
园艺学报 ›› 2022, Vol. 49 ›› Issue (1): 100-116.doi: 10.16420/j.issn.0513-353x.2020-0850
谢思艺1,3, 周承哲1,2,3, 朱晨1,2,3, 詹冬梅1,3, 陈兰1,3, 吴祖春1,3, 赖钟雄1,2, 郭玉琼1,3,*( )
)
收稿日期:2021-04-23
修回日期:2021-07-26
出版日期:2022-01-25
发布日期:2022-01-24
通讯作者:
郭玉琼
E-mail:guoyq828@163.com
基金资助:
XIE Siyi1,3, ZHOU Chengzhe1,2,3, ZHU Chen1,2,3, ZHAN Dongmei1,3, CHEN Lan1,3, WU Zuchun1,3, LAI Zhongxiong1,2, GUO Yuqiong1,3,*( )
)
Received:2021-04-23
Revised:2021-07-26
Online:2022-01-25
Published:2022-01-24
Contact:
GUO Yuqiong
E-mail:guoyq828@163.com
摘要:
TIFY转录因子是陆地植物特有的转录因子,按照结构域特征共分为4个亚家族TIFY、PPD、JAZ、ZML,调控植物的生长发育过程。以茶树TIFY转录因子为研究对象,从茶树基因组数据库中共鉴定了22个CsTIFY转录因子家族成员,都包含了TIFY结构域。基因结构分析发现,大部分CsTIFY家族成员外显子数在5到12之间,同一亚家族所含Motif和外显子数相似。系统进化树表明,6个物种122个TIFY家族基因分为4个组,其中茶树与葡萄和杨树亲缘关系更近。启动子顺式作用元件分析发现CsTIFY家族含有大量响应SA、ABA、低温和干旱等的元件。亚细胞定位试验结果表明CsTIFY1、CsJAZ1和CsJAZ5定位于细胞核。不同茶树品种的组织表达量分析发现,大部分CsTIFY在成熟叶、根和果实中相对表达量较高。在外源ABA、SA和MeJA处理下,大部分CsJAZ上调表达,而CsZML的表达量下降;低温和干旱处理下,CsZML1表达量都极显著上升。
中图分类号:
谢思艺, 周承哲, 朱晨, 詹冬梅, 陈兰, 吴祖春, 赖钟雄, 郭玉琼. 茶树CsTIFY家族全基因组鉴定及非生物胁迫和激素处理中主要基因表达分析[J]. 园艺学报, 2022, 49(1): 100-116.
XIE Siyi, ZHOU Chengzhe, ZHU Chen, ZHAN Dongmei, CHEN Lan, WU Zuchun, LAI Zhongxiong, GUO Yuqiong. Genome-wide Identification and Expression Analysis of CsTIFY Transcription Factor Family Under Abiotic Stress and Hormone Treatments in Camellia sinensis[J]. Acta Horticulturae Sinica, 2022, 49(1): 100-116.
| 用途 Purpose | 基因 Gene | 上游引物序列(5′-3′) Forward primer | 下游引物序列(5′-3′) Reverse primer |
|---|---|---|---|
| qRT-PCR | CsTIFY1 | TGAGTAACCGATTCGCTGG | GCTTGTGGAAACCCATCTCTG |
| CsTIFY2 | TCAACGAGGAAGAGGCTATGG | GGGTCCTCATCCTCAACCATT | |
| CsTIFY3 | GCCTCTGTTGGTGGTTCTTC | CGGTTATTCATCTCAGTCCCT | |
| CsTIFY4 | TCTCCGCCACTTCTGATCTG | GCGAATCGGTTACTCATCTCAG | |
| CsPPD1 | CCTACCAGATGTGCGAGAATG | TCGCCTTCATCCAAGCTGAT | |
| CsPPD2 | GCAAATTCTCCATTGCTCGC | TCCACGACGGTCTTCTCATC | |
| CsJAZ1 | TCCTCCAAACCTCTTCCCAAT | CACAGAGCCAGAAGCAATCAC | |
| CsJAZ2 | TCATTGATCGCCGAAGAAGC | GAATAGAGAGGCAAAGCAGG | |
| CsJAZ4 | TGACCCACTTGCTGCTTCT | GAACGCCACCTTGTCCAT | |
| CsJAZ5 | TGTCGAGTTCGTCTGGTTCT | CCGAATGTGCCTTTCTTCTC | |
| CsJAZ6 | CGGTGGCTGTCAAAGAAGGTATT | CTCTAATCGCCATTTCAGTCCCT | |
| CsJAZ7 | TCATTGATCGCCGAAGAAGC | GAATAGAGAGGCAAAGCAGGAG | |
| CsJAZ8 | CGGATTACAACCACCACCAC | TCGTTAGCGGCTGTTCCTGT | |
| CsZML1 | GGAGGGAGGAATCTTTGTTTGG | CTGCCAGTTTCTCCTGTTACAAC | |
| CsZML2 | CTAAACGTTCCCAACCACTTCAC | GTCGTTGAGAGAGTGAGTTTCGT | |
| CsZML3 | TGCAGCACATGAGAAATGG | GAACACATAAACCTGACCCTGG | |
| CsZML4 | CCAGAGACGAATCACCAATCC | CTCACCACCCACAACTTCATC | |
| CsZML5 | CCGATTCCACTTCAAGCAAG | TCCTCCTCCTCCGTCTTCAT | |
| CsZML6 | TCGATGAAGACGGAGGAGG | GGGTTGTTATTCACAGGGAGG | |
| CsZML7 | CGAATCCACAGCCACTTCAAG | TCCGCCTCATCCATAGCATC | |
| CsZML8 | CTGAGACCAACCACGAAGCT | TCGAATCGGATCTGAGGGTT | |
| 内参基因 | β-ACTNI | GCCATCTTTGATTGGAATGG | GGTGCCACAACCTTGATCTT |
| Reference gene | SAND | CCAATTGCCCCCTTAATGAC | GCAATCATTTCCTTCGTGGAG |
| 亚细胞定位 | CsTIFY1-1302 | ACTCTTGACCATGGTAGATCTATGATAGATAGACAGGTTGCAAATCAT | ATCTGATCCACCTTTACTAGTCTAAAAAATTGCCTTGTTCGGG |
| Subcellular | |||
| localization | CsJAZ1-1302 | ACTCTTGACCATGGTAGATCTATGGCATCGAGATCAGCTGTTG | ATCTGATCCACCTTTACTAGTCTAATCTTCATATCTTGATGTATTCTTCCC |
| CsJAZ5-1302 | ACTCTTGACCATGGTAGATCTATGTCGAGTTCGTCTGGTTCTGC | ATCTGATCCACCTTTACTAGTCTACAAATGGTGCTCAAACTGCA |
表1 茶树CsTIFY基因家族引物序列
Table 1 CsTIFY gene family primer sequences
| 用途 Purpose | 基因 Gene | 上游引物序列(5′-3′) Forward primer | 下游引物序列(5′-3′) Reverse primer |
|---|---|---|---|
| qRT-PCR | CsTIFY1 | TGAGTAACCGATTCGCTGG | GCTTGTGGAAACCCATCTCTG |
| CsTIFY2 | TCAACGAGGAAGAGGCTATGG | GGGTCCTCATCCTCAACCATT | |
| CsTIFY3 | GCCTCTGTTGGTGGTTCTTC | CGGTTATTCATCTCAGTCCCT | |
| CsTIFY4 | TCTCCGCCACTTCTGATCTG | GCGAATCGGTTACTCATCTCAG | |
| CsPPD1 | CCTACCAGATGTGCGAGAATG | TCGCCTTCATCCAAGCTGAT | |
| CsPPD2 | GCAAATTCTCCATTGCTCGC | TCCACGACGGTCTTCTCATC | |
| CsJAZ1 | TCCTCCAAACCTCTTCCCAAT | CACAGAGCCAGAAGCAATCAC | |
| CsJAZ2 | TCATTGATCGCCGAAGAAGC | GAATAGAGAGGCAAAGCAGG | |
| CsJAZ4 | TGACCCACTTGCTGCTTCT | GAACGCCACCTTGTCCAT | |
| CsJAZ5 | TGTCGAGTTCGTCTGGTTCT | CCGAATGTGCCTTTCTTCTC | |
| CsJAZ6 | CGGTGGCTGTCAAAGAAGGTATT | CTCTAATCGCCATTTCAGTCCCT | |
| CsJAZ7 | TCATTGATCGCCGAAGAAGC | GAATAGAGAGGCAAAGCAGGAG | |
| CsJAZ8 | CGGATTACAACCACCACCAC | TCGTTAGCGGCTGTTCCTGT | |
| CsZML1 | GGAGGGAGGAATCTTTGTTTGG | CTGCCAGTTTCTCCTGTTACAAC | |
| CsZML2 | CTAAACGTTCCCAACCACTTCAC | GTCGTTGAGAGAGTGAGTTTCGT | |
| CsZML3 | TGCAGCACATGAGAAATGG | GAACACATAAACCTGACCCTGG | |
| CsZML4 | CCAGAGACGAATCACCAATCC | CTCACCACCCACAACTTCATC | |
| CsZML5 | CCGATTCCACTTCAAGCAAG | TCCTCCTCCTCCGTCTTCAT | |
| CsZML6 | TCGATGAAGACGGAGGAGG | GGGTTGTTATTCACAGGGAGG | |
| CsZML7 | CGAATCCACAGCCACTTCAAG | TCCGCCTCATCCATAGCATC | |
| CsZML8 | CTGAGACCAACCACGAAGCT | TCGAATCGGATCTGAGGGTT | |
| 内参基因 | β-ACTNI | GCCATCTTTGATTGGAATGG | GGTGCCACAACCTTGATCTT |
| Reference gene | SAND | CCAATTGCCCCCTTAATGAC | GCAATCATTTCCTTCGTGGAG |
| 亚细胞定位 | CsTIFY1-1302 | ACTCTTGACCATGGTAGATCTATGATAGATAGACAGGTTGCAAATCAT | ATCTGATCCACCTTTACTAGTCTAAAAAATTGCCTTGTTCGGG |
| Subcellular | |||
| localization | CsJAZ1-1302 | ACTCTTGACCATGGTAGATCTATGGCATCGAGATCAGCTGTTG | ATCTGATCCACCTTTACTAGTCTAATCTTCATATCTTGATGTATTCTTCCC |
| CsJAZ5-1302 | ACTCTTGACCATGGTAGATCTATGTCGAGTTCGTCTGGTTCTGC | ATCTGATCCACCTTTACTAGTCTACAAATGGTGCTCAAACTGCA |
| 基因名称 Gene name | ID | 氨基酸 Amino acid | 分子量/kD Molecular weight | 等电点 Isoelectric point | 内含子 Intron | 外显子 Exons | 不稳定指数 Instability index | 脂肪族指数 Aliphatic index | 平均疏水性 GRAVY | 亚细胞定位 Subcellular localization |
|---|---|---|---|---|---|---|---|---|---|---|
| CsTIFY1 | TEA007552.1 | 250 | 26.77771 | 9.90 | 1 | 2 | 52.44 | 55.04 | -0.694 | 细胞核 |
| Cell nucleus | ||||||||||
| CsTTFY2 | TEA012041.1 | 934 | 103.1778 | 5.57 | 11 | 12 | 49.30 | 84.03 | -0.140 | 叶绿体 |
| Chloroplast | ||||||||||
| CsTIFY3 | TEA032457.1 | 352 | 38.90351 | 10.25 | 4 | 5 | 53.41 | 72.81 | -0.461 | 液泡 Vacuole |
| CsTIFY4 | TEA009327.1 | 393 | 41.65471 | 9.40 | 5 | 6 | 46.36 | 65.29 | -0.437 | 细胞核 |
| Cell nucleus | ||||||||||
| CsPPD1 | TEA005947.1 | 1 820 | 204.7527 | 6.14 | 16 | 17 | 41.51 | 92.61 | -0.120 | 胞间连丝 |
| Plasmodesma | ||||||||||
| CsPPD2 | TEA011549.1 | 1 114 | 125.3216 | 8.23 | 12 | 13 | 50.60 | 88.93 | -0.299 | 细胞核 |
| Cell nucleus | ||||||||||
| CsJAZ1 | TEA027049.1 | 212 | 23.87791 | 9.15 | 5 | 6 | 55.42 | 67.08 | -0.632 | 细胞核 |
| Cell nucleus | ||||||||||
| CsJAZ2 | TEA030826.1 | 148 | 16.39844 | 7.84 | 0 | 1 | 48.12 | 58.65 | -0.564 | 细胞核 |
| Cell nucleus | ||||||||||
| CsJAZ3 | TEA001501.1 | 261 | 29.26615 | 9.17 | 4 | 5 | 57.65 | 70.69 | -0.666 | 细胞核 |
| Cell nucleus | ||||||||||
| CsJAZ4 | TEA001681.1 | 410 | 42.84663 | 9.34 | 7 | 8 | 35.36 | 66.44 | -0.310 | 细胞核 |
| Cell nucleus | ||||||||||
| CsJAZ5 | TEA030190.1 | 294 | 31.53145 | 9.46 | 4 | 5 | 55.85 | 70.37 | -0.395 | 细胞核 |
| Cell nucleus | ||||||||||
| CsJAZ6 | TEA001821.1 | 436 | 46.20707 | 9.58 | 7 | 8 | 48.77 | 71.40 | -0.343 | 细胞质 Cytoplasm |
| CsJAZ7 | TEA032228.1 | 220 | 24.45353 | 9.18 | 5 | 6 | 55.48 | 67.36 | -0.557 | 细胞核 |
| Cell nucleus | ||||||||||
| CsJAZ8 | TEA004474.1 | 136 | 15.60498 | 9.93 | 2 | 3 | 84.54 | 75.29 | -0.721 | 细胞核 |
| Cell nucleus | ||||||||||
| CsZML1 | TEA033832.1 | 385 | 42.00050 | 4.88 | 10 | 11 | 42.88 | 70.08 | -0.625 | 细胞核 |
| Cell nucleus | ||||||||||
| CsZML2 | TEA033836.1 | 271 | 30.51872 | 4.57 | 4 | 5 | 46.48 | 78.04 | -0.601 | 细胞核 |
| Cell nucleus | ||||||||||
| CsZML3 | TEA013465.1 | 303 | 33.49678 | 6.19 | 6 | 7 | 31.87 | 60.79 | -0.916 | 细胞核 |
| Cell nucleus | ||||||||||
| CsZML4 | TEA014550.1 | 234 | 25.20185 | 5.00 | 3 | 4 | 50.61 | 71.24 | -0.550 | 细胞核 |
| Cell nucleus | ||||||||||
| CsZML5 | TEA014541.1 | 352 | 38.21245 | 4.85 | 10 | 11 | 50.11 | 72.30 | -0.574 | 细胞核 |
| Cell nucleus | ||||||||||
| CsZML6 | TEA011504.1 | 352 | 38.21245 | 4.85 | 10 | 11 | 50.11 | 72.30 | -0.574 | 细胞核 |
| Cell nucleus | ||||||||||
| CsZML7 | TEA002032.1 | 383 | 41.64484 | 4.76 | 10 | 11 | 54.89 | 63.92 | -0.795 | 细胞核 |
| Cell nucleus | ||||||||||
| CsZML8 | TEA001414.1 | 461 | 49.38947 | 5.75 | 11 | 12 | 41.39 | 73.19 | -0.375 | 细胞核 |
| Cell nucleus |
表2 茶树CsTIFY家族蛋白的理化特性
Table 2 Physicochemical properties of CsTIFY family proteins of tea tree
| 基因名称 Gene name | ID | 氨基酸 Amino acid | 分子量/kD Molecular weight | 等电点 Isoelectric point | 内含子 Intron | 外显子 Exons | 不稳定指数 Instability index | 脂肪族指数 Aliphatic index | 平均疏水性 GRAVY | 亚细胞定位 Subcellular localization |
|---|---|---|---|---|---|---|---|---|---|---|
| CsTIFY1 | TEA007552.1 | 250 | 26.77771 | 9.90 | 1 | 2 | 52.44 | 55.04 | -0.694 | 细胞核 |
| Cell nucleus | ||||||||||
| CsTTFY2 | TEA012041.1 | 934 | 103.1778 | 5.57 | 11 | 12 | 49.30 | 84.03 | -0.140 | 叶绿体 |
| Chloroplast | ||||||||||
| CsTIFY3 | TEA032457.1 | 352 | 38.90351 | 10.25 | 4 | 5 | 53.41 | 72.81 | -0.461 | 液泡 Vacuole |
| CsTIFY4 | TEA009327.1 | 393 | 41.65471 | 9.40 | 5 | 6 | 46.36 | 65.29 | -0.437 | 细胞核 |
| Cell nucleus | ||||||||||
| CsPPD1 | TEA005947.1 | 1 820 | 204.7527 | 6.14 | 16 | 17 | 41.51 | 92.61 | -0.120 | 胞间连丝 |
| Plasmodesma | ||||||||||
| CsPPD2 | TEA011549.1 | 1 114 | 125.3216 | 8.23 | 12 | 13 | 50.60 | 88.93 | -0.299 | 细胞核 |
| Cell nucleus | ||||||||||
| CsJAZ1 | TEA027049.1 | 212 | 23.87791 | 9.15 | 5 | 6 | 55.42 | 67.08 | -0.632 | 细胞核 |
| Cell nucleus | ||||||||||
| CsJAZ2 | TEA030826.1 | 148 | 16.39844 | 7.84 | 0 | 1 | 48.12 | 58.65 | -0.564 | 细胞核 |
| Cell nucleus | ||||||||||
| CsJAZ3 | TEA001501.1 | 261 | 29.26615 | 9.17 | 4 | 5 | 57.65 | 70.69 | -0.666 | 细胞核 |
| Cell nucleus | ||||||||||
| CsJAZ4 | TEA001681.1 | 410 | 42.84663 | 9.34 | 7 | 8 | 35.36 | 66.44 | -0.310 | 细胞核 |
| Cell nucleus | ||||||||||
| CsJAZ5 | TEA030190.1 | 294 | 31.53145 | 9.46 | 4 | 5 | 55.85 | 70.37 | -0.395 | 细胞核 |
| Cell nucleus | ||||||||||
| CsJAZ6 | TEA001821.1 | 436 | 46.20707 | 9.58 | 7 | 8 | 48.77 | 71.40 | -0.343 | 细胞质 Cytoplasm |
| CsJAZ7 | TEA032228.1 | 220 | 24.45353 | 9.18 | 5 | 6 | 55.48 | 67.36 | -0.557 | 细胞核 |
| Cell nucleus | ||||||||||
| CsJAZ8 | TEA004474.1 | 136 | 15.60498 | 9.93 | 2 | 3 | 84.54 | 75.29 | -0.721 | 细胞核 |
| Cell nucleus | ||||||||||
| CsZML1 | TEA033832.1 | 385 | 42.00050 | 4.88 | 10 | 11 | 42.88 | 70.08 | -0.625 | 细胞核 |
| Cell nucleus | ||||||||||
| CsZML2 | TEA033836.1 | 271 | 30.51872 | 4.57 | 4 | 5 | 46.48 | 78.04 | -0.601 | 细胞核 |
| Cell nucleus | ||||||||||
| CsZML3 | TEA013465.1 | 303 | 33.49678 | 6.19 | 6 | 7 | 31.87 | 60.79 | -0.916 | 细胞核 |
| Cell nucleus | ||||||||||
| CsZML4 | TEA014550.1 | 234 | 25.20185 | 5.00 | 3 | 4 | 50.61 | 71.24 | -0.550 | 细胞核 |
| Cell nucleus | ||||||||||
| CsZML5 | TEA014541.1 | 352 | 38.21245 | 4.85 | 10 | 11 | 50.11 | 72.30 | -0.574 | 细胞核 |
| Cell nucleus | ||||||||||
| CsZML6 | TEA011504.1 | 352 | 38.21245 | 4.85 | 10 | 11 | 50.11 | 72.30 | -0.574 | 细胞核 |
| Cell nucleus | ||||||||||
| CsZML7 | TEA002032.1 | 383 | 41.64484 | 4.76 | 10 | 11 | 54.89 | 63.92 | -0.795 | 细胞核 |
| Cell nucleus | ||||||||||
| CsZML8 | TEA001414.1 | 461 | 49.38947 | 5.75 | 11 | 12 | 41.39 | 73.19 | -0.375 | 细胞核 |
| Cell nucleus |

图1 茶树(Cs)、葡萄(Vv)、拟南芥(At)、杨树(Pt)、水稻(Os)和二穗短柄草(Bd)的TIFY蛋白进化树
Fig. 1 TIFY protein phylogenetic tree of tea plant(Cs),grape(Vv),Arabidopsis(At),poplar(Pt), rice(Os)and Brachypodium distachyon(Bd)
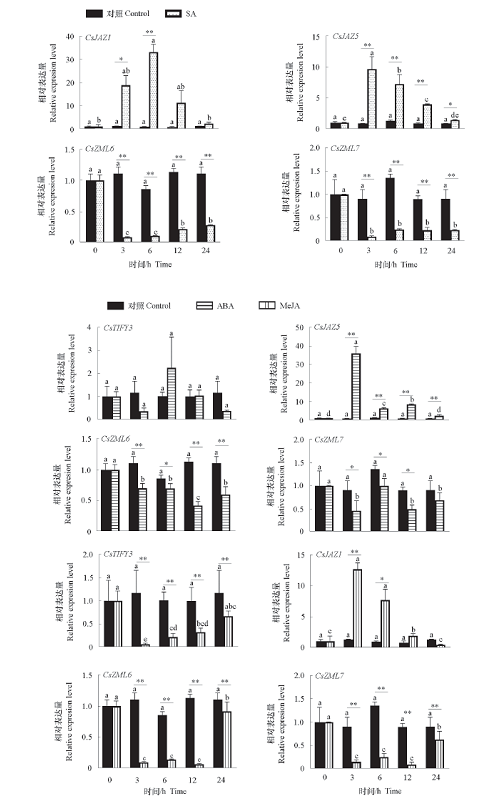
图5 外源SA、ABA、MeJA处理下部分茶树CsTIFY基因的表达 *表示同一时间点差异显著(α = 0.05),**为极显著(α = 0.01);不同小写字母表示不同时间点差异显著(P < 0.05)。下同。
Fig. 5 Expression of CsTIFY genes in tea plants under exogenous SA,ABA,MeJA stresses * indicates significant differences at the same time point(α = 0.05),** indicates extremely significant(α = 0.01);Different lowercase letters indicate significant differences at different points in time(P < 0.05). The same below.
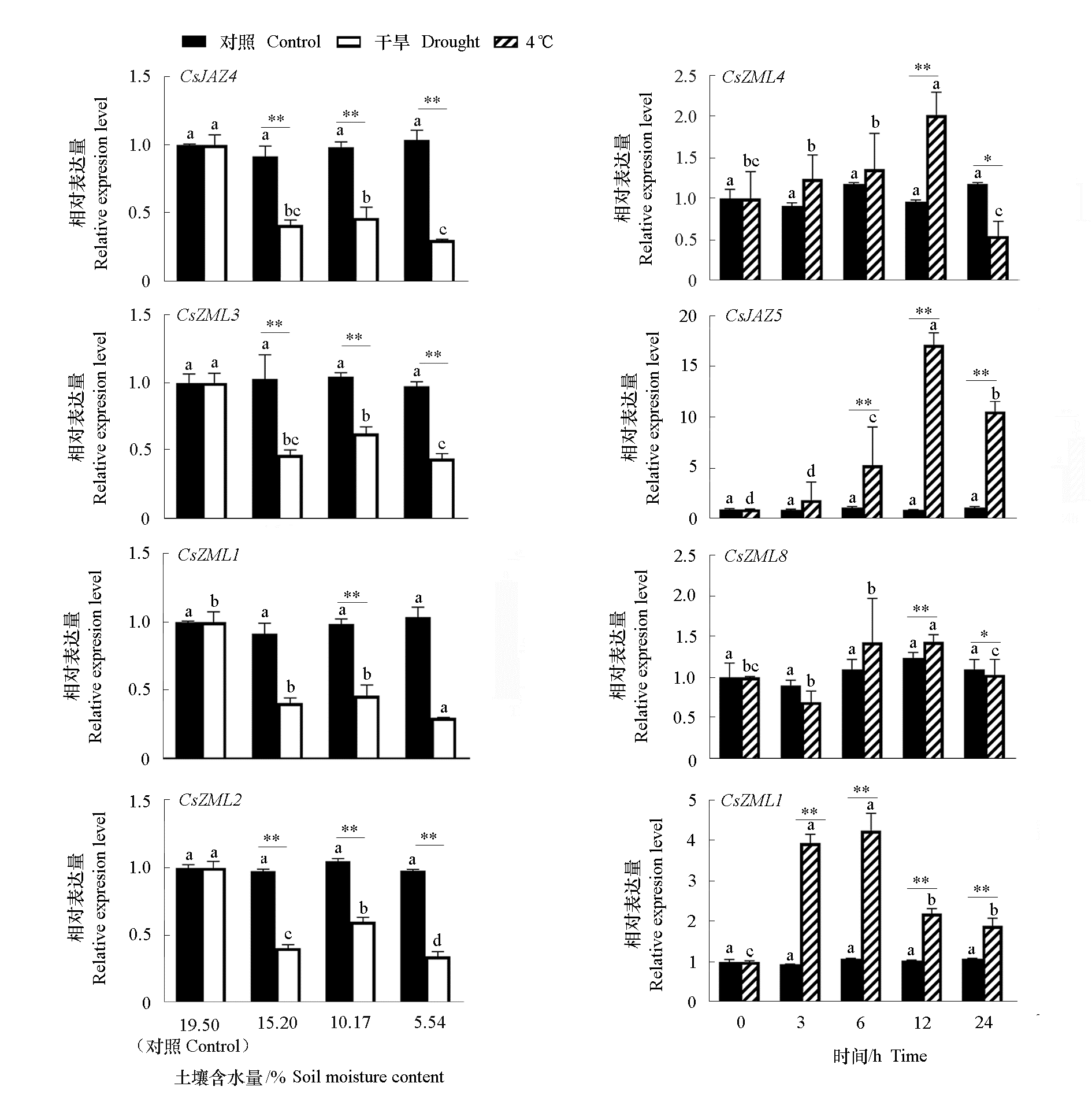
图6 干旱和4 ℃低温处理下部分茶树CsTIFY的表达
Fig. 6 CsTIFY gene expression of tea under abiotic stress drought and 4 ℃ treatments CsTIFY gene expression of tea under abiotic stress drought and 4 ℃ treatments
| [1] | Bartel V, Wim G, Alex B, akayuki K, Gheysen, Godelieve. 2007. The tify family previously known as ZIM. Trends in Plant Science, 12 (6):239-244. |
| [2] | Ding Y, Wang Y, Qiu C, Qian W, Xie H, Ding Z. 2020. Alternative splicing in tea plants was extensively triggered by drought,heat and their combined stresses. PeerJ:e8258. |
| [3] | Dong Taoxing, Cai Kunzheng, Zhang Jingxin, Rong Hui, Xie Guozheng, Zeng Rensen. 2007. The physiological roles of methyl jasmonate(MeJA) in drought resistance of rice seedlings. Ecology and Environmental Sciences,(4):1261-1265. (in Chinese) |
| 董桃杏, 蔡昆争, 张景欣, 荣辉, 谢国政, 曾任森. 2007. 茉莉酸甲酯(MeJA)对水稻幼苗的抗旱生理效应. 生态环境,(4):1261-1265. | |
| [4] |
Ebel C, BenFeki A, Hanin M, Solano R, Chini A. 2018. Characterization of wheat(Triticum aestivum)TIFY family and role of Triticum durum TdTIFY11a in salt stress tolerance. PLoS ONE, 13 (7):e0200566.
doi: 10.1371/journal.pone.0200566 URL |
| [5] |
Guo Y, Zhao S, Zhu C, Chang X, Yue C, Wang Z, Lin Y, Lai Z. 2017. Identification of drought-responsive miRNAs and physiological characterization of tea plant(Camellia sinensis L.)under drought stress. BMC Plant Biology, 17 (1):211.
doi: 10.1186/s12870-017-1172-6 URL |
| [6] |
Hause B, Wasternack C. 2013. Jasmonates:biosynthesis,perception,signal transduction and action in plant stress response,growth and development. An update to the 2007 review in Annals of Botany. Annals of Botany,(6):1021-1058.
doi: 10.1093/aob/mct067 pmid: 23558912 |
| [7] | Hong G, Xue X, Mao Y. 2012. Arabidopsis MYC 2 interacts with DELLA proteins in regulating sesquiterpene synthase gene expression. Plant Cell, (6):2635-2648. |
| [8] | Huang Ying. 2017. Analysis of TIFY gene family and its interaction protein screening in Salvia miltiorrhiza[Ph. D. Dissertation]. Xi'an: Shaanxi Normal University. (in Chinese) |
| 黄英. 2017. 丹参TIFY基因家族分析及其互作蛋白筛选[博士论文]. 西安: 陕西师范大学. | |
| [9] | Huang Z, Jin S, Guo H, Zhong X, He J, Li X, Jiang M, Yu X, Long H, Ma M, Chen Q. 2016. Genome-wide identification and characterization of TIFY family genes in Moso bamboo(Phyllostachys edulis)and expression profiling analysis under dehydration and cold stresses. PeerJ:4. |
| [10] |
Li N, Yue C, Cao H, Qian W, Hao X, Wang Y, Wang L, Ding C, Wang X, Yang Y. 2018. Transcriptome sequencing dissection of the mechanisms underlying differential cold sensitivity in young and mature leaves of the tea plan(Camellia sinensis). Journal of Plant Physiology, 224-225:144-155.
doi: 10.1016/j.jplph.2018.03.017 URL |
| [11] | Li Shuyu, Li Chuanyou. 2016. Developmental plasticity of plant roots. China Basic Science, 18 (2):14-21. (in Chinese) |
| 李淑钰, 李传友. 2016. 植物根系可塑性发育的研究进展与展望. 中国基础科学, 18 (2):14-21. | |
| [12] | Li X, Yin X, Wang H, Li J, Guo C, Gao H, Zheng Y, Fan C, Wang X. 2015. Genome-wide identification and analysis of the apple(Malus × domestica Borkh.)TIFY gene family. Tree Genetics & Genomes,(1):1-13. |
| [13] | Mei Chuang, Zhang Xiaoyan, Yan Peng, Aisajan Mamat, Feng Beibei, Ma Kai, Han Liqun, Dong Lianxin, Wang Jixun. 2021. Identification of TIFY family in apple and their expression analysis under insect stress. Acta Horticulturae Sinica, 48 (2):233-242. (in Chinese) |
| 梅闯, 张小燕, 闫鹏, 艾沙江 · 买买提, 冯贝贝, 马凯, 韩立群, 董连新, 王继勋. 2021. 苹果TIFY 家族基因鉴定及其在虫害胁迫下的表达分析. 园艺学报, 48 (2):233-242. | |
| [14] |
Meng L, Zhang T, Geng S, Scott P B, Li H, Chen S. 2019. Comparative proteomics and metabolomics of JAZ7-mediated drought tolerance in Arabidopsis. Journal of Proteomics: 196:81-91.
doi: S1874-3919(19)30036-3 pmid: 30731210 |
| [15] | Nishii A, Takemura M, Fujita H, Shikata M, Yokota A, Kohchi T. 2000. Characterization of a novel gene encoding a putative single Zinc-finger protein,ZIM,expressed during the reproductive phase in Arabidopsis thaliana. Bioscience,Biotechnology,and Biochemistry,(7):1402-1409. |
| [16] | Qing Z, Muchen C, Xiaomin Y, Lishan W, Chunfang G, Ray M, Jisen Z. 2017. Transcriptome dynamics of Camellia sinensis in response to continuous salinity and drought stress. Tree Genetics & Genomes,(4):1-17. |
| [17] |
Sarfraz H S, Akhtar K M, Muhammad A. 2011. Transcription factors as tools to engineer enhanced drought stress tolerance in plants. Biotechnology Progress, 27 (2):297-306.
doi: 10.1002/btpr.514 pmid: 21302367 |
| [18] |
Shikata M, Matsuda Y, Ando K, Nishii A, Takemura M, Yokota A, Kohchi T. 2004. Characterization of Arabidopsis ZIM,a member of a novel plant-specific GATA factor gene family. Journal of Experimental Botany, 55 (397):631-639.
doi: 10.1093/jxb/erh078 URL |
| [19] | Shuichi Y. 1998. Transcription factors in plants:physiological functions and regulation of expression. Journal of Plant Research,(1103):363-371. |
| [20] | Sirhindi G A, Sharma P A, Arya P B C, Goel P B C, Kumar G B C, Acharya V B C, Singh A K B C. 2016. Genome-wide characterization and expression profiling of TIFY gene family in pigeonpea[Cajanus cajan(L.)Mill sp.] under copper stress(Article). Journal of Plant Biochemistry and Biotechnology,(3):301-310. |
| [21] | Song Yun, Li Linxuan, Zhuo Fengping, Zhang Xueyan, Ren Maozhi, Li Fuguang. 2015. Progress on jasmonic acid signaling in plant stress resistant. Journal of Agricultural Science and Technology, 17 (2):17-24. (in Chinese) |
| 宋云, 李林宣, 卓凤萍, 张雪妍, 任茂智, 李付广. 2015. 茉莉酸信号传导在植物抗逆性方面研究进展. 中国农业科技导报, 17 (2): 17-24. | |
| [22] | Su Wenbing, Jiang Yuanyuan, Bai Yunlu, Gan Xiaoqing, Liu Yuexue, Lin Shunquan. 2019. Journal of Agricultural Biotechnology, 27 (5):919-926. (in Chinese) |
| 苏文炳, 蒋园园, 白昀鹭, 甘小清, 刘月学, 林顺权. 2019. 转录因子调控植物萜类化合物生物合成研究进展. 农业生物技术学报, 27 (5): 919-926. | |
| [23] | Sun Kaiwen. 2019. Regulation of the plant hormone abscisic acid on the defense response mediated by jasmonic acid[Ph. D. Dissertation]. Nanjing: Nanjing Normal University. (in Chinese) |
| 孙凯文. 2019. 植物激素脱落酸对茉莉酸介导的防御反应的调控[博士论文]. 南京: 南京师范大学. | |
| [24] |
Vanholme B, Grunewald W, Bateman A, Kohchi T, Gheysen G. 2007. The TIFY family previously known as ZIM. Trends in Plant Science, 12 (6):239-244..
pmid: 17499004 |
| [25] | Wang Linhua, Liang Shurong, Lü Shumin, Zhao Huijie, Qu Xiaofei, Liu Weiwei. 2010. Protective effect of SA on photosynthetic apparatus of wheat leaves under heat and high irradiance stress. Journal of Henan Agricultural Sciences,(4):21-25. (in Chinese) |
| 王林华, 梁书荣, 吕淑敏, 赵会杰, 曲小菲, 刘魏魏. 2010. 水杨酸对高温强光胁迫下小麦叶片光合机构的保护作用. 河南农业科学,(4):21-25. | |
| [26] | Wang Niyan, Jiang De’an. 2002. Jasmonic acid and its methyl esters and plant induced disease resistance. Plant Physiology Journal,(3):279-284. (in Chinese) |
| 王妮妍, 蒋德安. 2002. 茉莉酸及其甲酯与植物诱导抗病性. 植物生理学通讯,(3):279-284. | |
| [27] |
Wang Y, Pan F, Chen D, Chu W, Liu H, Xiang Y. 2017. Genome-wide identification and analysis of the Populus trichocarpa TIFY gene family. Plant Physiology and Biochemistry, 115:360-371.
doi: S0981-9428(17)30135-3 pmid: 28431355 |
| [28] | White D W R. 2006. PEAPOD regulates lamina size and curvature in Arabidopsis. Proceedings of the National Academy of Sciences of the United States of America, 103 (35):13238-13243. |
| [29] | Wu Yanyan. 2019. Effects of exogenous ABA on physiological characteristics of Machilus machinei seedlings under drought stress[Ph. D. Dissertation]. Chongqing: Southwest University. (in Chinese) |
| 吴琰琰. 2019. 外源ABA对干旱胁迫下桢楠幼苗生理特性的影响[博士论文]. 重庆: 西南大学. | |
| [30] | Xia E H, Li F D, Tong W, Li P H, Wu Q, Zhao H J, Ge R H, Li R P, Li Y Y, Zhang Z Z, Wei C L, Wan X C. 2019. Tea plant information archive:a comprehensive genomics and bioinformatics platform for tea plant. Plant Biotechnology Journal,(10):1938-1953. |
| [31] | Xu G, Guo C, Shan H. 2012. Divergence of duplicate genes in exon-intron structure. Proceedings of The National Academy of Sciences of The United States of America,(4):1187-1192. |
| [32] |
Yang Ruijia, Zhang Zhongbao, Wu Zhongyi. 2020. Progress of the structural and functional analysis of plant transcription factor TIFY protein family. Biotechnology Bulletin, 36 (12):121-128. (in Chinese)
doi: 10.13560/j.cnki.biotech.bull.1985.2020-0307 |
|
杨锐佳, 张中保, 吴忠义. 2020. 植物转录因子TIFY家族蛋白结构和功能的研究进展. 生物技术通报, 36 (12):121-128.
doi: 10.13560/j.cnki.biotech.bull.1985.2020-0307 |
|
| [33] | Yang Y, Ahammed G J, Wan C, Liu H, Chen R, Zhou Y. 2019. Comprehensive analysis of TIFY transcription factors and their expression profiles under jasmonic acid and abiotic stresses in watermelon. International Journal of Genomics, 10.1155/2019/6813086. |
| [34] | Ye H, Du H, Tang N, Li X, Xiong L. 2009. Identification and expression profiling analysis of TIFY family genes involved in stress and phytohormone responses in rice. Plant Molecular Biology,(3):291-305. |
| [35] | Yin Chao, Li Mei, Liu Yuhui. 2013. The role of the nucleus in plant defense response. Current Biotechnology, 3 (1):12-17. (in Chinese) |
| 尹超, 李梅, 刘昱辉. 2013. 细胞核在植物防御反应中的作用. 生物技术进展, 3 (1):12-17. | |
| [36] |
Yu Hong, Wang Xianyun, Yang Xiaohui, Li Haitao, Xiao Shuguang, Du Huatang, Hou Zhenghui. 2009. Relationship between microclimatic factors and yield of spring tea Camellia sinensis(L.)O. Kuntze in Henan Jigongshan Nature Reserve. Chinese Journal of Eco-Agriculture, 17 (1):94-99. (in Chinese)
doi: 10.3724/SP.J.1011.2009.00094 URL |
| 喻泓, 王贤赟, 杨晓晖, 李海涛, 肖曙光, 杜化堂, 侯正辉. 2009. 河南鸡公山茶园春茶产量与小气候关系研究. 中国生态农业学报, 17 (1):94-99. | |
| [37] | Zhang L, You J, Chan Z. 2015. Identification and characterization of TIFY family genes in Brachypodium distachyon. Journal of Plant Research,(6):995-1005. |
| [38] | Zhang Y, Gao M, Singer S D, Fei Z, Wang H, Wang X. 2012. Genome-wide identification and analysis of the TIFY gene family in grape. PLoS ONE,(9):1-13. |
| [39] | Zhao Xiaoxiao, Xie Kunliang, Zhang Shumeng, Zhang Chao, Xi Yajun, Sun Fengli. 2019. Identification and analysis of TIFY gene family in Switchgrass. Acta Agrestia Sinica, 27 (5):1126-1137. (in Chinese) |
| 赵晓晓, 谢坤良, 张舒梦, 张超, 奚亚军, 孙风丽. 2019. 柳枝稷TIFY基因家族的鉴定与分析. 草地学报, 27 (5):1126-1137. | |
| [40] | Zheng Y, Chen X, Wang P, Sun Y, Yue C, Ye N. 2020. Genome-wide and expression pattern analysis of JAZ family involved in stress responses and postharvest processing treatments in Camellia sinensis. Scientific Reports,(1):2792. |
| [41] |
Zhou C, Zhu C, Fu H, Li X, Chen L, Lin Y, Lai Z, Guo Y. 2019. Genome-wide investigation of superoxide dismutase(SOD)gene family and their regulatory miRNAs reveal the involvement in abiotic stress and hormone response in tea plant(Camellia sinensis). PLoS ONE,doi: 10.1371/journal.pone.0223609.
doi: 10.1371/journal.pone.0223609 |
| [42] |
Zhu D, Bai X, Chen C, Chen Q, Cai H, Li Y, Ji W, Zhai H, Lv D, Luo X, Zhu Y. 2011. GsTIFY10,a novel positive regulator of plant tolerance to bicarbonate stress and a repressor of jasmonate signaling. Plant Molecular Biology, 77 (3):285-297.
doi: 10.1007/s11103-011-9810-0 URL |
| [1] | 张 欣, 漆艳香, 曾凡云, 王艳玮, 谢培兰, 谢艺贤, 彭 军. 香蕉枯萎病菌Dicer-like基因的功能分析[J]. 园艺学报, 2023, 50(2): 279-294. |
| [2] | 程庆华, 张志鹏, 吴艳萍, 万宇鹤, 陈应娟. 苦参碱对茶树炭疽菌的抑菌作用研究[J]. 园艺学报, 2023, 50(2): 432-440. |
| [3] | 翟含含, 翟宇杰, 田义, 张叶, 杨丽, 温陟良, 陈海江. 桃SAUR家族基因分析及PpSAUR5功能鉴定[J]. 园艺学报, 2023, 50(1): 1-14. |
| [4] | 袁馨, 徐云鹤, 张雨培, 单楠, 陈楚英, 万春鹏, 开文斌, 翟夏琬, 陈金印, 甘增宇. 猕猴桃后熟过程中ABA响应结合因子AcAREB1调控AcGH3.1的表达[J]. 园艺学报, 2023, 50(1): 53-64. |
| [5] | 岳翠男, 王治会, 杨普香, 李文金, 彭 华, 陈罗军, 周汉中. 茶树新品种‘宁州早1号’[J]. 园艺学报, 2022, 49(S2): 281-282. |
| [6] | 李兰英, 胥亚琼, 刘东娜, 尧 渝, 龚雪蛟, 罗 晟, 罗 凡, . 茶树新品种‘金凤2号’[J]. 园艺学报, 2022, 49(S2): 283-284. |
| [7] | 李兰英, 王 强, 龚雪蛟, 刘东娜, 尧 渝, 王迎春, 胥亚琼, 罗 凡, . 茶树新品种‘甘露1号’[J]. 园艺学报, 2022, 49(S2): 285-286. |
| [8] | 任志红, 吴焕焕, 肖文敏, 张 虹, 杨圣祥, 孙海伟, 王 健, 高文星. 抗寒茶树新品种‘岱鼎御丰’[J]. 园艺学报, 2022, 49(S2): 287-288. |
| [9] | 王治会, 杨普香, 彭 华, 李文金, 王胜利, 鲍润元, 江新凤. 茶树新品种‘浮梁槠叶1号’[J]. 园艺学报, 2022, 49(S1): 191-192. |
| [10] | 彭 华, 李文金, 杨普香, 王治会, 岳翠男, 李延升, 谢小群, 李 琛. 早生持嫩性强茶树新品种‘婺绿1号’[J]. 园艺学报, 2022, 49(S1): 193-194. |
| [11] | 王治会, 彭 华, 岳翠男, 李文金, 杨普香, 陈年生, 李延升, 蔡海兰, 江新凤. 茶树新品种‘赣茶4号’[J]. 园艺学报, 2022, 49(S1): 195-196. |
| [12] | 马伟伟, 王 云, 李春华, 刘 晓, 唐晓波, 张 厅, 甘 勇, 王小萍, . 茶树新品种‘彝黄1号’[J]. 园艺学报, 2022, 49(S1): 197-198. |
| [13] | 马伟伟, 王 云, 李春华, 熊元元, 刘 晓, 王小萍, 李 鑫. 茶树新品种‘天府5号’[J]. 园艺学报, 2022, 49(S1): 199-200. |
| [14] | 徐小萍, 曹清影, 蔡柔荻, 官庆栩, 张梓浩, 陈裕坤, 徐涵, 林玉玲, 赖钟雄. 龙眼miR408与DlLAC12克隆及其在球形胚发生和非生物胁迫下的表达分析[J]. 园艺学报, 2022, 49(9): 1866-1882. |
| [15] | 王沙, 张心慧, 赵玉洁, 李变变, 招雪晴, 沈雨, 董建梅, 苑兆和. 石榴花青苷合成相关基因PgMYB111的克隆与功能分析[J]. 园艺学报, 2022, 49(9): 1883-1894. |
| 阅读次数 | ||||||
|
全文 |
|
|||||
|
摘要 |
|
|||||
版权所有 © 2012 《园艺学报》编辑部 京ICP备10030308号-2 国际联网备案号 11010802023439
编辑部地址: 北京市海淀区中关村南大街12号中国农业科学院蔬菜花卉研究所 邮编: 100081
电话: 010-82109523 E-Mail: yuanyixuebao@126.com
技术支持:北京玛格泰克科技发展有限公司