGGCAAG | AGCAGCAGGAGAAGTGGGATCAG |
| MdPOD42 | MDP0000142485 | TTTGAGGCTCTTCTTCCACGACTG | AATCCATCTCCAGCAAGCGAAAGG |
| MdPOD43 | MDP0000943804 | CAATCCTCTTCCCGCCGTCAAAG | TGGCAGTGTGGCAGTAAGTGTAAG |
| MdPOD44 | MDP0000169497 | GCGGGAGGAGTGGTGGTAGAC | TCGGCGTTGTATGTTCCAGCATC |
| MdPOD45 | MDP0000918790 | GGTGTCTCATACGCCGACTTGTTC | GACGTAGATGATCAGCAGCGAAGG |
| MdPOD46 | MDP0000612477 | CTGATCCTCGTAATGCCGCCTTG | CCTGTCAGCGTGATTGTGTCCTC |
| MdPOD47 | MDP0000849735 | TGTGCTCCATGATCGTCGTTGC | TCGGTCCTTCTCTACAGCCTTGG |
| MdPOD48 | MDP0000292744 | AGCATTTGGGTTGAAGGGCAAGG | GCAGCAGGAGAAGTAGGATCAGTG |
| MdPOD49 | MDP0000567775 | GGAAGAGGCAAGCCACTCGATTAC | CATCCTGGTGTCGTTCTTGAGAGC |
| MdPOD50 | MDP0000220665 | ACGGCAACGGAGATACATTGAACG | AGCACCTGATCGGTCTGGAGAAG |
| MdPOD51 | MDP0000208213 | AGGCAGAGGCTCTACAACCAGTC | GTAGGGCTCACAAAGTCCATCACG |
| MdGAPDH | XM_008363855.1 | GAGCTCGCAGGTATCCTTTCT | TACCAAGCAATGACCTTGACC |
表1 苹果POD表达分析的实时荧光定量引物
Table 1 qRT-PCR primers for expression analysis of POD in apple
| 基因 Gene | 基因ID Gene ID | 上游引物(5′-3′) Forward primer | 下游引物(5′-3′) Reverse primer |
|---|---|---|---|
| MdPOD01 | MDP0000207215 | CCTGCTGCTCTGCTTATCGTCTTC | TCCTTCTGTTGCCAACCAAGTTCC |
| MdPOD02 | MDP0000449389 | ACCAAATGCTCTAAGCCGAACAGG | CCGATCCAAACACGCCCTTACC |
| MdPOD03 | MDP0000451182 | CGCATGTCCACACACTGTTTCTTG | CTTCCGTCTTTCCTCCCCTTTAGC |
| MdPOD04 | MDP0000226365 | GGGAAGGAGAGACAGTAGGACAGC | TGTATGGGCACCTGATAACGCAAC |
| MdPOD05 | MDP0000272643 | AGGGAGGCAACAAAGCTGAGAAAG | ATGAAACGACACCAGGGCAAGAAG |
| MdPOD06 | MDP0000234905 | GACAAGGCGTTGGTGGAGGATC | CGGAGCCAAGAAGGTTGAGTGAAG |
| MdPOD07 | MDP0000151864 | CGTGATGGACTTTGTGAGCCCTAC | CACCAACTGCTTCGACACCTGAC |
| MdPOD08 | MDP0000488361 | CAACCCACCACCACCTTCATCATC | GTCCACAGTTCACGCCCACAC |
| MdPOD09 | MDP0000021998 | GCCCCGCCATTGATATTTCTCCAG | CCTTGTCAGACAGCCCCATCATTC |
| MdPOD10 | MDP0000611163 | CGGAGCGGAGATACGTTGAATGAC | AGCACCTGATCGGTCTGGAGAAG |
| MdPOD11 | MDP0000229817 | TCAGCAGCTTTTCAGTGGTGGATC | GTCGTTCCAGTAAGCGGGCTAAG |
| MdPOD12 | MDP0000241173 | GCCTCATCGCCGAGAAGAACTG | TGCCATTCCCACACAAACAGAGAG |
| MdPOD13 | MDP0000261341 | GGCCTTCTTCCTGATGCTACCAAG | GATCGCTCCTTGTGGCACCTTC |
| MdPOD14 | MDP0000678562 | TGAAGAGGCTAGGAACACCGTAGG | TTTGGGACGCTGCATGGAACG |
| MdPOD15 | MDP0000209189 | CCAAAGGGCCTATTGCACTCTGAC | TCGGTTCTGAAGGTTGCTGGATTG |
| MdPOD16 | MDP0000727958 | CGTACTAAGCGGTGGACCTGATTG | AGAGATCGGGTTTGTGCTGGAATG |
| MdPOD17 | MDP0000283650 | ACACATTCCGCTCACGCATCTAC | TGTCGTCATTTCCGCTTCTTGGG |
| MdPOD18 | MDP0000194474 | AGTAATCAGCACGGCGGCAATG | TTGAGAGTGGCAGGAGGAAGTGG |
| MdPOD19 | MDP0000316890 | GCTTGCGGGTGTTATTGCAGTTG | ACCTCGTTTAGCATCTGGAAGTCG |
| MdPOD20 | MDP0000292921 | AGAGACATTCCAGCACCACTACCC | TGACGAACGAACACTTAGCGAACC |
| MdPOD21 | MDP0000151342 | GTCATACACTGGGAAGGGCACATC | CTCGGACTCCTCGTTCAACAGTTC |
| MdPOD22 | MDP0000214851 | GCTGATTCCGACGAAGGAGATACC | ACTCCCACTTCTCTCCCTACCTTG |
| MdPOD23 | MDP0000143123 | GCATGATTGGCCTTCCCTCAGAC | GGTCCTCGGCATACTTTGTGATCC |
| MdPOD24 | MDP0000903820 | GCCGCTGATGTTGGTGGACTC | CAGGACACTGGGTTGACTGCATC |
| MdPOD25 | MDP0000248823 | CTCTCCCAATCCTCCTCCTCACTC | GGGTCGGAAGCCTGGCATTTG |
| MdPOD26 | MDP0000171732 | GCGAACGAGAGGAGGGCTTTTG | TGGTTGCGGCAGTGAGAATGTC |
| MdPOD27 | MDP0000509183 | TCAGCCACTGGAATCACGAAGTTG | CAGCATCGTAGTCGCCACCATG |
| MdPOD28 | MDP0000199034 | TGCCACTCCATCGCTGCTAGG | GCCGTCTTGTACTCTTCGCTCAC |
| MdPOD29 | MDP0000136412 | ACAGGACGCAGGGATGGGATG | GCTCCAAGAAGGGCAACAAGATCC |
| MdPOD30 | MDP0000598572 | CTCCACCCACTTTGAATGCCTCTG | ACGAGCACCTGGCTACACCTAG |
| MdPOD31 | MDP0000126107 | CGGATCATCTGAGGGCTGTCTTTG | GAACGCTCCTTGTGGCACCTAC |
| MdPOD32 | MDP0000301828 | TGATTCGAGGATTGGTGCAAGCC | CGAGCGGAGTTGGTGTTTGGAG |
| MdPOD33 | MDP0000210077 | GCTGGCTGGAGTTGTTGCTGTAG | CGACCTTCTGGTGGTGGTTCTTG |
| MdPOD34 | MDP0000399965 | GTGTTGGTGCTCTAGGTTCGGATC | AATCCAGAACGCTCCTTGTGACAC |
| MdPOD35 | MDP0000319048 | CGTTGCTGCTCGGGACTCTATTG | GCTTGCTGTGGTGGAGTCTCTTC |
| MdPOD36 | MDP0000179781 | TCGGCAGAAGAGACTCCACCAC | GCAACTAGGTCCTTGGCAGTGAAG |
| MdPOD37 | MDP0000672440 | TTCCACCACAGCAAGCCAAACC | CCTAAGTCGCCAAATCGCTCTCTC |
| MdPOD38 | MDP0000264519 | TGGCAGTGTGGCAGTAAGTGTAAG | GCCTAAGTCGCCAAATCGCTCTC |
| MdPOD39 | MDP0000305310 | GATTCACATCAAAGACGGCAAGGC | ACAACTGGCACGCACGGATTAC |
| MdPOD40 | MDP0000208152 | TGCGATGGATCAATTCTGCTGGAC | GACGGCGGGACAAGCATTCTC |
| MdPOD41 | MDP0000124622 | CGGCATTTGTGTTGAAGGGCAAG | AGCAGCAGGAGAAGTGGGATCAG |
| MdPOD42 | MDP0000142485 | TTTGAGGCTCTTCTTCCACGACTG | AATCCATCTCCAGCAAGCGAAAGG |
| MdPOD43 | MDP0000943804 | CAATCCTCTTCCCGCCGTCAAAG | TGGCAGTGTGGCAGTAAGTGTAAG |
| MdPOD44 | MDP0000169497 | GCGGGAGGAGTGGTGGTAGAC | TCGGCGTTGTATGTTCCAGCATC |
| MdPOD45 | MDP0000918790 | GGTGTCTCATACGCCGACTTGTTC | GACGTAGATGATCAGCAGCGAAGG |
| MdPOD46 | MDP0000612477 | CTGATCCTCGTAATGCCGCCTTG | CCTGTCAGCGTGATTGTGTCCTC |
| MdPOD47 | MDP0000849735 | TGTGCTCCATGATCGTCGTTGC | TCGGTCCTTCTCTACAGCCTTGG |
| MdPOD48 | MDP0000292744 | AGCATTTGGGTTGAAGGGCAAGG | GCAGCAGGAGAAGTAGGATCAGTG |
| MdPOD49 | MDP0000567775 | GGAAGAGGCAAGCCACTCGATTAC | CATCCTGGTGTCGTTCTTGAGAGC |
| MdPOD50 | MDP0000220665 | ACGGCAACGGAGATACATTGAACG | AGCACCTGATCGGTCTGGAGAAG |
| MdPOD51 | MDP0000208213 | AGGCAGAGGCTCTACAACCAGTC | GTAGGGCTCACAAAGTCCATCACG |
| MdGAPDH | XM_008363855.1 | GAGCTCGCAGGTATCCTTTCT | TACCAAGCAATGACCTTGACC |
| 基因 Gene | 登录号 Accession No. | 染色体 定位 Chr. | CDS/bp | 全长/bp Full length | 氨基酸数 Amino acid number | 分子量/kD Molecular weight | 等电点 Isoelectric point | 不稳定 系数 Instability index | 亚细胞定位 Subcellular location |
|---|---|---|---|---|---|---|---|---|---|
| MdPOD01 | MDP0000207215 | 1 | 1 662 | 6 251 | 553 | 61.10 | 9.30 | 31.26 | Nucl、Plas |
| MdPOD02 | MDP0000449389 | 1 | 1 254 | 6 838 | 417 | 45.98 | 9.23 | 45.17 | Nucl、Chlo |
| MdPOD03 | MDP0000451182 | 1 | 987 | 1 849 | 328 | 36.13 | 8.36 | 41.02 | Nucl、Chlo |
| MdPOD04 | MDP0000226365 | 2 | 567 | 1 833 | 188 | 21.18 | 9.01 | 30.38 | Nucl、Cyto、Chlo |
| MdPOD05 | MDP0000272643 | 2 | 957 | 1 309 | 318 | 34.31 | 9.26 | 43.27 | Plas |
| MdPOD06 | MDP0000234905 | 2 | 1 386 | 5 052 | 461 | 51.35 | 6.46 | 43.26 | Nucl、Cyto、Chlo、Plas |
| MdPOD07 | MDP0000151864 | 2 | 2 355 | 6 866 | 784 | 86.10 | 8.75 | 44.08 | Nucl、Chlo、Plas |
| MdPOD08 | MDP0000488361 | 2 | 1 236 | 3 723 | 411 | 44.62 | 7.50 | 39.84 | Nucl、Chlo |
| MdPOD09 | MDP0000021998 | 2 | 657 | 2 995 | 219 | 24.53 | 6.63 | 33.13 | Nucl、Cyto、Plas |
| MdPOD10 | MDP0000611163 | 3 | 975 | 1 731 | 324 | 34.92 | 4.58 | 35.61 | Nucl |
| MdPOD11 | MDP0000229817 | 3 | 378 | 378 | 125 | 13.58 | 8.91 | 36.09 | Nucl、Chlo |
| MdPOD12 | MDP0000241173 | 3 | 1 011 | 5 864 | 336 | 37.44 | 6.46 | 40.32 | Nucl、Cyto、Chlo、Plas |
| MdPOD13 | MDP0000261341 | 3 | 816 | 4 458 | 271 | 29.99 | 7.61 | 36.85 | Cyto、Chlo、Plas |
| MdPOD14 | MDP0000678562 | 3 | 1 551 | 8 349 | 516 | 57.07 | 7.83 | 46.50 | Nucl、Cyto、Chlo、Plas |
| MdPOD15 | MDP0000209189 | 4 | 996 | 2 332 | 331 | 34.61 | 7.51 | 32.31 | Cyto、Plas |
| MdPOD16 | MDP0000727958 | 4 | 900 | 1 531 | 299 | 32.47 | 8.10 | 44.42 | Nucl、Cyto、Chlo、Plas |
| MdPOD17 | MDP0000283650 | 5 | 2 775 | 15 847 | 924 | 100.30 | 6.47 | 38.37 | Nucl、Cyto、Chlo |
| MdPOD18 | MDP0000194474 | 5 | 4 284 | 14 538 | 1 427 | 159.79 | 8.55 | 48.54 | Plas |
| MdPOD19 | MDP0000316890 | 7 | 711 | 4 729 | 236 | 27.53 | 8.66 | 23.08 | Plas |
| MdPOD20 | MDP0000292921 | 8 | 864 | 3 498 | 287 | 31.19 | 9.03 | 44.06 | Nucl、Chlo |
| MdPOD21 | MDP0000151342 | 8 | 1 086 | 4 365 | 361 | 39.72 | 8.70 | 42.18 | Cyto、Chlo |
| MdPOD22 | MDP0000214851 | 8 | 477 | 1 800 | 160 | 18.45 | 9.90 | 69.36 | Nucl、Cyto、Chlo、Plas |
| MdPOD23 | MDP0000143123 | 8 | 396 | 2 232 | 131 | 14.31 | 6.55 | 22.87 | Nucl、Cyto、Chlo、Plas |
| MdPOD24 | MDP0000903820 | 8 | 288 | 1 247 | 95 | 10.18 | 4.78 | 62.56 | Cyto、Chlo |
| MdPOD25 | MDP0000248823 | 8 | 1 572 | 7 408 | 523 | 57.88 | 9.60 | 54.49 | Nucl、Cyto、Chlo |
| MdPOD26 | MDP0000171732 | 10 | 1 044 | 1 281 | 347 | 38.61 | 9.21 | 28.25 | Chlo、Plas |
| MdPOD27 | MDP0000509183 | 11 | 2 931 | 6 860 | 976 | 107.17 | 5.55 | 41.43 | Nucl、Cyto、Chlo |
| MdPOD28 | MDP0000199034 | 11 | 1 335 | 3 358 | 444 | 49.05 | 6.46 | 39.05 | Nucl、Cyto、Chlo |
| MdPOD29 | MDP0000136412 | 11 | 999 | 1 557 | 332 | 35.70 | 6.43 | 32.14 | Chlo |
| MdPOD30 | MDP0000598572 | 11 | 1 122 | 6 014 | 373 | 40.81 | 6.79 | 43.72 | Nucl、Cyto |
| MdPOD31 | MDP0000126107 | 12 | 618 | 1 604 | 205 | 22.85 | 6.07 | 48.97 | Nucl、Cyto |
| MdPOD32 | MDP0000301828 | 12 | 1 020 | 2 911 | 339 | 35.50 | 4.97 | 41.53 | Nucl、Chlo |
| MdPOD33 | MDP0000210077 | 12 | 1 026 | 2 915 | 341 | 37.69 | 5.70 | 38.27 | Nucl、Cyto |
| MdPOD34 | MDP0000399965 | 12 | 366 | 840 | 121 | 13.30 | 4.74 | 30.47 | Nucl、Cyto、Chlo |
| 基因 Gene | 登录号 Accession No. | 染色体 定位 Chr. | CDS/bp | 全长/bp Full length | 氨基酸数 Amino acid number | 分子量/kD Molecular weight | 等电点 Isoelectric point | 不稳定 系数 Instability index | 亚细胞定位 Subcellular location |
| MdPOD35 | MDP0000319048 | 12 | 1 386 | 9 973 | 461 | 50.42 | 9.17 | 45.08 | Chlo |
| MdPOD36 | MDP0000179781 | 12 | 726 | 1 839 | 241 | 25.58 | 8.55 | 33.29 | Nucl、Cyto、Chlo |
| MdPOD37 | MDP0000672440 | 13 | 774 | 963 | 257 | 27.37 | 5.32 | 31.33 | Nucl、Cyto、Chlo |
| MdPOD38 | MDP0000264519 | 13 | 987 | 1 159 | 328 | 35.21 | 6.89 | 34.90 | Nucl、Cyto、Chlo |
| MdPOD39 | MDP0000305310 | 13 | 4 332 | 16 763 | 1 443 | 161.02 | 5.45 | 45.26 | Plas |
| MdPOD40 | MDP0000208152 | 13 | 1 110 | 2 364 | 369 | 39.39 | 4.36 | 40.36 | Chlo |
| MdPOD41 | MDP0000124622 | 13 | 975 | 1 457 | 324 | 35.03 | 5.64 | 39.51 | Nucl、Cyto |
| MdPOD42 | MDP0000142485 | 13 | 981 | 1 522 | 326 | 35.78 | 8.94 | 36.02 | Nucl、Chlo |
| MdPOD43 | MDP0000943804 | 15 | 885 | 2 506 | 294 | 31.35 | 5.63 | 37.80 | Chlo |
| MdPOD44 | MDP0000169497 | 15 | 867 | 2 104 | 288 | 31.47 | 6.41 | 31.21 | Cyto、Chlo、Plas |
| MdPOD45 | MDP0000918790 | 15 | 876 | 2 073 | 291 | 32.23 | 5.61 | 41.77 | Nucl、Cyto、Chlo |
| MdPOD46 | MDP0000612477 | 16 | 1 026 | 1 622 | 341 | 37.15 | 4.70 | 32.21 | Cyto |
| MdPOD47 | MDP0000849735 | 16 | 1 011 | 1 150 | 336 | 36.71 | 8.32 | 36.40 | Nucl、Chlo |
| MdPOD48 | MDP0000292744 | 16 | 975 | 1 419 | 324 | 34.97 | 5.39 | 39.86 | Nucl、Chlo |
| MdPOD49 | MDP0000567775 | 未知 Unknown | 2 001 | 9 053 | 666 | 73.84 | 8.62 | 48.97 | Nucl、Cyto |
| MdPOD50 | MDP0000220665 | 未知 Unknown | 810 | 1 688 | 269 | 29.11 | 4.90 | 30.04 | Chlo |
| MdPOD51 | MDP0000208213 | 未知 Unknown | 876 | 2 685 | 291 | 32.20 | 9.54 | 44.18 | Nucl、Cyto、Chlo |
表2 苹果POD基因家族蛋白质理化性质
Table 2 Physicochemical properties POD gene family protein in apple
| 基因 Gene | 登录号 Accession No. | 染色体 定位 Chr. | CDS/bp | 全长/bp Full length | 氨基酸数 Amino acid number | 分子量/kD Molecular weight | 等电点 Isoelectric point | 不稳定 系数 Instability index | 亚细胞定位 Subcellular location |
|---|---|---|---|---|---|---|---|---|---|
| MdPOD01 | MDP0000207215 | 1 | 1 662 | 6 251 | 553 | 61.10 | 9.30 | 31.26 | Nucl、Plas |
| MdPOD02 | MDP0000449389 | 1 | 1 254 | 6 838 | 417 | 45.98 | 9.23 | 45.17 | Nucl、Chlo |
| MdPOD03 | MDP0000451182 | 1 | 987 | 1 849 | 328 | 36.13 | 8.36 | 41.02 | Nucl、Chlo |
| MdPOD04 | MDP0000226365 | 2 | 567 | 1 833 | 188 | 21.18 | 9.01 | 30.38 | Nucl、Cyto、Chlo |
| MdPOD05 | MDP0000272643 | 2 | 957 | 1 309 | 318 | 34.31 | 9.26 | 43.27 | Plas |
| MdPOD06 | MDP0000234905 | 2 | 1 386 | 5 052 | 461 | 51.35 | 6.46 | 43.26 | Nucl、Cyto、Chlo、Plas |
| MdPOD07 | MDP0000151864 | 2 | 2 355 | 6 866 | 784 | 86.10 | 8.75 | 44.08 | Nucl、Chlo、Plas |
| MdPOD08 | MDP0000488361 | 2 | 1 236 | 3 723 | 411 | 44.62 | 7.50 | 39.84 | Nucl、Chlo |
| MdPOD09 | MDP0000021998 | 2 | 657 | 2 995 | 219 | 24.53 | 6.63 | 33.13 | Nucl、Cyto、Plas |
| MdPOD10 | MDP0000611163 | 3 | 975 | 1 731 | 324 | 34.92 | 4.58 | 35.61 | Nucl |
| MdPOD11 | MDP0000229817 | 3 | 378 | 378 | 125 | 13.58 | 8.91 | 36.09 | Nucl、Chlo |
| MdPOD12 | MDP0000241173 | 3 | 1 011 | 5 864 | 336 | 37.44 | 6.46 | 40.32 | Nucl、Cyto、Chlo、Plas |
| MdPOD13 | MDP0000261341 | 3 | 816 | 4 458 | 271 | 29.99 | 7.61 | 36.85 | Cyto、Chlo、Plas |
| MdPOD14 | MDP0000678562 | 3 | 1 551 | 8 349 | 516 | 57.07 | 7.83 | 46.50 | Nucl、Cyto、Chlo、Plas |
| MdPOD15 | MDP0000209189 | 4 | 996 | 2 332 | 331 | 34.61 | 7.51 | 32.31 | Cyto、Plas |
| MdPOD16 | MDP0000727958 | 4 | 900 | 1 531 | 299 | 32.47 | 8.10 | 44.42 | Nucl、Cyto、Chlo、Plas |
| MdPOD17 | MDP0000283650 | 5 | 2 775 | 15 847 | 924 | 100.30 | 6.47 | 38.37 | Nucl、Cyto、Chlo |
| MdPOD18 | MDP0000194474 | 5 | 4 284 | 14 538 | 1 427 | 159.79 | 8.55 | 48.54 | Plas |
| MdPOD19 | MDP0000316890 | 7 | 711 | 4 729 | 236 | 27.53 | 8.66 | 23.08 | Plas |
| MdPOD20 | MDP0000292921 | 8 | 864 | 3 498 | 287 | 31.19 | 9.03 | 44.06 | Nucl、Chlo |
| MdPOD21 | MDP0000151342 | 8 | 1 086 | 4 365 | 361 | 39.72 | 8.70 | 42.18 | Cyto、Chlo |
| MdPOD22 | MDP0000214851 | 8 | 477 | 1 800 | 160 | 18.45 | 9.90 | 69.36 | Nucl、Cyto、Chlo、Plas |
| MdPOD23 | MDP0000143123 | 8 | 396 | 2 232 | 131 | 14.31 | 6.55 | 22.87 | Nucl、Cyto、Chlo、Plas |
| MdPOD24 | MDP0000903820 | 8 | 288 | 1 247 | 95 | 10.18 | 4.78 | 62.56 | Cyto、Chlo |
| MdPOD25 | MDP0000248823 | 8 | 1 572 | 7 408 | 523 | 57.88 | 9.60 | 54.49 | Nucl、Cyto、Chlo |
| MdPOD26 | MDP0000171732 | 10 | 1 044 | 1 281 | 347 | 38.61 | 9.21 | 28.25 | Chlo、Plas |
| MdPOD27 | MDP0000509183 | 11 | 2 931 | 6 860 | 976 | 107.17 | 5.55 | 41.43 | Nucl、Cyto、Chlo |
| MdPOD28 | MDP0000199034 | 11 | 1 335 | 3 358 | 444 | 49.05 | 6.46 | 39.05 | Nucl、Cyto、Chlo |
| MdPOD29 | MDP0000136412 | 11 | 999 | 1 557 | 332 | 35.70 | 6.43 | 32.14 | Chlo |
| MdPOD30 | MDP0000598572 | 11 | 1 122 | 6 014 | 373 | 40.81 | 6.79 | 43.72 | Nucl、Cyto |
| MdPOD31 | MDP0000126107 | 12 | 618 | 1 604 | 205 | 22.85 | 6.07 | 48.97 | Nucl、Cyto |
| MdPOD32 | MDP0000301828 | 12 | 1 020 | 2 911 | 339 | 35.50 | 4.97 | 41.53 | Nucl、Chlo |
| MdPOD33 | MDP0000210077 | 12 | 1 026 | 2 915 | 341 | 37.69 | 5.70 | 38.27 | Nucl、Cyto |
| MdPOD34 | MDP0000399965 | 12 | 366 | 840 | 121 | 13.30 | 4.74 | 30.47 | Nucl、Cyto、Chlo |
| 基因 Gene | 登录号 Accession No. | 染色体 定位 Chr. | CDS/bp | 全长/bp Full length | 氨基酸数 Amino acid number | 分子量/kD Molecular weight | 等电点 Isoelectric point | 不稳定 系数 Instability index | 亚细胞定位 Subcellular location |
| MdPOD35 | MDP0000319048 | 12 | 1 386 | 9 973 | 461 | 50.42 | 9.17 | 45.08 | Chlo |
| MdPOD36 | MDP0000179781 | 12 | 726 | 1 839 | 241 | 25.58 | 8.55 | 33.29 | Nucl、Cyto、Chlo |
| MdPOD37 | MDP0000672440 | 13 | 774 | 963 | 257 | 27.37 | 5.32 | 31.33 | Nucl、Cyto、Chlo |
| MdPOD38 | MDP0000264519 | 13 | 987 | 1 159 | 328 | 35.21 | 6.89 | 34.90 | Nucl、Cyto、Chlo |
| MdPOD39 | MDP0000305310 | 13 | 4 332 | 16 763 | 1 443 | 161.02 | 5.45 | 45.26 | Plas |
| MdPOD40 | MDP0000208152 | 13 | 1 110 | 2 364 | 369 | 39.39 | 4.36 | 40.36 | Chlo |
| MdPOD41 | MDP0000124622 | 13 | 975 | 1 457 | 324 | 35.03 | 5.64 | 39.51 | Nucl、Cyto |
| MdPOD42 | MDP0000142485 | 13 | 981 | 1 522 | 326 | 35.78 | 8.94 | 36.02 | Nucl、Chlo |
| MdPOD43 | MDP0000943804 | 15 | 885 | 2 506 | 294 | 31.35 | 5.63 | 37.80 | Chlo |
| MdPOD44 | MDP0000169497 | 15 | 867 | 2 104 | 288 | 31.47 | 6.41 | 31.21 | Cyto、Chlo、Plas |
| MdPOD45 | MDP0000918790 | 15 | 876 | 2 073 | 291 | 32.23 | 5.61 | 41.77 | Nucl、Cyto、Chlo |
| MdPOD46 | MDP0000612477 | 16 | 1 026 | 1 622 | 341 | 37.15 | 4.70 | 32.21 | Cyto |
| MdPOD47 | MDP0000849735 | 16 | 1 011 | 1 150 | 336 | 36.71 | 8.32 | 36.40 | Nucl、Chlo |
| MdPOD48 | MDP0000292744 | 16 | 975 | 1 419 | 324 | 34.97 | 5.39 | 39.86 | Nucl、Chlo |
| MdPOD49 | MDP0000567775 | 未知 Unknown | 2 001 | 9 053 | 666 | 73.84 | 8.62 | 48.97 | Nucl、Cyto |
| MdPOD50 | MDP0000220665 | 未知 Unknown | 810 | 1 688 | 269 | 29.11 | 4.90 | 30.04 | Chlo |
| MdPOD51 | MDP0000208213 | 未知 Unknown | 876 | 2 685 | 291 | 32.20 | 9.54 | 44.18 | Nucl、Cyto、Chlo |
| 基序 Motif | 序列 Sequence | 氨基酸数 Amino acid number | 基序 Motif | 序列 Sequence | 氨基酸数 Amino acid number |
|---|---|---|---|---|---|
| Motif1 | ACPGVVSCADILALAARDAVVLTGGPSWTVPLGRRDSKTAS | 41 | Motif9 | CTTFRSRJYNFSGTGNPDPTL | 21 |
| Motif2 | SNFYDSTCPNVESIVRSAVEQAVQKDPRIAASLJRLHFHDCFVNGCDASI | 50 | Motif10 | AEGNLPAPTENLSQLITKFSR | 21 |
| Motif3 | KGLSDKDJVALSGAHTJGRAR | 21 | Motif11 | CAPIMLRLAWHDAGTYDAKTKTGGPNGSI | 29 |
| Motif4 | YAABQSAFFADFAEAMVKLGNJG | 23 | Motif12 | SICPRSGGDBNLAPLDLVTPT | 21 |
| Motif5 | FDNSYYKNLLKGKGLLHSDQVLFSDSRTD | 29 | Motif13 | EGLLKLPTDKALLEDPVFRPY | 21 |
| Motif6 | PNNNSLRGFEVIDDIKAKLEK | 21 | Motif14 | ELSHGANNGLKIAVRLLEEVK | 21 |
| Motif7 | TGTNGEIRKNCRRVN | 15 | Motif15 | LLDDTPNFTGEKTA | 14 |
| Motif8 | RSGFEGPWTPNPLIFDNSYFK | 21 |
表3 苹果POD蛋白氨基酸保守序列
Table 3 Conserved motifs of POD proteins in apple
| 基序 Motif | 序列 Sequence | 氨基酸数 Amino acid number | 基序 Motif | 序列 Sequence | 氨基酸数 Amino acid number |
|---|---|---|---|---|---|
| Motif1 | ACPGVVSCADILALAARDAVVLTGGPSWTVPLGRRDSKTAS | 41 | Motif9 | CTTFRSRJYNFSGTGNPDPTL | 21 |
| Motif2 | SNFYDSTCPNVESIVRSAVEQAVQKDPRIAASLJRLHFHDCFVNGCDASI | 50 | Motif10 | AEGNLPAPTENLSQLITKFSR | 21 |
| Motif3 | KGLSDKDJVALSGAHTJGRAR | 21 | Motif11 | CAPIMLRLAWHDAGTYDAKTKTGGPNGSI | 29 |
| Motif4 | YAABQSAFFADFAEAMVKLGNJG | 23 | Motif12 | SICPRSGGDBNLAPLDLVTPT | 21 |
| Motif5 | FDNSYYKNLLKGKGLLHSDQVLFSDSRTD | 29 | Motif13 | EGLLKLPTDKALLEDPVFRPY | 21 |
| Motif6 | PNNNSLRGFEVIDDIKAKLEK | 21 | Motif14 | ELSHGANNGLKIAVRLLEEVK | 21 |
| Motif7 | TGTNGEIRKNCRRVN | 15 | Motif15 | LLDDTPNFTGEKTA | 14 |
| Motif8 | RSGFEGPWTPNPLIFDNSYFK | 21 |
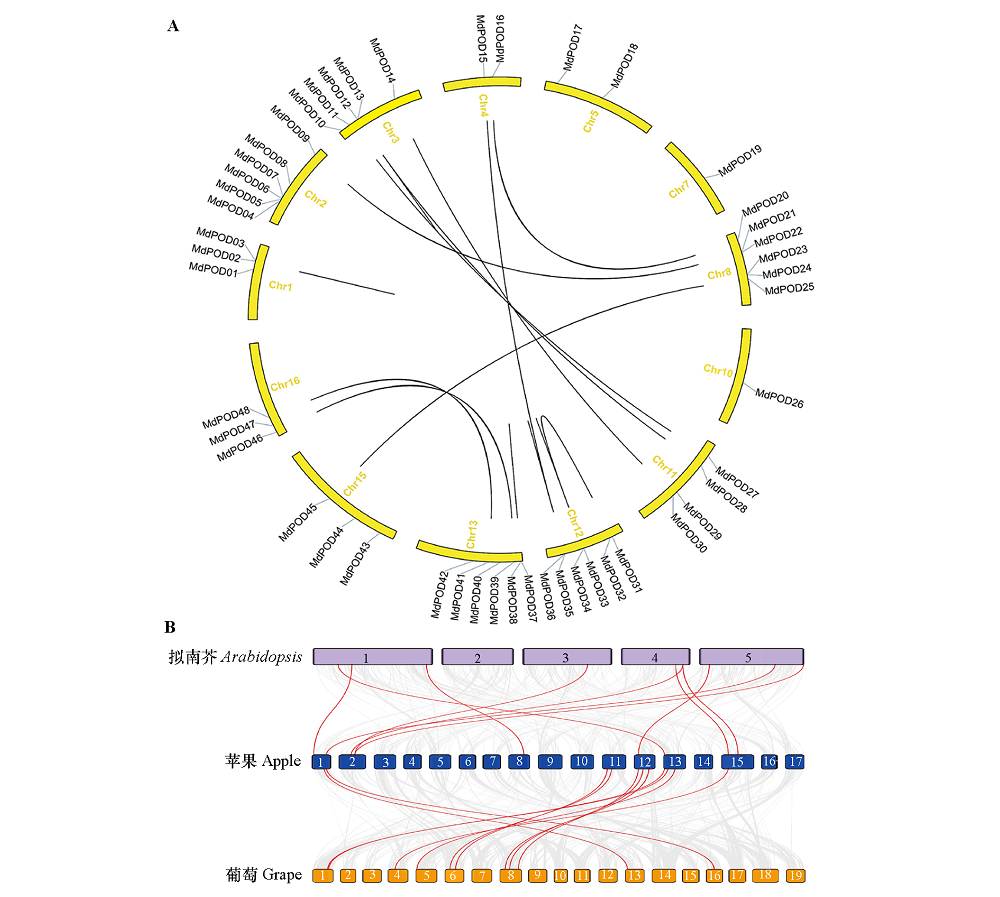
图3 苹果POD家族成员之间的共线性(A)和拟南芥、葡萄与苹果之间的POD基因组共线性(B)
Fig. 3 Collinearity between members of apple POD family(A)and POD genome collinearity between Arabidopsis,grape and apple(B)
| 基因对 Gene pairs | 非同义突变率(Ka) Non-synonymous | 同义突变频率(Ks) Synonymous | 突变频率比值 Ka/Ks | |
|---|---|---|---|---|
| MdPOD14 | MdPOD30 | 0.1262 | 0.2773 | 0.4552 |
| MdPOD03 | MdPOD02 | 0.1752 | 0.2348 | 0.7459 |
| MdPOD46 | MdPOD39 | 0.0415 | 0.1552 | 0.2675 |
| MdPOD16 | MdPOD20 | 0.0583 | 0.1686 | 0.3459 |
| MdPOD37 | MdPOD38 | 0.0170 | 0.0668 | 0.2550 |
| MdPOD45 | MdPOD25 | 0.1070 | 0.3620 | 0.2955 |
| MdPOD27 | MdPOD11 | 0.0336 | 0.1404 | 0.2392 |
| MdPOD10 | MdPOD50 | 0.0247 | 0.1430 | 0.1728 |
| MdPOD17 | MdPOD01 | 0.3099 | 0.4489 | 0.6904 |
| MdPOD48 | MdPOD41 | 0.0379 | 0.1334 | 0.2841 |
| MdPOD21 | MdPOD09 | 0.0424 | 0.1714 | 0.2473 |
| MdPOD31 | MdPOD33 | 0.0220 | 0.1853 | 0.1187 |
| MdPOD34 | MdPOD33 | 0.0399 | 0.0699 | 0.5706 |
| MdPOD12 | MdPOD28 | 0.3017 | 0.5232 | 0.5766 |
| MdPOD12 | MdPOD13 | 0.2964 | 0.3145 | 0.9422 |
| MdPOD13 | MdPOD28 | 0.5137 | 0.7140 | 0.7195 |
| MdPOD15 | MdPOD35 | 0.1918 | 0.4364 | 0.4396 |
| MdPOD15 | MdPOD36 | 0.0829 | 0.2345 | 0.3534 |
| MdPOD35 | MdPOD36 | 0.0802 | 0.1849 | 0.4340 |
| MdPOD51 | MdPOD07 | 0.0170 | 0.0729 | 0.2335 |
表4 苹果POD同源基因Ka/Ks分析
Table 4 Ka/Ks analysis of PODs homologous gene in apple
| 基因对 Gene pairs | 非同义突变率(Ka) Non-synonymous | 同义突变频率(Ks) Synonymous | 突变频率比值 Ka/Ks | |
|---|---|---|---|---|
| MdPOD14 | MdPOD30 | 0.1262 | 0.2773 | 0.4552 |
| MdPOD03 | MdPOD02 | 0.1752 | 0.2348 | 0.7459 |
| MdPOD46 | MdPOD39 | 0.0415 | 0.1552 | 0.2675 |
| MdPOD16 | MdPOD20 | 0.0583 | 0.1686 | 0.3459 |
| MdPOD37 | MdPOD38 | 0.0170 | 0.0668 | 0.2550 |
| MdPOD45 | MdPOD25 | 0.1070 | 0.3620 | 0.2955 |
| MdPOD27 | MdPOD11 | 0.0336 | 0.1404 | 0.2392 |
| MdPOD10 | MdPOD50 | 0.0247 | 0.1430 | 0.1728 |
| MdPOD17 | MdPOD01 | 0.3099 | 0.4489 | 0.6904 |
| MdPOD48 | MdPOD41 | 0.0379 | 0.1334 | 0.2841 |
| MdPOD21 | MdPOD09 | 0.0424 | 0.1714 | 0.2473 |
| MdPOD31 | MdPOD33 | 0.0220 | 0.1853 | 0.1187 |
| MdPOD34 | MdPOD33 | 0.0399 | 0.0699 | 0.5706 |
| MdPOD12 | MdPOD28 | 0.3017 | 0.5232 | 0.5766 |
| MdPOD12 | MdPOD13 | 0.2964 | 0.3145 | 0.9422 |
| MdPOD13 | MdPOD28 | 0.5137 | 0.7140 | 0.7195 |
| MdPOD15 | MdPOD35 | 0.1918 | 0.4364 | 0.4396 |
| MdPOD15 | MdPOD36 | 0.0829 | 0.2345 | 0.3534 |
| MdPOD35 | MdPOD36 | 0.0802 | 0.1849 | 0.4340 |
| MdPOD51 | MdPOD07 | 0.0170 | 0.0729 | 0.2335 |
| [1] | Cao Y P, Han Y H, Meng D D, Li D H, Jin Q, Lin Y, Cai Y P. 2016. Structural,evolutionary,and functional analysis of the class Ⅲ peroxidase gene family in Chinese pear(Pyrus bretschneideri). Frontiers in Plant Science, 7:1874. |
| [2] |
Cao Y Y, Yang M T, Chen S Y, Zhou Z Q, Li X, Wang X J, Bai J G. 2015. Exogenous sucrose influences antioxidant enzyme activities and reduces lipid peroxidation in water-stressed cucumber leaves. Biologia Plantarum, 59 (1):147-153.
doi: 10.1007/s10535-014-0469-7 URL |
| [3] | Chao Jin-quan, Yang Shu-guang, Chen Yue-yi, Tian Wei-min. 2018. Identification and expression of APX gene family members in Hevea brasiliensis. Genomics and Applied Biology, 37 (11):4769-4774. (in Chinese) |
| 晁金泉, 杨署光, 陈月异, 田维敏. 2018. 巴西橡胶树APX基因家族成员的鉴定及表达分析. 基因组学与应用生物学, 37 (11):4769-4774. | |
| [4] | Chen Guo-hu, Wang Hao, Li Guang, Tang Xiao-yan, Wang Cheng-gang, Zhang Lei, Hou Jin-feng, Yuan Ling-yun. 2020. Genome-wide identification and bioinformatics analysis of PRX gene family in Brassica rapa. Journal of Zhejiang University(Agriculture & Life Sciences), 46 (6):677-686. (in Chinese) |
| 陈国户, 王浩, 李广, 唐小燕, 汪承刚, 张磊, 侯金锋, 袁凌云. 2020. 白菜PRX基因家族的鉴定与生物信息学分析. 浙江大学学报(农业与生命科学版), 46 (6):677-686. | |
| [5] |
Claudia C, Christophe D. 2009. Specific functions of individual class Ⅲ peroxidase genes. Journal of Experimental Botany, 60 (2):391-408.
doi: 10.1093/jxb/ern318 pmid: 19088338 |
| [6] |
Coego A, Ramirez V, Ellul P, Mayda E, Vera P. 2010. The H2O2-regulated ep5c gene encodes a peroxidase required for bacterial speck susceptibility in tomato. Plant Journal, 42 (2),283-293.
doi: 10.1111/j.1365-313X.2005.02372.x URL |
| [7] |
Corpet F. 1988. Multiple sequence alignment with hierarchical clustering. Nucleic Acids Res, 16 (22):10881-10890.
doi: 10.1093/nar/16.22.10881 pmid: 2849754 |
| [8] |
Daudi A, Cheng Z, O’brien J A, Mammarella N, Khan S, Ausubel F M, Bolwell G P. 2012. The apoplastic oxidative burst peroxidase in Arabidopsis is a major component of pattern-triggered immunity. Plant Cell, 24 (1):275-287.
doi: 10.1105/tpc.111.093039 URL |
| [9] |
Duan P F, Wang G, Chao M N, Zhang Z Y, Zhang B H. 2019. Genome-wide identification and analysis of class III peroxidases in allotetraploid cotton(Gossypium hirsutum L.)and their responses to PK deficiency. Genes, 10 (6):473.
doi: 10.3390/genes10060473 URL |
| [10] | Feng Li-juan, Shi Zuo-ya, Yang Xue-mei, Tang Hai-xia, Jiao Qi-qing, Yin Yan-lei. 2020. Identification and bioinformatics analysis of peroxidase gene family in pomegranate. Molecular Plant Breeding, 18 (10):3172-3184. (in Chinese) |
| 冯立娟, 史作亚, 杨雪梅, 唐海霞, 焦其庆, 尹燕雷. 2020. 石榴POD基因家族鉴定及生物信息学分析. 分子植物育种, 18 (10):3172-3184. | |
| [11] |
Gabaldón C, López-serrano M, Pedreño M A, Barceló A R. 2005. Cloning and molecular characterization of the basic peroxidase isoenzyme from Zinnia elegans,an enzyme involved in lignin biosynthesis. Plant Physiology, 139 (3):1138-1154.
pmid: 16258008 |
| [12] |
Gao C Q, Wang Y C, Liu G F, Wang C, Jiang J, Yang C P. 2010. Cloning of ten peroxidase(POD)genes from Tamarix hispida and characterization of their responses to abiotic stress. Plant Molecular Biology Reporter, 28 (1):77-89.
doi: 10.1007/s11105-009-0129-9 URL |
| [13] | Gao He-ning. 2020. Effects of salt stress and its alleviation by calcium chloride in Ligustrum obtusifolium Sieb. et Zucc seedlings[M. D. Dissertation]. Shenyang: Shenyang Agricultural University. (in Chinese) |
| 高鹤宁. 2020. 盐胁迫对水蜡幼苗的影响及氯化钙对盐胁迫的缓解[硕士论文]. 沈阳: 沈阳农业大学. | |
| [14] |
Gong Z Z, Xiong L M, Shi H Z, Yang S H, Herrera-Estrella L R, Xu G H, Chao D Y, Li J R, Wang P Y, Qin F, Li J J, Ding Y L, Shi Y T, Wang Y, Yang Y Q, Guo Y, Zhu J K. 2020. Plant abiotic stress response and nutrient use efficiency. Science China Life Sciences, 63 (5):635-674.
doi: 10.1007/s11427-020-1683-x URL |
| [15] | Guo Xiao-hong. 2009. Sequence and function analysis of peroxidase in Tamarix hispida[Ph. D. Dissertation]. Harbin:Northeast Forestry University. (in Chinese) |
| 郭晓红. 2009. 柽柳过氧化物酶基因的序列分析及功能验证[博士论文]. 哈尔滨: 东北林业大学. | |
| [16] | Guo Li-ping. 2020. Transcriptome analysis revealing flower coloration of Sophora japonica Linn. and function analysis for key genes involved in anthocyanin accumulation[Ph. D. Dissertation]. Beijing:Beijing Forestry University. (in Chinese) |
| 郭丽萍. 2020. 国槐花色转录组学分析与花青苷积累关键基因功能验证[博士论文]. 北京: 北京林业大学. | |
| [17] | Hiraga S, Ichinose C, Onogi T, Niki H, Yamazoe M. 2000. Bidirectional migration of SeqA-bound hemimethylated DNA clusters and pairing of oriC copies in Escherichia coli. Genes to Cells:Devoted to Molecular & Cellular Mechanisms, 5 (5):327-341. |
| [18] | Hiraga S, Sasaki K, Ito H, Ohashi Y, Matsui H. 2001. A large family of class III plant peroxidases. Plant & Cell Physiology, 42 (5):462-468. |
| [19] |
Jourda C, Dunand C, Mathé C, Barre A. 2010. Evolution and expression of class III peroxidases. Archives of Biochemistry and Biophysics, 500 (1):58-65.
doi: 10.1016/j.abb.2010.04.007 URL |
| [20] |
Juretic N. 2005. The evolutionary fate of MULE-mediated duplications of host gene fragments in rice. Genome Research, 15 (9):1292-1297.
doi: 10.1101/gr.4064205 URL |
| [21] |
Kumar S, Jaggi M, Sinha A K. 2012. Ectopic overexpression of vacuolar and apoplastic Catharanthus roseus peroxidases confers differential tolerance to salt and dehydration stress in transgenic tobacco. Protoplasma, 249 (2):423-432.
doi: 10.1007/s00709-011-0294-1 URL |
| [22] |
Li Q, Dou W F, Qi J J, Qin X J, Chen S C, He Y R. 2020. Genomewide analysis of the CIII peroxidase family in sweet orange(Citrus sinensis)and expression profiles induced by Xanthomonas citri subsp. citri and hormones. Journal of Genetics, 99 (1):10.
doi: 10.1007/s12041-019-1163-5 URL |
| [23] |
Li Xuan, Yue Hong, Wang Sheng, Huang Lu-qi, Ma Jiong, Guo Lan-ping. 2013. Research of different effects on activity of plant antioxidant enzymes. China Journal of Chinese Materia Medica, 38 (7):973-978. (in Chinese)
pmid: 23847940 |
|
李璇, 岳红, 王升, 黄璐琦, 马炯, 郭兰萍. 2013. 影响植物抗氧化酶活性的因素及其研究热点和现状. 中国中药杂志, 38 (7):973-978.
pmid: 23847940 |
|
| [24] | Liang Guo-ping. 2019. Expression of genes related to sugar metabolism under low-temperature acclimation of Vitis amurensis and functional identification of Vv AGPS1 gene[M. D. Dissertation]. Lanzhou: Gansu Agricultural University. (in Chinese) |
| 梁国平. 2019. 山葡萄低温锻炼期糖代谢相关基因的表达及VvAGPS1基因的功能鉴定[硕士论文]. 兰州: 甘肃农业大学. | |
| [25] |
Llorente F, Rosa M, Catala R, Martinez-zapater J M, Salinas J. 2002. A novel cold-inducible gene from Arabidopsis,RCI3,encodes a peroxidase that constitutes a component for stress tolerance. Plant Journal, 32 (1):13-24.
doi: 10.1046/j.1365-313X.2002.01398.x URL |
| [26] | Lü Jie, Wang Chao, Liu Lu, Xu Hong-wei. 2019. Advance in response of antioxidant enzyme system in plant to UV-B stress. Jilin Normal University Journal(Natural Science Edition), 40 (4):113-117. (in Chinese) |
| 吕杰, 王超, 刘璐, 徐洪伟. 2019. UV-B胁迫对植物抗氧化酶活性的影响. 吉林师范大学学报(自然科学版), 40 (4):113-117. | |
| [27] |
Lysak M A. 2005. Chromosome triplication found across the tribe Brassiceae. Genome Research, 15 (4):516-525.
doi: 10.1101/gr.3531105 URL |
| [28] |
Maksymiec W, Krupa Z. 2006. The effects of short-term exposition to Cd,excess Cu ions and jasmonate on oxidative stress appearing in Arabidopsis thaliana. Environ Exp Bot, 57:187-194.
doi: 10.1016/j.envexpbot.2005.05.006 URL |
| [29] |
Marc B, Sylvain L, Jean-francois H, Gea G. 2015. Analysis of cell wall-related genes in organs of Medicago sativa L. under different abiotic stresses. International Journal of Molecular Sciences, 16 (7):16104-16124.
doi: 10.3390/ijms160716104 pmid: 26193255 |
| [30] |
Mount S, Henikoff S. 1993. Gene organization:nested genes take flight. Curr Biol, 3:372-374.
pmid: 15335735 |
| [31] |
Niedzwiedz-siegien I, Bogatek-leszczynska R, Côme D, Corbineau F. 2004. Effects of drying rate on dehydration sensitivity of excised wheat seedling shoots as related to sucrose metabolism and antioxidant enzyme activities. Plant Science, 167 (4):879-888.
doi: 10.1016/j.plantsci.2004.05.042 URL |
| [32] |
Ostergaard L, Teilum K, Mirza O, Mattsson O, Petersen M, Welinder K G, Mundy J, Gajhede M, Henriksen A. 2000. Arabidopsis ATP A 2 peroxidase. expression and high-resolution structure of a plant peroxidase with implications for lignification. Plant Molecular Biology, 44 (2):231-243.
pmid: 11117266 |
| [33] |
Passardi F, Longet D, Penel C, Dunand C. 2004a. The class III peroxidase multigenic family in rice and its evolution in land plants. Phytochemistry, 65 (13):1879-1893.
doi: 10.1016/j.phytochem.2004.06.023 URL |
| [34] |
Passardi F, Penel C, Dunand C. 2004b. Performing the paradoxical:how plant peroxidases modify the cell wall. Trends in Plant science, 9 (11):534-540.
doi: 10.1016/j.tplants.2004.09.002 URL |
| [35] |
Ren L L, Liu Y J, Liu H J. 2014. Subcellular relocalization and positive selection play key roles in the retention of duplicate genes of populus class III peroxidase family. The Plant Cell, 26 (6):2404-2419.
doi: 10.1105/tpc.114.124750 URL |
| [36] |
Rogozin I B, Wolf Y I, Sorokin A V, Mirkin B G, Koonin E V. 2003. Remarkable interkingdom conservation of intron positions and massive,lineage-specific intron loss and gain in eukaryotic evolution. Current Biology, 13 (17):1512-1517.
pmid: 12956953 |
| [37] |
Shannon L M, Kay E, Lew J Y. 1966. Peroxidase isozymes from horseradish roots. I. Isolation and physical properties. Journal of Biological Chemistry, 241 (9):2166-2172.
pmid: 5946638 |
| [38] |
Tognolli M, Penel C, Greppin H, Greppin H, Simon P. 2002. Analysis and expression of the class III peroxidase large gene family in Arabidopsis thaliana. Gene, 288 (1-2):129-138.
pmid: 12034502 |
| [39] |
Velasco R, Zharkikh A, Affourtit J, Dhingra A, Cestaro A, Kalyanaraman A, Fontana P, Bhatnagar S K, Troggio M, Pruss D. 2010. The genome of the domesticated apple(Malus domestica Borkh.). Nature Genetics, 42:833-839.
doi: 10.1038/ng.654 URL |
| [40] | Wang Jing, Liu Lun, Yang Yong, Hu Sheng, Li Li-qin. 2019. Cloning and expression analysis of tobacco peroxidase gene NtPOD2. Biotechnology Bulletin, 35 (12):24-30. (in Chinese) |
|
王静, 刘仑, 杨勇, 胡圣, 李立芹. 2019. 烟草过氧化物酶基因NtPOD2的克隆及表达分析. 生物技术通报, 35 (12):24-30.
doi: 10.13560/j.cnki.biotech.bull.1985.2019-0395 |
|
| [41] | Wang Le. 2009. Research on salt tolerant peroxidases in wheat and arsenic resistant transgenic plant[M. D. Dissertation]. Ji’nan: Shandong University. (in Chinese) |
| 王乐. 2009. 山融3号小麦耐盐相关过氧化物酶基因及植物抗砷基因工程初步研究[硕士论文]. 济南: 山东大学. | |
| [42] | Wang Wei. 2019. Functional analysis of reactive oxygen species metabolism related genes in upland cotton under salt stress[Ph. D. Dissertation]. Tai’an:Shandong Agricultural University. (in Chinese) |
| 王维. 2019. 盐胁迫条件下陆地棉活性氧代谢相关基因的功能研究[博士论文]. 泰安: 山东农业大学. | |
| [43] |
Wang Y, Wang Q Q, Zhao Y, Han G M, Zhu S W. 2015a. Systematic analysis of maize class III peroxidase gene family reveals a conserved subfamily involved in abiotic stress response. Gene, 566 (1):95-108.
doi: 10.1016/j.gene.2015.04.041 URL |
| [44] |
Wang Y Y, Feng L, Zhu Y X, Li Y, Yan H W, Xiang Y. 2015b. Comparative genomic analysis of the WRKY III gene family in populus,grape,Arabidopsis and rice. Biology Direct, 10:48.
doi: 10.1186/s13062-015-0076-3 URL |
| [45] | Wei Jin-feng, Guo Jing-gong, Zhang Qing-de, Li Kun, Bai Ling. 2009. Effect of ultraviolet radiation on the antioxidant enzymes and the membrane systems of Arabidopsis thaliana leaves. Journal of Henan University(Natural Science), 39 (5):500-504. (in Chinese) |
| 魏金凤, 郭敬功, 张清德, 李坤, 白玲. 2009. UV辐射对拟南芥叶片抗氧化酶系统和膜系统的影响. 河南大学学报(自然科学版), 39 (5):500-504. | |
| [46] | Wei Xia, Li Yun-feng, Xiang Jia, Ling Ying-hua, Yang Zheng-lin, He Guang-hua, Zhao Fang-ming. 2015. Effect of drought stress on physiological characters of rice variant line R21 by introgression sorghum DNA. Southwest China Journal of Agricultural Sciences, 28 (3):930-934. (in Chinese) |
| 魏霞, 李云峰, 向佳, 凌英华, 杨正林, 何光华, 赵芳明. 2015. 干旱胁迫对导入高粱DNA的水稻变异系R21生理的影响. 西南农业学报, 28 (3):930-934. | |
| [47] |
Welinder K G. 1992. Superfamily of plant,fungal and bacterial peroxidases. Curr Opin Struct Biol, 2 (3):388-393.
doi: 10.1016/0959-440X(92)90230-5 URL |
| [48] |
Wu C L, Ding X P, Ding Z H, Tie W W, Yan Y, Wang Y, Yang H, Hu W. 2019. The class III peroxidase(POD)gene family in cassava:identification,phylogeny,duplication and expression. International Journal of Molecular Sciences, 20 (11):2730.
doi: 10.3390/ijms20112730 URL |
| [49] |
Wu Y S, Yang Z L, How J Y, Xu H N, Chen L M, Li K Z. 2017. Overexpression of a peroxidase gene(AtPrx64) of Arabidopsis thaliana in tobacco improves plant’s tolerance to aluminum stress. Plant Molecular Biology,Doi: 10.1007/s11103-017-0644-2.
doi: 10.1007/s11103-017-0644-2 URL |
| [50] |
Xiao H L, Wang C P, Khan N, Chen M X, Fu W H, Guan L, Leng X P. 2020. Genome-wide identification of the class III POD gene family and their expression profiling in grapevine(Vitis vinifera L.). BMC Genomics, 21 (1):1-13.
doi: 10.1186/s12864-019-6419-1 URL |
| [51] | Zhang Tingting, Kang Hui, Fu Lulu, You Chunxiang, Wang Xiaofei, Hao Yujin. 2019. Expression analysis of apple MdCYP707A family genes and functional characterization of MdCYP707A1. Acta Horticulturae Sinica, 46 (8):1429-1444. (in Chinese) |
| 张婷婷, 康慧, 付璐璐, 由春香, 王小非, 郝玉金. 2019. 苹果MdCYP707A家族基因表达分析和MdCYP707A1的功能鉴定. 园艺学报, 46 (8):1429-1444. | |
| [52] | Zhang Wei-jing, Lu Hai, Du Xi-hua. 2008. The function of ascorbate peroxidases in plant resistance to oxidative stress. Journal of Shandong Normal University(Natural Science), 23 (4):113-115. (in Chinese) |
| 张维静, 陆海, 杜希华. 2008. 抗坏血酸过氧化物酶在植物抵抗氧化胁迫中的作用. 山东师范大学学报(自然科学版), 23 (4):113-115. | |
| [53] | Zhang Yu-gang, Cheng Jian-hong, Han Zhen-hai, Xu Xue-feng, Li Tian-zhong. 2005. Comparison of methods for total RNA isolation from Malus xiaojinensis and cDNA LD-PCR amplication. Biotechnology Bulletin,(4):50-53. (in Chinese) |
| 张玉刚, 成建红, 韩振海, 许雪峰, 李天忠. 2005. 小金海棠总 RNA 提取方法比较及cDNA的LD-PCR扩增. 生物技术通报,(4):50-53. | |
| [54] | Zhong Zaofa, Zhang Lijuan, Gao Sisi, Peng Ting. 2021. Leaf cytological characteristics and resistance comparison of four citrus rootstocks under drought stress. Acta Horticulturae Sinica, 48 (8):1579-1588. (in Chinese) |
| 钟灶发, 张利娟, 高思思, 彭婷. 2021. 干旱胁迫下4种柑橘砧木叶片细胞学特征及抗旱性比较. 园艺学报, 48 (8):1579-1588. | |
| [55] | Zhou Zhe, Zhang Cai-xia, Zhang Li-yi, Wang Qiang, Li Wu-xing, Tian Yi, Cong Pei-hua. 2014. Bioinformatics and expression analysis of the LysM gene family in apple. Scientia Agricultura Sinica, 47 (13):2602-2612. (in Chinese) |
| 周喆, 张彩霞, 张利义, 王强, 李武兴, 田义, 丛佩华. 2014. 苹果LysM基因家族的生物信息学及表达分析. 中国农业科学, 47 (13):2602-2612. |
| [1] | 于婷婷, 李 欢, 宁源生, 宋建飞, 彭璐琳, 贾竣淇, 张玮玮, 杨洪强. 苹果GRAS全基因组鉴定及其对生长素的响应分析[J]. 园艺学报, 2023, 50(2): 397-409. |
| [2] | 赵雪艳, 王琪, 王莉, 王方圆, 王庆, 李艳. 基于比较转录组的延胡索组织差异性表达分析[J]. 园艺学报, 2023, 50(1): 177-187. |
| [3] | 韩晓蕾, 张彩霞, 刘 锴, 杨 安, 严家帝, 李武兴, 康立群, 丛佩华. 中熟苹果新品种‘中苹优蕾’[J]. 园艺学报, 2022, 49(S2): 1-2. |
| [4] | 韩晓蕾, 张彩霞, 刘 锴, 严家帝, 李武兴, 康立群, 丛佩华. 中熟苹果新品种‘苹优2号’[J]. 园艺学报, 2022, 49(S1): 1-2. |
| [5] | 王 强, 丛佩华, 刘肖烽. 晚熟苹果新品种‘华优甜娃’[J]. 园艺学报, 2022, 49(S1): 3-4. |
| [6] | 王 强, 丛佩华, 刘肖烽. 中熟苹果新品种‘华优宝蜜’[J]. 园艺学报, 2022, 49(S1): 5-6. |
| [7] | 杨 玲, 丛佩华, 王 强, 李武兴, 康立群. 中熟鲜食苹果新品种‘华丰’[J]. 园艺学报, 2022, 49(S1): 7-8. |
| [8] | 徐小萍, 曹清影, 蔡柔荻, 官庆栩, 张梓浩, 陈裕坤, 徐涵, 林玉玲, 赖钟雄. 龙眼miR408与DlLAC12克隆及其在球形胚发生和非生物胁迫下的表达分析[J]. 园艺学报, 2022, 49(9): 1866-1882. |
| [9] | 贾鑫, 曾臻, 陈月, 冯慧, 吕英民, 赵世伟. 月季‘月月粉’RcDREB2A的克隆与表达分析[J]. 园艺学报, 2022, 49(9): 1945-1956. |
| [10] | 丁志杰, 包金波, 柔鲜古丽, 朱甜甜, 李雪丽, 苗浩宇, 田新民. 新疆野苹果与‘元帅’‘金冠’的叶绿体基因组比对研究[J]. 园艺学报, 2022, 49(9): 1977-1990. |
| [11] | 高彦龙, 吴玉霞, 张仲兴, 王双成, 张瑞, 张德, 王延秀. 苹果ELO家族基因鉴定及其在低温胁迫下的表达分析[J]. 园艺学报, 2022, 49(8): 1621-1636. |
| [12] | 邱子文, 刘林敏, 林永盛, 林晓洁, 李永裕, 吴少华, 杨超. 千层金MbEGS基因的克隆与功能分析[J]. 园艺学报, 2022, 49(8): 1747-1760. |
| [13] | 郑晓东, 袭祥利, 李玉琪, 孙志娟, 马长青, 韩明三, 李少旋, 田义轲, 王彩虹. 油菜素内酯对盐碱胁迫下平邑甜茶幼苗生长的影响及调控机理研究[J]. 园艺学报, 2022, 49(7): 1401-1414. |
| [14] | 夏炎, 黄松, 武雪莉, 刘一琪, 王苗苗, 宋春晖, 白团辉, 宋尚伟, 庞宏光, 焦健, 郑先波. 基于宏病毒组测序技术的苹果病毒病鉴定与分析[J]. 园艺学报, 2022, 49(7): 1415-1428. |
| [15] | 郑林, 王帅, 刘语诺, 杜美霞, 彭爱红, 何永睿, 陈善春, 邹修平. 柑橘响应黄龙病菌侵染的NAC基因的克隆及表达分析[J]. 园艺学报, 2022, 49(7): 1441-1457. |
| 阅读次数 | ||||||
|
全文 |
|
|||||
|
摘要 |
|
|||||
版权所有 © 2012 《园艺学报》编辑部 京ICP备10030308号-2 国际联网备案号 11010802023439
编辑部地址: 北京市海淀区中关村南大街12号中国农业科学院蔬菜花卉研究所 邮编: 100081
电话: 010-82109523 E-Mail: yuanyixuebao@126.com
技术支持:北京玛格泰克科技发展有限公司