
园艺学报 ›› 2022, Vol. 49 ›› Issue (11): 2489-2501.doi: 10.16420/j.issn.0513-353x.2021-0763
侯天泽1, 易双双2,3, 张志群2,3, 王健1,*( ), 李崇晖2,3,*
), 李崇晖2,3,*
收稿日期:2021-10-15
修回日期:2022-04-30
出版日期:2022-11-25
发布日期:2022-11-25
通讯作者:
王健,李崇晖
E-mail:blchh@sina.com
基金资助:
HOU Tianze1, YI Shuangshuang2,3, ZHANG Zhiqun2,3, WANG Jian1,*( ), LI Chonghui2,3,*
), LI Chonghui2,3,*
Received:2021-10-15
Revised:2022-04-30
Online:2022-11-25
Published:2022-11-25
Contact:
WANG Jian,LI Chonghui
E-mail:blchh@sina.com
摘要:
为了筛选出秋石斛中稳定表达的内参基因用于实时荧光定量PCR(RT-qPCR)分析,以不同品种和不同发育阶段花芽(包括花瓣、唇瓣和萼片)为材料,使用geNorm、NormFinder 和BestKeeper软件计算了常用的7个候选内参基因表达的稳定性,使用RefFinder对其稳定性进行综合评价,并对筛选出的内参基因进行了验证。结果表明,各内参基因在秋石斛花芽中的表达稳定性差异较大,在不同品种同一发育阶段的花芽中β-actin、TUA和GADPH表达最稳定;在花芽发育过程中β-actin、TUA和CYP表达最稳定;而对于花器官不同组织,β-actin、TUA和GADPH在萼片中,β-actin、CYP和PGK在花瓣中,CYP、TUA和PGK在唇瓣中表达最稳定。
中图分类号:
侯天泽, 易双双, 张志群, 王健, 李崇晖. 秋石斛RT-qPCR内参基因的筛选与验证[J]. 园艺学报, 2022, 49(11): 2489-2501.
HOU Tianze, YI Shuangshuang, ZHANG Zhiqun, WANG Jian, LI Chonghui. Selection and Validation of Reference Genes for RT-qPCR in Phalaenopsis- type Dendrobium Hybrid[J]. Acta Horticulturae Sinica, 2022, 49(11): 2489-2501.
| 基因 Gene | 引物序列(5′-3′) Primer sequences | 产物长 度/bp Product length | Tm/℃ (F/R) | E/% Amplifcation efficiency | r2 Correlation coefficient |
|---|---|---|---|---|---|
| PGK | F:ATCGGTGAGGAAGTTGAGAAAAC;R:GCCAATTTCTTAGCAAACTCTGG | 165 | 60.6/60.5 | 100 | 0.997 |
| TUA | F:CAAAGAAGATGCAGCCAACAAC;R:AAGACCAGTGCAGTTGTCAGCTA | 111 | 60.1/59.4 | 104 | 0.992 |
| TUB | F:TCACGGTGAGACGGACCTG;R:TATCCGGGCGAAAAATCTGA | 157 | 59.4/60.6 | 104 | 0.998 |
| EF1α | F:GATGGATGCGACCACACCC;R:TCGAGAAGAGTTGGTCCCTTG | 191 | 61.1/59.2 | 105 | 0.996 |
| CYP | F:TCTACGCCGACACGACTCCT;R:GGTGAAAGGTAGAGCCCTTGAA | 113 | 60.4/60.3 | 102 | 0.998 |
| GADPH | F:AGCTGCACAACCAACTGTTTG;R:GCTCTTCCACCCCTCCAGTC | 151 | 58.7/60.7 | 100 | 0.993 |
| β-actin | F:GTCAGGGACATCAAGGAGAAG;R:TGGGCACCTAAATCTCTCAGC | 153 | 60.5/60.6 | 95 | 0.993 |
表1 候选内参基因描述以及用于RT-qPCR引物序列
Table 1 Description of candidate reference genes and a list of primer sequences for RT-qPCR
| 基因 Gene | 引物序列(5′-3′) Primer sequences | 产物长 度/bp Product length | Tm/℃ (F/R) | E/% Amplifcation efficiency | r2 Correlation coefficient |
|---|---|---|---|---|---|
| PGK | F:ATCGGTGAGGAAGTTGAGAAAAC;R:GCCAATTTCTTAGCAAACTCTGG | 165 | 60.6/60.5 | 100 | 0.997 |
| TUA | F:CAAAGAAGATGCAGCCAACAAC;R:AAGACCAGTGCAGTTGTCAGCTA | 111 | 60.1/59.4 | 104 | 0.992 |
| TUB | F:TCACGGTGAGACGGACCTG;R:TATCCGGGCGAAAAATCTGA | 157 | 59.4/60.6 | 104 | 0.998 |
| EF1α | F:GATGGATGCGACCACACCC;R:TCGAGAAGAGTTGGTCCCTTG | 191 | 61.1/59.2 | 105 | 0.996 |
| CYP | F:TCTACGCCGACACGACTCCT;R:GGTGAAAGGTAGAGCCCTTGAA | 113 | 60.4/60.3 | 102 | 0.998 |
| GADPH | F:AGCTGCACAACCAACTGTTTG;R:GCTCTTCCACCCCTCCAGTC | 151 | 58.7/60.7 | 100 | 0.993 |
| β-actin | F:GTCAGGGACATCAAGGAGAAG;R:TGGGCACCTAAATCTCTCAGC | 153 | 60.5/60.6 | 95 | 0.993 |
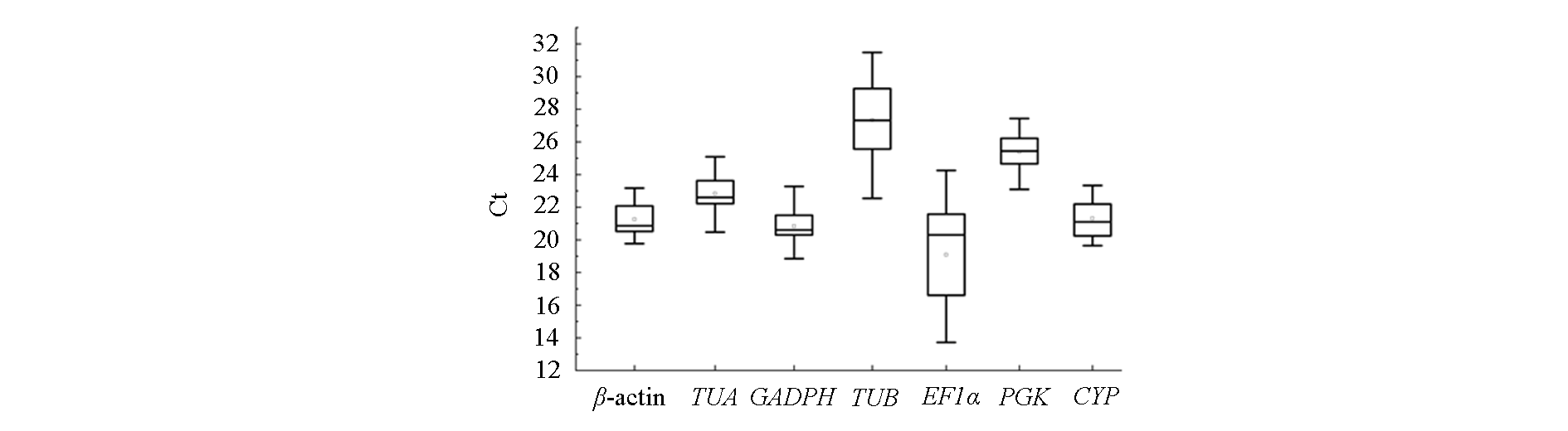
图3 本研究所用秋石斛试材中7个候选内参基因的RT-qPCR Ct值 箱线图表示第25和第75个百分位;框上的线表示中位数;中间的圆圈显示平均值。
Fig. 3 The RT-qPCR Ct values of seven candidate reference genes across Phalaenopsis-type Dendrobium hybrid test material used in this study Box graph indicates the 25th and 75th percentiles;the line across the box depicts the median;middle circle show the mean values.
| 排名 Rank | 萼片 Sepal | 花瓣 Petal | 唇瓣 Labellum | 不同品种花芽 Floral bud of different cultivars | 发育不同阶段花芽 Floral bud of different developmental stages | ||||||
|---|---|---|---|---|---|---|---|---|---|---|---|
| 内参基因 Reference gene | M值 M | 内参基因 Reference gene | M值 M | 内参基因 Reference gene | M值 M | 内参基因 Reference gene | M值 M | 内参基因 Reference gene | M值 M | ||
| 1 | TUA | 0.840 | β-actin | 0.759 | CYP | 0.499 | β-actin | 0.857 | β-actin | 1.334 | |
| 2 | GADPH | 0.840 | CYP | 0.759 | GADPH | 0.499 | TUA | 0.857 | TUA | 1.334 | |
| 3 | β-actin | 0.954 | TUA | 0.862 | β-actin | 0.587 | GADPH | 0.879 | CYP | 1.434 | |
| 4 | PGK | 1.225 | GADPH | 0.912 | TUA | 0.628 | CYP | 1.001 | GADPH | 1.461 | |
| 5 | CYP | 1.478 | PGK | 0.954 | PGK | 0.795 | PGK | 1.125 | PGK | 1.735 | |
| 6 | TUB | 1.813 | TUB | 1.180 | TUB | 1.142 | TUB | 1.441 | TUB | 2.040 | |
| 7 | EF1α | 2.254 | EF1α | 1.788 | EF1α | 1.699 | EF1α | 1.977 | EF1α | 2.234 | |
表2 根据geNorm 7个候选内参基因的排名
Table 2 Ranking of seven candidate reference genes according to geNorm
| 排名 Rank | 萼片 Sepal | 花瓣 Petal | 唇瓣 Labellum | 不同品种花芽 Floral bud of different cultivars | 发育不同阶段花芽 Floral bud of different developmental stages | ||||||
|---|---|---|---|---|---|---|---|---|---|---|---|
| 内参基因 Reference gene | M值 M | 内参基因 Reference gene | M值 M | 内参基因 Reference gene | M值 M | 内参基因 Reference gene | M值 M | 内参基因 Reference gene | M值 M | ||
| 1 | TUA | 0.840 | β-actin | 0.759 | CYP | 0.499 | β-actin | 0.857 | β-actin | 1.334 | |
| 2 | GADPH | 0.840 | CYP | 0.759 | GADPH | 0.499 | TUA | 0.857 | TUA | 1.334 | |
| 3 | β-actin | 0.954 | TUA | 0.862 | β-actin | 0.587 | GADPH | 0.879 | CYP | 1.434 | |
| 4 | PGK | 1.225 | GADPH | 0.912 | TUA | 0.628 | CYP | 1.001 | GADPH | 1.461 | |
| 5 | CYP | 1.478 | PGK | 0.954 | PGK | 0.795 | PGK | 1.125 | PGK | 1.735 | |
| 6 | TUB | 1.813 | TUB | 1.180 | TUB | 1.142 | TUB | 1.441 | TUB | 2.040 | |
| 7 | EF1α | 2.254 | EF1α | 1.788 | EF1α | 1.699 | EF1α | 1.977 | EF1α | 2.234 | |
| 排名 Rank | 萼片 Sepal | 花瓣 Petal | 唇瓣 Labellum | 不同品种花芽 Floral bud of different cultivars | 不同发育阶段花芽 Floral bud of different developmental stages | |||||
|---|---|---|---|---|---|---|---|---|---|---|
| 内参基因 Reference gene | 稳定值 Stability value | 内参基因 Reference gene | 稳定值 Stability value | 内参基因 Reference gene | 稳定值 Stability value | 内参基因 Reference gene | 稳定值 Stability value | 内参基因 Reference gene | 稳定值 Stability value | |
| 1 | β-actin | 0.022 | β-actin | 0.017 | β-actin | 0.021 | β-actin | 0.019 | β-actin | 0.013 |
| 2 | TUA | 0.022 | CYP | 0.032 | CYP | 0.022 | TUA | 0.032 | TUA | 0.034 |
| 3 | PGK | 0.042 | TUB | 0.032 | TUA | 0.030 | PGK | 0.038 | PGK | 0.058 |
| 4 | GADPH | 0.047 | PGK | 0.032 | GADPH | 0.036 | GADPH | 0.045 | CYP | 0.063 |
| 5 | TUB | 0.066 | TUA | 0.035 | TUB | 0.042 | TUB | 0.048 | GADPH | 0.074 |
| 6 | CYP | 0.100 | GADPH | 0.052 | PGK | 0.045 | CYP | 0.065 | TUB | 0.092 |
| 7 | EF1α | 0.179 | EF1α | 0.163 | EF1α | 0.163 | EF1α | 0.173 | EF1α | 0.121 |
表3 根据NormFinder 7个候选内参基因的排名
Table 3 Ranking of seven candidate reference genes according to NormFinder
| 排名 Rank | 萼片 Sepal | 花瓣 Petal | 唇瓣 Labellum | 不同品种花芽 Floral bud of different cultivars | 不同发育阶段花芽 Floral bud of different developmental stages | |||||
|---|---|---|---|---|---|---|---|---|---|---|
| 内参基因 Reference gene | 稳定值 Stability value | 内参基因 Reference gene | 稳定值 Stability value | 内参基因 Reference gene | 稳定值 Stability value | 内参基因 Reference gene | 稳定值 Stability value | 内参基因 Reference gene | 稳定值 Stability value | |
| 1 | β-actin | 0.022 | β-actin | 0.017 | β-actin | 0.021 | β-actin | 0.019 | β-actin | 0.013 |
| 2 | TUA | 0.022 | CYP | 0.032 | CYP | 0.022 | TUA | 0.032 | TUA | 0.034 |
| 3 | PGK | 0.042 | TUB | 0.032 | TUA | 0.030 | PGK | 0.038 | PGK | 0.058 |
| 4 | GADPH | 0.047 | PGK | 0.032 | GADPH | 0.036 | GADPH | 0.045 | CYP | 0.063 |
| 5 | TUB | 0.066 | TUA | 0.035 | TUB | 0.042 | TUB | 0.048 | GADPH | 0.074 |
| 6 | CYP | 0.100 | GADPH | 0.052 | PGK | 0.045 | CYP | 0.065 | TUB | 0.092 |
| 7 | EF1α | 0.179 | EF1α | 0.163 | EF1α | 0.163 | EF1α | 0.173 | EF1α | 0.121 |
| 排名 | 萼片Sepal | 花瓣Petal | 唇瓣Labellum | ||||||
|---|---|---|---|---|---|---|---|---|---|
| Rank | 基因 | 标准差 | 变异系数/% | 基因 | 标准差 | 变异系数/% | 基因 | 标准差 | 变异系数/% |
| Gene | SD | CV | Gene | SD | CV | Gene | SD | CV | |
| 1 | β-actin | 0.45 | 2.17 | PGK | 0.99 | 3.88 | PGK | 0.63 | 2.41 |
| 2 | GADPH | 0.6 | 2.95 | CYP | 0.99 | 4.65 | GADPH | 0.73 | 3.44 |
| 3 | TUA | 0.79 | 3.48 | β-actin | 1.01 | 4.67 | CYP | 0.89 | 4.2 |
| 4 | PGK | 0.9 | 3.61 | TUA | 1.04 | 4.56 | TUA | 1.09 | 4.76 |
| 5 | CYP | 1.33 | 6.2 | GADPH | 1.31 | 6.36 | β-actin | 1.16 | 5.43 |
| 6 | TUB | 1.87 | 7.11 | TUB | 1.76 | 6.27 | EF1α | 1.83 | 8.86 |
| 7 | EF1α | 2.3 | 13.03 | EF1α | 1.88 | 9.33 | TUB | 2.23 | 7.98 |
| 排名 | 不同品种花芽 | 不同发育阶段花芽 | |||||||
| Rank | Floral bud of different cultivars | Floral bud of different developmental stages | |||||||
| 基因 | 标准差 | 变异系数/% | 基因 | 标准差 | 变异系数/% | ||||
| Gene | SD | CV | Gene | SD | CV | ||||
| 1 | β-actin | 0.89 | 4.2 | β-actin | 0.85 | 4.02 | |||
| 2 | TUA | 0.91 | 3.97 | PGK | 0.91 | 3.64 | |||
| 3 | GADPH | 0.92 | 4.43 | GADPH | 0.99 | 4.92 | |||
| 4 | PGK | 0.95 | 3.73 | TUA | 1.04 | 4.59 | |||
| 5 | CYP | 1 | 4.7 | CYP | 1.14 | 5.44 | |||
| 6 | TUB | 1.95 | 7.15 | TUB | 1.52 | 5.31 | |||
| 7 | EF1α | 2.71 | 14.19 | EF1α | 1.77 | 8.39 | |||
表4 根据BestKeeper 7个候选内参基因的排名
Table 4 Ranking of seven candidate reference genes according to BestKeeper
| 排名 | 萼片Sepal | 花瓣Petal | 唇瓣Labellum | ||||||
|---|---|---|---|---|---|---|---|---|---|
| Rank | 基因 | 标准差 | 变异系数/% | 基因 | 标准差 | 变异系数/% | 基因 | 标准差 | 变异系数/% |
| Gene | SD | CV | Gene | SD | CV | Gene | SD | CV | |
| 1 | β-actin | 0.45 | 2.17 | PGK | 0.99 | 3.88 | PGK | 0.63 | 2.41 |
| 2 | GADPH | 0.6 | 2.95 | CYP | 0.99 | 4.65 | GADPH | 0.73 | 3.44 |
| 3 | TUA | 0.79 | 3.48 | β-actin | 1.01 | 4.67 | CYP | 0.89 | 4.2 |
| 4 | PGK | 0.9 | 3.61 | TUA | 1.04 | 4.56 | TUA | 1.09 | 4.76 |
| 5 | CYP | 1.33 | 6.2 | GADPH | 1.31 | 6.36 | β-actin | 1.16 | 5.43 |
| 6 | TUB | 1.87 | 7.11 | TUB | 1.76 | 6.27 | EF1α | 1.83 | 8.86 |
| 7 | EF1α | 2.3 | 13.03 | EF1α | 1.88 | 9.33 | TUB | 2.23 | 7.98 |
| 排名 | 不同品种花芽 | 不同发育阶段花芽 | |||||||
| Rank | Floral bud of different cultivars | Floral bud of different developmental stages | |||||||
| 基因 | 标准差 | 变异系数/% | 基因 | 标准差 | 变异系数/% | ||||
| Gene | SD | CV | Gene | SD | CV | ||||
| 1 | β-actin | 0.89 | 4.2 | β-actin | 0.85 | 4.02 | |||
| 2 | TUA | 0.91 | 3.97 | PGK | 0.91 | 3.64 | |||
| 3 | GADPH | 0.92 | 4.43 | GADPH | 0.99 | 4.92 | |||
| 4 | PGK | 0.95 | 3.73 | TUA | 1.04 | 4.59 | |||
| 5 | CYP | 1 | 4.7 | CYP | 1.14 | 5.44 | |||
| 6 | TUB | 1.95 | 7.15 | TUB | 1.52 | 5.31 | |||
| 7 | EF1α | 2.71 | 14.19 | EF1α | 1.77 | 8.39 | |||
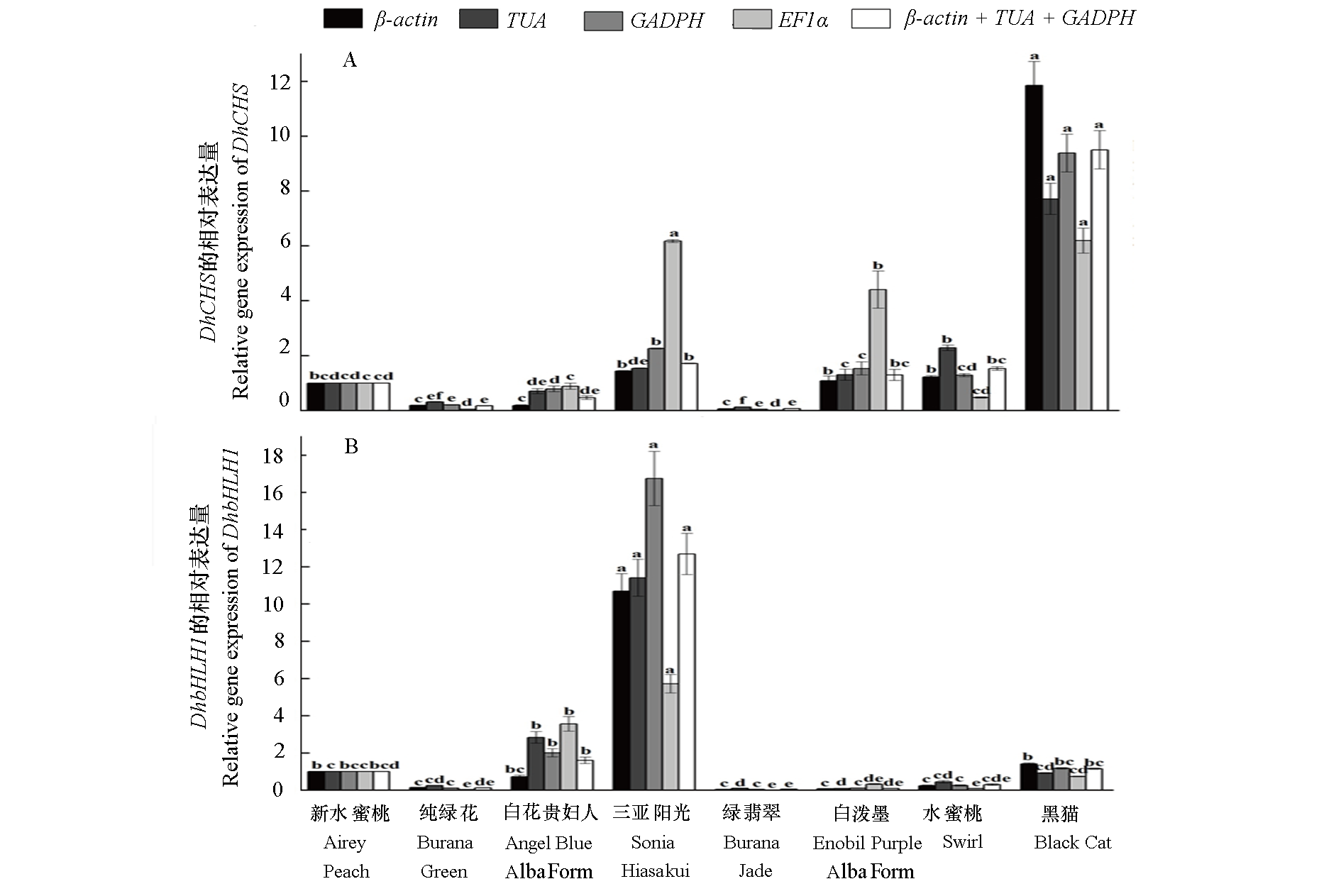
图5 使用选定的内参基因(包括最稳定或最不稳定的内参基因)标准化DhCHS(A)和DhbHLH1(B)在不同品种中的相对表达
Fig. 5 The relative expression of DhCHS(A)and DhbHLH1(B)in different cultivars normalize using the selected reference genes (including the most stable or unstable reference genes) P < 0.05.
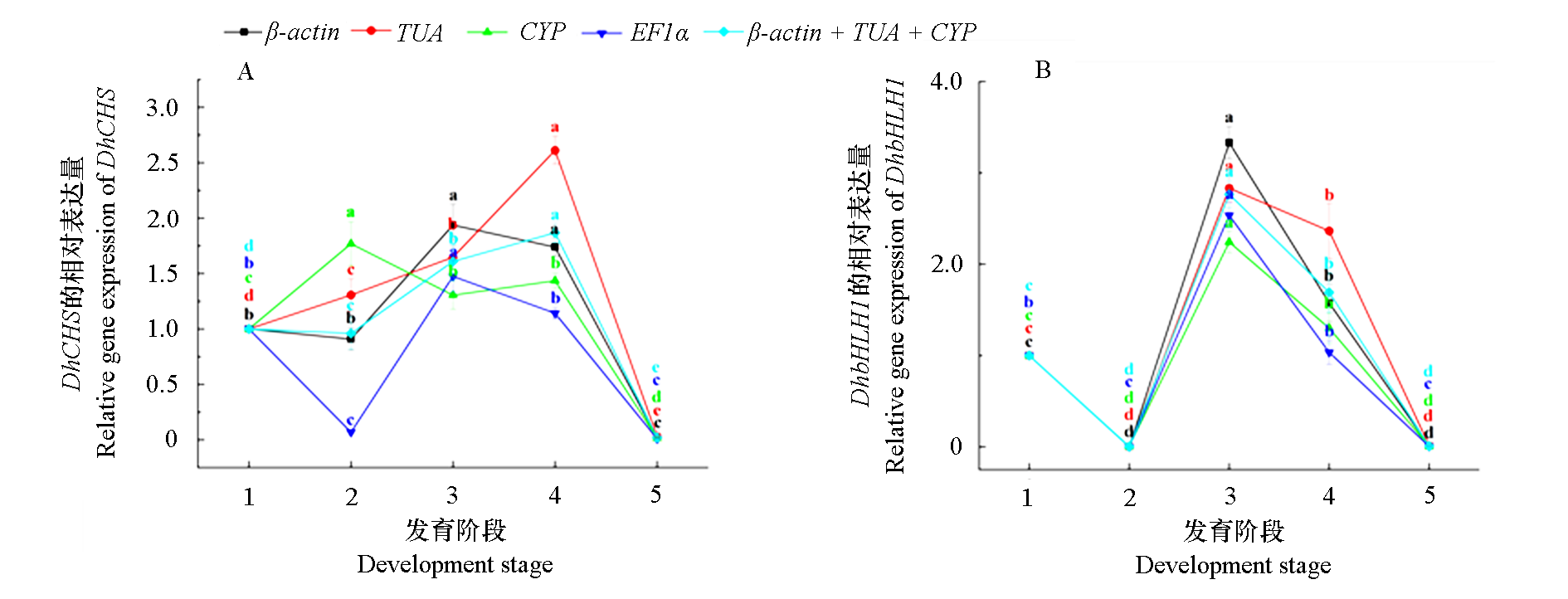
图6 使用选定的内参基因(包括最稳定或最不稳定的内参基因)标准化DhCHS(A)和DhbHLH1(B)在发育不同阶段的相对表达
Fig. 6 The relative expression of DhCHS(A)and DhbHLH1(B)at different developmental stages normalize using the selected reference genes(including the most stable or unstable reference genes) P < 0.05.
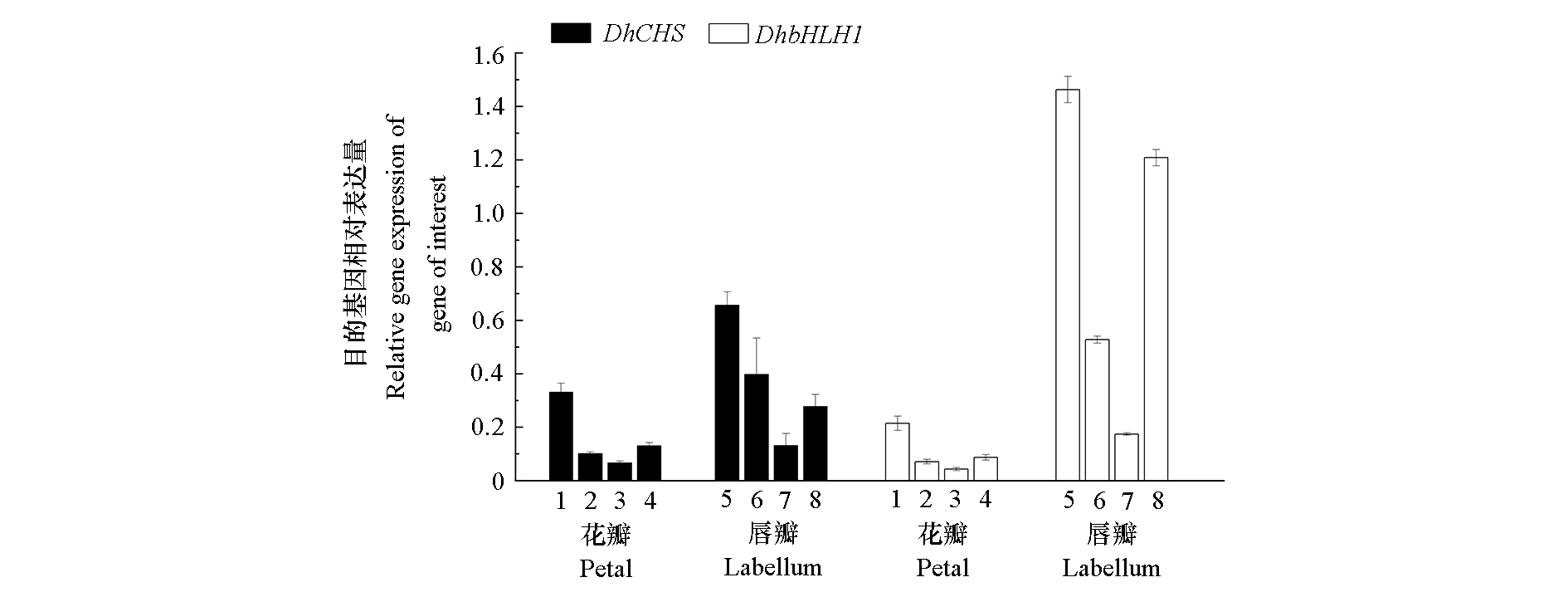
图7 使用选定的内参基因标准化DhCHS和DhbHLH1在不同组织中的相对表达
Fig. 7 The relative expression of DhCHS and DhbHLH1 in different tissues normalize using the selected reference genes (including the most stable or unstable reference genes) 1:β-actin;2:CYP;3:PGK;4:β-actin + CYP + PGK;5:CYP;6:TUA;7:PGK;8:CYP + TUA + PGK
| [1] |
Andersen C L, Jensen J L, Ørntoft T F. 2004. Normalization of real-time quantitative reverse transcription-PCR data:a model-based variance estimation approach to identify genes suited for normalization,applied to bladder and colon cancer data sets. Cancer Research, 64 (15):5245-5250.
doi: 10.1158/0008-5472.CAN-04-0496 URL |
| [2] |
Artico S, Nardeli S M, Brilhante O, Grossi-de-Sa M F, Alves-Ferreira M. 2010. Identification and evaluation of new reference genes in Gossypium hirsutum for accurate normalization of real-time quantitative RT-PCR data. BMC Plant Biology, 10 (1):49.
doi: 10.1186/1471-2229-10-49 URL |
| [3] |
Bustin S A, Nolan T. 2004. Pitfalls of quantitative real-time reverse-transcription polymerase chain reaction. Journal of Biomolecular Techniques, 15 (3):155-166.
pmid: 15331581 |
| [4] | Bustin S A, Vladimir B, Garson J A, Hellemans J, Huggett J, Kubista M, Wittwer C T. 2009. The MIQE guidelines:minimum information for publication of quantitative real-time PCR experiments. Clinical Chemistry,(4):611. |
| [5] | Davies K M, Marshall G B, Lewis D H, Winefield C S, Deroles S C, Boase M R, Bloor S J. 2003. Generation of new ornamental varieties through genetic modification of pigment biosynthesis. Acta Horticulturae, 624:435-447. |
| [6] |
Dheda K, Huggett J F, Bustin S A, Johnson M A, Rook G, Zumla A. 2004. Validation of housekeeping genes for normalizing RNA expression in real-time PCR. Biotechniques, 37 (1):112-119.
pmid: 15283208 |
| [7] |
Die J V, Román B, Nadal S, González-Verdejo C I. 2010. Evaluation of candidate reference genes for expression studies in Pisum sativum under different experimental conditions. Planta, 232 (1):145-153.
doi: 10.1007/s00425-010-1158-1 URL |
| [8] |
Derveaux S, Vandesompele J, Hellemans J. 2010. How to do successful gene expression analysis using real-time PCR. Methods, 50 (4):227-230.
doi: 10.1016/j.ymeth.2009.11.001 pmid: 19969088 |
| [9] | Enrico P, Tanzarella O A, Paolacci A R, Ciaffi M. 2009. Identification and validation of reference genes for quantitative RT-PCR normalization in wheat. BMCMolecular Biology, 10 (1):11. |
| [10] |
Fulvio F, Martinelli T, Paris R. 2021. Selection and validation of reference genes for RT-qPCR normalization in different tissues of milk thistle(Silybum marianum,Gaert.). Gene, 768:145272.
doi: 10.1016/j.gene.2020.145272 URL |
| [11] |
Garrido J, Aguilar M, Prieto P. 2020. Identification and validation of reference genes for RT-qPCR normalization in wheat meiosis. Scientific Reports, 10 (1):1-12.
doi: 10.1038/s41598-019-56847-4 URL |
| [12] |
Gutierrez L, Mauriat M, Guenin S, Pelloux J, Lefebvre J F, Louvet R, van Wuytswinkel O. 2008. The lack of a systematic validation of reference genes:a serious pitfall undervalued in reverse transcription-polymerase chain reaction(RT-PCR)analysis in plants. Plant Biotechnology Journal, 6 (6):609-618.
doi: 10.1111/j.1467-7652.2008.00346.x pmid: 18433420 |
| [13] |
Huang Y X, Tan H X, Yun J, Chen Y, Guo Z, Wang G, Diao Y. 2017. Stable internal reference genes for normalizing real-time quantitative PCR in Baphicacanthus cusia under hormonal stimuli and UV irradiation and in different plant organs. Frontiers in Plant Science, 8:668.
doi: 10.3389/fpls.2017.00668 URL |
| [14] | Huggett J, Dheda K, Bustin S, Zumla A. 2005. Real-time RT-PCR normalisation:strategies and considerations. Genes & Immunity, 6 (4):279-284. |
| [15] |
Jin X H, Fu J X, Dai S L, Sun Y, Hong Y. 2013. Reference gene selection for qPCR analysis in cineraria developing flowers. Scientia Horticulturae, 153 (1):64-70.
doi: 10.1016/j.scienta.2013.01.023 URL |
| [16] |
Kuehnle A R, Lewis D H, Markham K R, Mitchell K A, Davies K M, Jordan B R. 1997. Floral flavonoids and pH in Dendrobium orchid species and hybrids. Euphytica, 95 (2):187-194.
doi: 10.1023/A:1002945632713 URL |
| [17] |
Kriangphan N, Vuttipongchaikij S, Kittiwongwattana C, Suttangkakul A, Pinmanee P, Sakulsathaporn A, Apisitwanich S. 2015. Effects of sequence and expression of eight anthocyanin biosynthesis genes on floral coloration in four Dendrobium hybrids. The Horticulture Journal, 84 (1):83-92.
doi: 10.2503/hortj.MI-020 URL |
| [18] | Lee J M, Roche J R, Donaghy D J, Thrush A, Sathish P. 2010. Validation of reference genes for quantitative RT-PCR studies of gene expression in perennial ryegrass(Lolium perenne L.). Molecular Biology, 11 (1):1-14. |
| [19] |
Li C H, Qiu J, Ding L, Huang M, Huang S R, Yang G S, Yin J M. 2017a. Anthocyanin biosynthesis regulation of DhMYB2 and DhbHLH1 in Dendrobium hybrids petals. Plant Physiology and Biochemistry, 112:335-345.
doi: 10.1016/j.plaphy.2017.01.019 URL |
| [20] | Li Chong-hui, Ren Yu, Huang Su-rong, Huang Shao-hua, Yang Guang-sui. 2013. Floral colors of Phalaenopsis type dendrobium and their flavonoid composition. Acta Horticulture Sinica, 40 (1):107-116. (in Chinese) |
| 李崇晖, 任羽, 黄素荣, 黄少华, 杨光穗. 2013. 蝴蝶石斛兰花色表型及类黄酮成分分析. 园艺学报, 40 (1):107-116. | |
| [21] |
Li Chong-hui, Yin Jun-mei. 2019. Genetic engineering progress and breeding tactics on blue flowers. Biotechnology Bulletin, 35 (11):160-168. (in Chinese)
doi: 10.13560/j.cnki.biotech.bull.1985.2019-0450 |
|
李崇晖, 尹俊梅. 2019. 蓝色花形成的基因工程进展与育种策略. 生物技术通报, 35 (11):160-168.
doi: 10.13560/j.cnki.biotech.bull.1985.2019-0450 |
|
| [22] |
Li W G, Zhang L H, Zhang Y D, Wang G, Song D, Zhang Y. 2017b. Selection and validation of appropriate reference genes for quantitative real-time PCR normalization in staminate and perfect flowers of andromonoecious Taihangia rupestris. Frontiers in Plant Science, 8:729.
doi: 10.3389/fpls.2017.00729 URL |
| [23] | Liang Jin, Liu Hai-ting, Zhong Rong, Li Hao, Yin Dong-mei, Liu Xiang, Qin Qiao-ping, Zhang Zhi-guo, Duan Ke, Ni Di-an. 2020. Screening of reference genes for quantitative real-time PCR in different organs of Hemerocallis fulva. Plant Physiology Journal, 56 (9):1891-1898. (in Chinese) |
| 梁锦, 刘海婷, 钟荣, 李昊, 尹冬梅, 刘翔, 秦巧平, 张志国, 段可, 倪迪安. 2020. 萱草不同器官实时荧光定量PCR内参基因的筛选. 植物生理学报, 56 (9):1891-1898. | |
| [24] |
Liliana M, Andreia M, Ricardo C P, Miguel C. 2012. Reference gene selection for quantitative real-time PCR normalization in Quercus suber. PLoS ONE, 7 (4):e35113.
doi: 10.1371/journal.pone.0035113 URL |
| [25] | Ma Lulin, Duan Qing, Cui Guangfen, Du Wenwen, Jia Wenjie, Wang Xiangning, Wang Jihua, Chen Fadi. 2021. Selection and validation of reference genes for qRT-PCR analysis of the correlated genes in flower pigments biosynthesis pathway of Anemone obtusiloba. Acta Horticulturae Sinica, 48 (2):377-388. (in Chinese) |
| 马璐琳, 段青, 崔光芬, 杜文文, 贾文杰, 王祥宁, 王继华, 陈发棣. 2021. 钝裂银莲花花色素合成相关基因qRT-PCR内参基因的筛选. 园艺学报, 48 (2):377-388. | |
| [26] |
Nikalje G C, Srivastava A K, Sablok G, Pandey G K, Nikam T D, Suprasanna P. 2017. Identification and validation of reference genes for quantitative real-time PCR under salt stress in a halophyte,Sesuvium portulacastrum, Plant Gene, 13:18-24.
doi: 10.1016/j.plgene.2017.11.003 URL |
| [27] | Pan Li-jing, Cao You-pei, Xiao Yang, Fan Gan-qun, Chen Wei-ting. 2009. Review of research on breeding technology of Dendrobium. Guangdong Agricultural Sciences,(9):71-73. (in Chinese) |
| 潘丽晶, 曹友培, 肖杨, 范干群, 陈伟庭. 2009. 观赏石斛育种技术研究综述. 广东农业科学,(9):71-73. | |
| [28] |
Pfaffl M W, Tichopad A, Prgomet C, Neuvians T P. 2004. Determination of stable housekeeping genes differentially regulated target genes and sample integrity:BestKeeper-Excel-Based tool using pair-wise correlations. Biotechnology Letters, 26 (6):509-515.
doi: 10.1023/B:BILE.0000019559.84305.47 URL |
| [29] | Qiao Yong-gang, Wang Yong-fei, Cao Ya-ping, He Jia-xin, Jia Meng-jun, Li Zheng, Zhang Xin-rui, Song Yun. 2020. Reference genes selection and related genes expression analysis under low and high temperature stress in Taraxacum officinale. Acta Horticulturae Sinica, 47 (6):1153-1164. (in Chinese) |
| 乔永刚, 王勇飞, 曹亚萍, 贺嘉欣, 贾孟君, 李政, 张鑫瑞, 宋芸. 2020. 药用蒲公英低温和高温胁迫下内参基因筛选与相关基因表达分析. 园艺学报, 47 (6):1153-1164. | |
| [30] |
Rahul G, Rupwate S D, Tumaney A W. 2017. Selection and validation of appropriate reference genes for quantitative real-time PCR analysis in Salvia hispanica. PLoS ONE, 12 (11):e0186978.
doi: 10.1371/journal.pone.0186978 URL |
| [31] |
Ransbotyn V, Reusch T B H. 2006. Housekeeping gene selection for quantitative real-time PCR assays in the seagrass Zostera marina subjected to heat stress. Limnology and Oceanography Methods, 4 (10):367-373.
doi: 10.4319/lom.2006.4.367 URL |
| [32] |
Ratanasut K, Monmai C, Piluk P. 2015. Transient hairpin RNAi-induced silencing in floral tissues of Dendrobium Sonia‘Earsakul’by agroinfiltration for rapid assay of flower colour modification. Plant Cell Tissue and Organ Culture, 120 (2):643-654.
doi: 10.1007/s11240-014-0632-z URL |
| [33] | Reddy D S, Bhatnagar-Mathur P, Cindhuri K S, Sharma K K. 2013. Evaluation and validation of reference genes for normalization of quantitative real-time PCR based gene expression studies in peanut. PLoS ONE, 8 (10):1-14. |
| [34] |
Stéphanie G, Mélanie M, Jérôme P, van Wuytswinkel O, Bellini C, Gutierrez L. 2009. Normalization of qRT-PCR data:the necessity of adopting a systematic,experimental conditions-specific,validation of references. Journal of Experimental Botany, 60 (2):487-493.
doi: 10.1093/jxb/ern305 pmid: 19264760 |
| [35] | To K Y, Wang C K. 2006. Molecular breeding of flower color. Floriculture Ornamental and Plant Biotechnology,(1):300-310. |
| [36] | Vandesompele J, Preter K D, Pattyn F, Poppe B, van Roy N, de Paepe A, Speleman F. 2002. Accurate normalization of real-time quantitative RT-PCR data by geometric averaging of multiple internal control genes. Genome Biology, 3 (7):1-12. |
| [37] |
Wang L J, Wang Y C, Zhou P. 2013. Validation of reference genes for quantitative real-time PCR during Chinese wolfberry fruit development. Plant Physiology and Biochemistry, 70:304-310.
doi: 10.1016/j.plaphy.2013.05.038 pmid: 23811043 |
| [38] | Wu J Y, Zhang H G, Liu L Q, Li W, Wei Y, Shi S. 2016. Validation of reference genes for RT-qPCR studies of gene expression in preharvest and postharvest longan fruits under different experimental conditions. Frontiers in Plant Science, 7 (439):780. |
| [39] |
Xi L, Tang D Q, Shi Y M. 2018. Selection of reference genes for quantitative real-time PCR normalization in Narcissus pseudonarcissus in different cultivars and different organs. Heliyon, 4 (7):e00686.
doi: 10.1016/j.heliyon.2018.e00686 URL |
| [40] | Xiao X L, Ma J B, Wang J R, Wu X, Li P, Yao Y. 2015. Validation of suitable reference genes for gene expression analysis in the halophyte Salicornia europaea by real-time quantitative PCR. Frontiers in Plant Science, 5:788. |
| [41] |
Xu L, Xu H, Cao Y, Yang P, Feng Y, Tang Y, Ming J. 2017. Validation of reference genes for quantitative real-time PCR during bicolor tepal development in asiatic hybrid lilies(Lilium spp.). Frontiers in Plant Science, 8:669-688.
doi: 10.3389/fpls.2017.00669 URL |
| [42] | Yang Lan, Yang Guang-sui,Li Chong-hui,Niu Jun-hai,Yin Jun-mei. 2015. Screening of reference genes in Anthurium andraeanum spathes for qRT-PCR analysis. Journal of Tropical and Subtropical Botany, 23 (1):51-58. (in Chinese) |
| 杨澜, 杨光穗, 李崇晖, 牛俊海, 尹俊梅. 2015. 红掌佛焰苞基因qRT-PCR分析中内参基因的筛选. 热带亚热带植物学报, 23 (1):51-58. | |
| [43] | Yin Han-tai, Yin Jun-mei, Liao Yi, Lu Shun-jiao, Li Chong-hui. 2021. Phenotype classification based on flower color,pigment distribution and epidermal cell shape of Dendrobium hybrids. Acta Horticulturae Sinica, 48 (10):1907-1920. (in Chinese) |
| 殷涵泰, 尹俊梅, 廖易, 陆顺教, 李崇晖. 2021. 基于秋石斛花朵颜色、色素分布及表皮细胞形态的表型分类. 园艺学报, 48 (10):1907-1920. | |
| [44] | Zhang Ji-yu, Huang Sheng-nan, Wang Tao, Pan De-lin, Zhai Min, Guo Zhong-ren. 2018. Screening of reference genes for reverse transcription quantitative real-time PCR in Actinidia deliciosa. Acta Agriculture Shanghai, 34 (1):84-88. (in Chinese) |
| 张计育, 黄胜男, 王涛, 潘德林, 翟敏, 郭忠仁. 2018. 金魁猕猴桃RT-qPCR内参基因的筛选. 上海农业学报, 34 (1):84-88. | |
| [45] |
Zhao Y C, Luo J, Xu S, Wang W, Liu T, Han C, Kong L. 2016. Selection of reference genes for gene expression normalization in Peucedanum praeruptorum Dunn under abiotic stresses,hormone treatments and different tissues. PLoS ONE, 11 (3):e0152356.
doi: 10.1371/journal.pone.0152356 URL |
| [46] | Zheng X, Sun X B, Liu X Q, Li C, He L, Chen S, Su J. 2016. Selection of reliable reference genes for gene expression studies on Rhododendron molle G. Don. Frontiers in Plant Science, 7:e0141853. |
| [1] | 王晓晨, 聂子页, 刘先菊, 段 伟, 范培格, 梁振昌, . 脱落酸对‘京香玉’葡萄果实单萜物质合成的影响[J]. 园艺学报, 2023, 50(2): 237-249. |
| [2] | 叶子茂, 申晚霞, 刘梦雨, 王 彤, 张晓楠, 余 歆, 刘小丰, 赵晓春, . R2R3-MYB转录因子CitMYB21对柑橘类黄酮生物合成的影响[J]. 园艺学报, 2023, 50(2): 250-264. |
| [3] | 翟含含, 翟宇杰, 田义, 张叶, 杨丽, 温陟良, 陈海江. 桃SAUR家族基因分析及PpSAUR5功能鉴定[J]. 园艺学报, 2023, 50(1): 1-14. |
| [4] | 李镇希, 潘睿翾, 许美容, 郑正, 邓晓玲. 柑橘黄龙病菌双重实时荧光PCR检测方法的建立[J]. 园艺学报, 2023, 50(1): 188-196. |
| [5] | 张婉青, 张红晓, 廉小芳, 李昱莹, 郭丽丽, 侯小改. ‘凤丹’牡丹愈伤组织分化和生根诱导中的DNA甲基化分析[J]. 园艺学报, 2022, 49(8): 1735-1746. |
| [6] | 蔡建法, 莫雪莲, 管思聪, 陈栩, 薛程. 草莓FvYABBY5.1表达特性和功能分析[J]. 园艺学报, 2022, 49(7): 1458-1472. |
| [7] | 张秋悦, 刘昌来, 于晓晶, 杨甲定, 封超年. 盐胁迫条件下杜梨叶片差异表达基因qRT-PCR内参基因筛选[J]. 园艺学报, 2022, 49(7): 1557-1570. |
| [8] | 魏晓羽, 王跃进. 中国野生葡萄果皮解剖结构与白粉病抗性的相关性研究[J]. 园艺学报, 2022, 49(6): 1200-1212. |
| [9] | 邱可立, 王玉民, 何金铃, 俞红, 潘海发, 盛玉, 谢庆梅, 陈红莉, 周晖, 张金云. 桃漆酶家族基因鉴定及PpLAC21功能分析[J]. 园艺学报, 2022, 49(6): 1351-1362. |
| [10] | 李亚梅, 马福利, 张山奇, 黄锦秋, 陈梦婷, 周军永, 孙其宝, 孙俊. 酸枣愈伤组织转化体系构建及在ZjBRC1调控ZjYUCCA表达中的应用[J]. 园艺学报, 2022, 49(4): 749-757. |
| [11] | 张瑞, 张夏燚, 赵婷, 王双成, 张仲兴, 刘博, 张德, 王延秀. 基于转录组分析垂丝海棠响应盐碱胁迫的分子机制[J]. 园艺学报, 2022, 49(2): 237-251. |
| [12] | 周至铭, 杨佳宝, 张程, 曾令露, 孟晚秋, 孙黎. 向日葵LACS家族鉴定及响应非生物胁迫表达分析[J]. 园艺学报, 2022, 49(2): 352-364. |
| [13] | 乔军, 王利英, 刘婧, 李素文. 基于转录组测序的茄子萼下果色光敏相关基因表达分析[J]. 园艺学报, 2022, 49(11): 2347-2356. |
| [14] | 叶广继, 郑贞贞, 纳添仓, 王舰. 马铃薯资源糖苷生物碱含量评价及合成相关基因表达分析[J]. 园艺学报, 2022, 49(11): 2357-2366. |
| [15] | 李悦雅, 何栋, 陈简村, 李博, 白梦, 罗乐, 张启翔. 尖萼报春苣苔叶斑的观察与分析[J]. 园艺学报, 2022, 49(11): 2388-2394. |
| 阅读次数 | ||||||
|
全文 |
|
|||||
|
摘要 |
|
|||||
版权所有 © 2012 《园艺学报》编辑部 京ICP备10030308号-2 国际联网备案号 11010802023439
编辑部地址: 北京市海淀区中关村南大街12号中国农业科学院蔬菜花卉研究所 邮编: 100081
电话: 010-82109523 E-Mail: yuanyixuebao@126.com
技术支持:北京玛格泰克科技发展有限公司