
Acta Horticulturae Sinica ›› 2021, Vol. 48 ›› Issue (2): 233-242.doi: 10.16420/j.issn.0513-353x.2020-0350
• Research Papers • Previous Articles Next Articles
MEI Chuang1, ZHANG Xiaoyan2, YAN Peng1,*( ), Aisajan Mamat1, FENG Beibei1, MA Kai1, HAN Liqun1, DONG Lianxin2, WANG Jixun1,*(
), Aisajan Mamat1, FENG Beibei1, MA Kai1, HAN Liqun1, DONG Lianxin2, WANG Jixun1,*( )
)
Received:2020-11-29
Revised:2021-01-18
Online:2021-02-25
Published:2021-03-09
Contact:
YAN Peng,WANG Jixun
E-mail:xaasyysyp@163.com;ee_wjx@163.com
CLC Number:
MEI Chuang, ZHANG Xiaoyan, YAN Peng, Aisajan Mamat, FENG Beibei, MA Kai, HAN Liqun, DONG Lianxin, WANG Jixun. Identification of TIFY Family in Apple and Their Expression Analysis Under Insect Stress[J]. Acta Horticulturae Sinica, 2021, 48(2): 233-242.
Add to citation manager EndNote|Ris|BibTeX
URL: https://www.ahs.ac.cn/EN/10.16420/j.issn.0513-353x.2020-0350
| 物种 Species | 基因名 Gene name | 基因号 Gene ID | 物种 Species | 基因名 Gene name | 基因号 Gene ID |
|---|---|---|---|---|---|
| 拟南芥 Arabidopsis thaliana | AthTIFY11A | AT1G17380.1 | 番茄 Solanum lycoperiscon | SLTIFY5A | Solyc01g097060.2 |
| AthTIFY10A | AT1G19180.1 | SLTIFY10A | Solyc01g103600.2 | ||
| AthTIFY11B | AT1G72450.1 | SLTIFY10b | Solyc03g122190.2 | ||
| AthTIFY10B | AT1G74950.1 | SLTIFY8 | Solyc06g065650.2 | ||
| AthTIFY3A | AT3G43440.1 | SLTIFY10b-A | Solyc07g042170.2 | ||
| AthTIFY8 | AT4G32570.1 | SLTIFY5A-a | Solyc08g036620.2 | ||
| AthTIFY3B | AT5G20900.1 | SLTIFY5A-b | Solyc08g036640.2 | ||
| 水稻 Oryza sativa | OsTIFY11a | Os03t0180800-01 | SLTIFY5A-c | Solyc08g036660.2 | |
| OsTIFY11c | Os03t0180900-01 | SLTIFY10b-B | Solyc12g009220.1 | ||
| OsTIFY11b | Os03t0181100-01 | SLTIFY10b-C | Solyc12g049400.1 | ||
| OsTIFY11g | Os03t0396500-00 | 葡萄 Vitis vinifera | VvTIFY5A | VIT_04s0008g00110.t01 | |
| OsTIFY10a | Os03t0402800-01 | VvTIFY8 | VIT_04s0008g04950.t01 | ||
| OsTIFY3 | Os04t0653000-01 | VvTIFY10A | VIT_09s0002g00890.t01 | ||
| OsTIFY5 | Os07t0153000-00 | VvTIFY5A-a | VIT_10s0003g03790.t01 | ||
| OsTIFY10b | Os07t0615200-01 | VvTIFY5A-b | VIT_10s0003g03800.t01 | ||
| OsTIFY10c | Os09t0439200-01 | VvTIFY5A-c | VIT_10s0003g03810.t01 | ||
| OsTIFY11e | Os10t0391400-01 | VvTIFY5A-d | VIT_01s0011g05560.t01 | ||
| OsTIFY11d | Os10t0392400-01 | VvTIFY10A-a | VIT_11s0016g00710.t01 | ||
| VvTIFY10A-b | VIT_12s0035g00900.t01 |
Table 1 Gene ID information in phylogenetic analysis
| 物种 Species | 基因名 Gene name | 基因号 Gene ID | 物种 Species | 基因名 Gene name | 基因号 Gene ID |
|---|---|---|---|---|---|
| 拟南芥 Arabidopsis thaliana | AthTIFY11A | AT1G17380.1 | 番茄 Solanum lycoperiscon | SLTIFY5A | Solyc01g097060.2 |
| AthTIFY10A | AT1G19180.1 | SLTIFY10A | Solyc01g103600.2 | ||
| AthTIFY11B | AT1G72450.1 | SLTIFY10b | Solyc03g122190.2 | ||
| AthTIFY10B | AT1G74950.1 | SLTIFY8 | Solyc06g065650.2 | ||
| AthTIFY3A | AT3G43440.1 | SLTIFY10b-A | Solyc07g042170.2 | ||
| AthTIFY8 | AT4G32570.1 | SLTIFY5A-a | Solyc08g036620.2 | ||
| AthTIFY3B | AT5G20900.1 | SLTIFY5A-b | Solyc08g036640.2 | ||
| 水稻 Oryza sativa | OsTIFY11a | Os03t0180800-01 | SLTIFY5A-c | Solyc08g036660.2 | |
| OsTIFY11c | Os03t0180900-01 | SLTIFY10b-B | Solyc12g009220.1 | ||
| OsTIFY11b | Os03t0181100-01 | SLTIFY10b-C | Solyc12g049400.1 | ||
| OsTIFY11g | Os03t0396500-00 | 葡萄 Vitis vinifera | VvTIFY5A | VIT_04s0008g00110.t01 | |
| OsTIFY10a | Os03t0402800-01 | VvTIFY8 | VIT_04s0008g04950.t01 | ||
| OsTIFY3 | Os04t0653000-01 | VvTIFY10A | VIT_09s0002g00890.t01 | ||
| OsTIFY5 | Os07t0153000-00 | VvTIFY5A-a | VIT_10s0003g03790.t01 | ||
| OsTIFY10b | Os07t0615200-01 | VvTIFY5A-b | VIT_10s0003g03800.t01 | ||
| OsTIFY10c | Os09t0439200-01 | VvTIFY5A-c | VIT_10s0003g03810.t01 | ||
| OsTIFY11e | Os10t0391400-01 | VvTIFY5A-d | VIT_01s0011g05560.t01 | ||
| OsTIFY11d | Os10t0392400-01 | VvTIFY10A-a | VIT_11s0016g00710.t01 | ||
| VvTIFY10A-b | VIT_12s0035g00900.t01 |
| 基因 Gene | 基因号 Gene ID | 基因组位置 Genomic position | 开放阅读框/bp ORF | 蛋白大小/aa Protein size | 外显子 Number of exons |
|---|---|---|---|---|---|
| MdTIFY1 | MD02G1096100 | Chr02:7600659..7602728+ | 843 | 280 | 5 |
| MdTIFY2 | MD15G1220400 | Chr15:17843310..17845336+ | 837 | 278 | 5 |
| MdTIFY3 | MD14G1238100 | Chr14:31732049..31736399- | 1 134 | 377 | 7 |
| MdTIFY4 | MD16G1020800 | Chr16:1492513..1504489+ | 1 128 | 375 | 7 |
| MdTIFY5 | MD06G1228900 | Chr06:35860218..35864674- | 1 188 | 395 | 7 |
| MdTIFY6 | MD15G1225800 | Chr15:18345401..18348150+ | 621 | 206 | 5 |
| MdTIFY7 | MD02G1105900 | Chr02:8591750 8594168 | 633 | 210 | 5 |
| MdTIFY8 | MD13G1030000 | Chr13:2164003..2166858+ | 1 149 | 382 | 7 |
| MdTIFY9 | MD09G1178600 | Chr09:15274688..15275998- | 615 | 204 | 2 |
| MdTIFY10 | MD15G1434400 | Chr15:53470255..53474071+ | 810 | 269 | 5 |
| MdTIFY11 | MD16G1199200 | Chr16:18010085..18014820+ | 1 026 | 341 | 9 |
| MdTIFY12 | MD13G1199500 | Chr13:17721679..17726448+ | 1 050 | 349 | 9 |
| MdTIFY13 | MD08G1137500 | Chr08:13179781..13184749+ | 1 338 | 445 | 8 |
| MdTIFY14 | MD15G1116100 | Chr15:8188285..8192990+ | 1 350 | 449 | 7 |
| MdTIFY15 | MD16G1127400 | Chr16:9274777..9277069+ | 636 | 211 | 4 |
| MdTIFY16 | MD13G1127100 | Chr15:9515820..9518542+ | 630 | 209 | 4 |
Table 2 Basic information of theTIFY family in apple plant
| 基因 Gene | 基因号 Gene ID | 基因组位置 Genomic position | 开放阅读框/bp ORF | 蛋白大小/aa Protein size | 外显子 Number of exons |
|---|---|---|---|---|---|
| MdTIFY1 | MD02G1096100 | Chr02:7600659..7602728+ | 843 | 280 | 5 |
| MdTIFY2 | MD15G1220400 | Chr15:17843310..17845336+ | 837 | 278 | 5 |
| MdTIFY3 | MD14G1238100 | Chr14:31732049..31736399- | 1 134 | 377 | 7 |
| MdTIFY4 | MD16G1020800 | Chr16:1492513..1504489+ | 1 128 | 375 | 7 |
| MdTIFY5 | MD06G1228900 | Chr06:35860218..35864674- | 1 188 | 395 | 7 |
| MdTIFY6 | MD15G1225800 | Chr15:18345401..18348150+ | 621 | 206 | 5 |
| MdTIFY7 | MD02G1105900 | Chr02:8591750 8594168 | 633 | 210 | 5 |
| MdTIFY8 | MD13G1030000 | Chr13:2164003..2166858+ | 1 149 | 382 | 7 |
| MdTIFY9 | MD09G1178600 | Chr09:15274688..15275998- | 615 | 204 | 2 |
| MdTIFY10 | MD15G1434400 | Chr15:53470255..53474071+ | 810 | 269 | 5 |
| MdTIFY11 | MD16G1199200 | Chr16:18010085..18014820+ | 1 026 | 341 | 9 |
| MdTIFY12 | MD13G1199500 | Chr13:17721679..17726448+ | 1 050 | 349 | 9 |
| MdTIFY13 | MD08G1137500 | Chr08:13179781..13184749+ | 1 338 | 445 | 8 |
| MdTIFY14 | MD15G1116100 | Chr15:8188285..8192990+ | 1 350 | 449 | 7 |
| MdTIFY15 | MD16G1127400 | Chr16:9274777..9277069+ | 636 | 211 | 4 |
| MdTIFY16 | MD13G1127100 | Chr15:9515820..9518542+ | 630 | 209 | 4 |
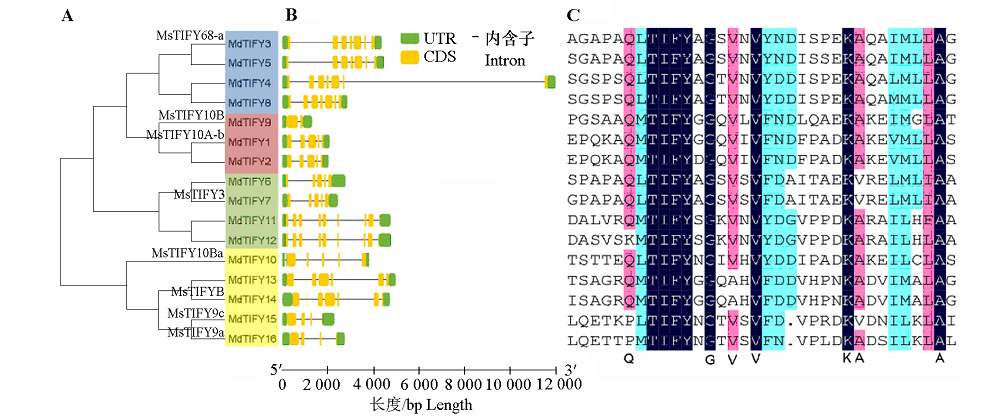
Fig. 2 Analysis of TIFY family in apple A:Phylogenetic analysis,different colors represent a clustered group;B:The exon-intron structure of the genes;C:Alignment analysis of amino acid sequence.
| [1] |
Bai Y, Meng Y, Huang D, Qi Y, Chen M. 2011. Origin and evolutionary analysis of the plant-specific TIFY transcription factor family. Genomics, 98(2):128-136.
doi: 10.1016/j.ygeno.2011.05.002 URL |
| [2] |
Barah P, Bones A M. 2015. Multidimensional approaches for studying plant defence against insects:from ecology to omics and synthetic biology. Journal of Experimental Botany, 66(2):479-493.
doi: 10.1093/jxb/eru489 URL |
| [3] |
Daccord N, Celton J M, Linsmith G, Becker C, Choisne N, Schijlen E, van de Geest H, Bianco L, Micheletti D, Velasco R, Di Pierro E A, Gouzy J, Rees D J G, Guerif P, Muranty H, Durel C E, Laurens F, Lespinasse Y, Gaillard S, Aubourg S, Quesneville H, Weigel D, van de Weg E, Troggio M, Bucher E. 2017. High-quality de novo assembly of the apple genome and methylome dynamics of early fruit development. Nature Genetics, 49(7):1099-1106.
doi: 10.1038/ng.3886 URL |
| [4] |
Dahl C C V, Baldwin I T. 2004. Methyl jasmonate and cis-jasmone do not dispose of the herbivore-induced jasmonate burst in Nicotiana attenuata. Physiologia Plantarum, 120(3):474-481.
doi: 10.1111/ppl.2004.120.issue-3 URL |
| [5] |
Dong Q, Zhao S, Duan D, Tian Y, Wang Y, Mao K, Zhou Z, Ma F. 2018. Structural and functional analyses of genes encoding VQ proteins in apple. Plant Science, 272:208-219.
doi: 10.1016/j.plantsci.2018.04.029 URL |
| [6] |
Ebel C, BenFeki A, Hanin M, Solano R, Chini A. 2018. Characterization of wheat(Triticum aestivum)TIFY family and role of Triticum durum TdTIFY11a in salt stress tolerance. PLoS ONE, 13(7):e0200566.
doi: 10.1371/journal.pone.0200566 URL |
| [7] | Engelberth J, Seidl-Adams I, Schultz J C, Tumlinson J H. 2007. Insect elicitors and exposure to green leafy volatiles differentially upregulate major octadecanoids and transcripts of 12-Oxo phytodienoic Acid Reductases in Zea mays. Molecular Plant, 20(6):707-716. |
| [8] |
Howe G A, Jander G. 2008. Plant immunity to insect herbivores. Annual Review of Plant Biology, 59:41-66.
doi: 10.1146/annurev.arplant.59.032607.092825 URL |
| [9] |
Hu C, Wei C, Ma Q, Dong H, Shi K, Zhou Y, Foyer C H, Yu J. 2021. Eethylene response factor 15 and 16 trigger jasmonate biosynthesis in tomato during herbivore resistance. Plant Physiology,Doi: 10.1093/plphys/kiaa089.
doi: 10.1093/plphys/kiaa089 |
| [10] |
Koo A J, Gao X, Jones A D, Howe G A. 2009. A rapid wound signal activates the systemic synthesis of bioactive jasmonates in Arabidopsis. Plant Journal, 59(6):974-986.
doi: 10.1111/tpj.2009.59.issue-6 URL |
| [11] | Li R, Jin J, Xu J, Wang L, Li J, Lou Y, Baldwin I T. 2020. Long non‐coding RNAs associate with jasmonate‐mediated plant defence against herbivores. Plant,Cell & Environment, pce: 13952. |
| [12] | Li X, Yin X, Wang H, Li J, Guo C, Gao H, Zheng Y, Fan C, Wang X. 2014. Genome-wide identification and analysis of the apple(Malus × domestica Borkh.)TIFY gene family. Tree Genetics & Genomes, 11(1):808. |
| [13] |
Liu Q, Wang X, Tzin V, Romeis J, Peng Y, Li Y. 2016. Combined transcriptome and metabolome analyses to understand the dynamic responses of rice plants to attack by the rice stem borer Chilo suppressalis(Lepidoptera:Crambidae). BMC Plant Biology, 16(1):259.
doi: 10.1186/s12870-016-0946-6 URL |
| [14] |
Mao K, Dong Q, Li C, Liu C, Ma F. 2017a. Genome wide identification and characterization of apple bHLH transcription factors and expression analysis in response to drought and salt stress. Frontiers in Plant Science, 8:480.
doi: 10.3389/fpls.2017.00480 pmid: 28443104 |
| [15] |
Mao Y B, Liu Y Q, Chen D Y, Chen F Y, Fang X, Hong G J, Wang L J, Wang J W, Chen X Y. 2017b. Jasmonate response decay and defense metabolite accumulation contributes to age-regulated dynamics of plant insect resistance. Nature Communications, 8:13925.
doi: 10.1038/ncomms13925 URL |
| [16] | Mei Chuang, Yan Peng, Ai Shajiang, Ma Kai, Han Liqun, Xu Zheng, Zhong Haixia, Diao Yongqiang, Wang Jixun. 2016. The relationship between bark thickness and branch roughnesson Agrilus mali damage in Xinjiang wild apple. Journal of Agricultural Science and Technology, 18(4):24-30. (in Chinese) |
| 梅闯, 闫鹏, 艾沙江·买买提, 马凯, 韩立群, 许正, 钟海霞, 刁永强, 王继勋. 2016. 新疆野苹果(Malus sieversii)受苹小吉丁虫危害程度与树皮厚度、径阶的关系. 中国农业科技导报, 18(4):24-30. | |
| [17] | Mei Chuang, Yan Peng, Ma Kai, Han Liqun, Xu Zheng, Lu Chunsheng, Fan Dingyu, Ai Shajiang, Wang Jixun. 2015. Agrilus mali Matsumura resistance of different type of Xinjiang wild apple. Xinjiang Agricultural Sciences, 52(10):1859-1865. (in Chinese). |
| 梅闯, 闫鹏, 马凯, 韩立群, 许正, 卢春生, 樊丁宇, 艾沙江, 王继勋. 2015. 新疆野苹果不同类型单株对苹果小吉丁虫的抗性差异. 新疆农业科学, 52(10):1859-1865. | |
| [18] |
Mei C, Yang J, Yan P, Li N, Ma K, Mamat A, Han L, Dong Q, Mao K, Ma F, Wang J. 2020. Full-length transcriptome and targeted metabolome analyses provide insights into defense mechanisms of Malus sieversii against Agrilus mali. PeerJ, 8:e8992.
doi: 10.7717/peerj.8992 URL |
| [19] |
Staswick P E. 2008. JAZing up jasmonate signaling. Trends in Plant Science, 13(2):66-71.
doi: 10.1016/j.tplants.2007.11.011 URL |
| [20] |
Tamura K, Peterson D, Peterson N, Stecher G, Nei M, Kumar S. 2011. MEGA5:molecular evolutionary genetics analysis using maximum likelihood,evolutionary distance,and maximum parsimony methods. Molecular Biology and Evolution, 28(10):2731-2739.
doi: 10.1093/molbev/msr121 URL |
| [21] |
Thines B, Katsir L, Melotto M, Niu Y, Mandaokar A, Liu G, Nomura K, He S Y, Howe G A, Browse J. 2007. JAZ repressor proteins are targets of the SCF(COI1)complex during jasmonate signalling. Nature, 448(7154):661-665.
pmid: 17637677 |
| [22] |
Thireault C, Shyu C, Yoshida Y, St Aubin B, Campos M L, Howe G A. 2015. Repression of jasmonate signaling by a non-TIFY JAZ protein in Arabidopsis. The Plant journal, 82(4):669-679.
doi: 10.1111/tpj.12841 pmid: 25846245 |
| [23] |
Vanholme B, Grunewald W, Bateman A, Kohchi T, Gheysen G. 2007. The tify family previously known as ZIM. Trends in Plant Science, 12(6):239-244.
doi: 10.1016/j.tplants.2007.04.004 URL |
| [24] |
Velasco R, Zharkikh A, Affourtit J, Dhingra A, Cestaro A, Kalyanaraman A, Fontana P, Bhatnagar S K, Troggio M, Pruss D, Salvi S, Pindo M, Baldi P, Castelletti S, Cavaiuolo M, Coppola G, Costa F, Cova V, Dal Ri A, Goremykin V, Komjanc M, Longhi S, Magnago P, Malacarne G, Malnoy M, Micheletti D, Moretto M, Perazzolli M, Si-Ammour A, Vezzulli S, Zini E, Eldredge G, Fitzgerald L M, Gutin N, Lanchbury J, Macalma T, Mitchell J T, Reid J, Wardell B, Kodira C, Chen Z, Desany B, Niazi F, Palmer M, Koepke T, Jiwan D, Schaeffer S, Krishnan V, Wu C, Chu V T, King S T, Vick J, Tao Q, Mraz A, Stormo A, Stormo K, Bogden R, Ederle D, Stella A, Vecchietti A, Kater M M, Masiero S, Lasserre P, Lespinasse Y, Allan A C, Bus V, Chagne D, Crowhurst R N, Gleave A P, Lavezzo E, Fawcett J A, Proost S, Rouze P, Sterck L, Toppo S, Lazzari B, Hellens R P, Durel C E, Gutin A, Bumgarner R E, Gardiner S E, Skolnick M, Egholm M, Van de Peer Y, Salamini F, Viola R. 2010. The genome of the domesticated apple(Malus × domestica Borkh.). Nature Genetics, 42(10):833-839.
doi: 10.1038/ng.654 URL |
| [25] | Wang Ping, Lu Shixiong, Liang Guoping, Ma Zonghuan, Li Wenfang, Mao Juan, Chen Baihong. 2019. Bioinformatics identification and expression analysis of Trihelix transcription factor family in apple. Acta Horticulturae Sinica, 46(11):2082-2098. (in Chinese). |
| 王萍, 卢世雄, 梁国平, 马宗桓, 李文芳, 毛娟, 陈佰鸿. 2019. 苹果Trihelix转录因子家族生物信息学鉴定与基因表达分析. 园艺学报, 46(11):2082-2098. | |
| [26] |
Wang Y, Pan F, Chen D M, Chu W Y, Liu H L, Xiang Y. 2017. Genome-wide identification and analysis of the Populus trichocarpa TIFY gene family. Plant Physiology and Biochemistry, 115:360-371.
doi: 10.1016/j.plaphy.2017.04.015 URL |
| [27] |
War A R, Paulraj M G, Ahmad T, Buhroo A A, Hussain B, Ignacimuthu S, Sharma H C. 2012. Mechanisms of plant defense against insect herbivores. Plant Signal Behav, 7(10):1306-1320.
doi: 10.4161/psb.21663 URL |
| [28] | Xia W X, Yu H Y, Cao P, Luo J, Wang N. 2017. Identification of TIFY family genes and analysis of their expression profiles in response to phytohormone treatments and Melampsora larici-populina infection in poplar. Frontiers in Plant Science, 8:493. |
| [29] |
Xie Y, Chen P, Yan Y, Bao C, Li X, Wang L, Shen X, Li H, Liu X, Niu C, Zhu C, Fang N, Shao Y, Zhao T, Yu J, Zhu J, Xu L, van Nocker S, Ma F, Guan Q. 2018. An atypical R2R3 MYB transcription factor increases cold hardiness by CBF-dependent and CBF-independent pathways in apple. The New Phytologist, 218(1):201-218.
doi: 10.1111/nph.14952 URL |
| [30] | Xu G X, Guo C C, Shan H Y, Kong H Z. 2012. Divergence of duplicate genes in exon-intron structure. Proceedings of the National Academy of Sciences of the United States of America, 109(4):1187-1192. |
| [31] |
Ye H Y, Du H, Tang N, Li X H, Xiong L Z. 2009. Identification and expression profiling analysis of TIFY family genes involved in stress and phytohormone responses in rice. Plant Molecular Biology, 71(3):291-305.
doi: 10.1007/s11103-009-9524-8 URL |
| [32] |
Zhang Y, Gao M, Singer S D, Fei Z, Wang H, Wang X. 2012. Genome-wide identification and analysis of the TIFY gene family in grape. PLoS ONE, 7(9):e44465.
doi: 10.1371/journal.pone.0044465 URL |
| [1] | YU Tingting, LI Huan, NING Yuansheng, SONG Jianfei, PENG Lulin, JIA Junqi, ZHANG Weiwei, and YANG Hongqiang. Genome-wide Identification of GRAS Gene Family in Apple and Expression Analysis of Its Response to Auxin [J]. Acta Horticulturae Sinica, 2023, 50(2): 397-409. |
| [2] | ZHAI Hanhan, ZHAI Yujie, TIAN Yi, ZHANG Ye, YANG Li, WEN Zhiliang, CHEN Haijiang. Genome-wide Identification of Peach SAUR Gene Family and Characterization of PpSAUR5 Gene [J]. Acta Horticulturae Sinica, 2023, 50(1): 1-14. |
| [3] | ZHAO Xueyan, WANG Qi, WANG Li, WANG Fangyuan, WANG Qing, LI Yan. Comparative Transcriptome Analysis of Differential Expression in Different Tissues of Corydalis yanhusuo [J]. Acta Horticulturae Sinica, 2023, 50(1): 177-187. |
| [4] | DING Zhijie, BAO Jinbo, ROUXIAN Guli, ZHU Tiantian, LI Xueli, MIAO Haoyu, TIAN Xinmin. Comparative Chloroplast Genome Study of Mallus servisii‘Red Delicious’and‘Golden Delicious’ [J]. Acta Horticulturae Sinica, 2022, 49(9): 1977-1990. |
| [5] | GAO Yanlong, WU Yuxia, ZHANG Zhongxing, WANG Shuangcheng, ZHANG Rui, ZHANG De, WANG Yanxiu. Bioinformatics Analysis of Apple ELO Gene Family and Its Expression Analysis Under Low Temperature Stress [J]. Acta Horticulturae Sinica, 2022, 49(8): 1621-1636. |
| [6] | LIU Jinming, GUO Caihua, YUAN Xing, KANG Chao, QUAN Shaowen, NIU Jianxin. Genome-wide Identification of Dof Family Genes and Expression Analysis Sepal Persistent and Abscission in Pear [J]. Acta Horticulturae Sinica, 2022, 49(8): 1637-1649. |
| [7] | QIU Ziwen, LIU Linmin, LIN Yongsheng, LIN Xiaojie, LI Yongyu, WU Shaohua, YANG Chao. Cloning and Functional Analysis of the MbEGS Gene from Melaleuca bracteata [J]. Acta Horticulturae Sinica, 2022, 49(8): 1747-1760. |
| [8] | ZHENG Lin, WANG Shuai, LIU Yunuo, DU Meixia, PENG Aihong, HE Yongrui, CHEN Shanchun, ZOU Xiuping. Gene Cloning and Expression Analysis of NAC Gene in Citrus in Response to Huanglongbing [J]. Acta Horticulturae Sinica, 2022, 49(7): 1441-1457. |
| [9] | MA Weifeng, LI Yanmei, MA Zonghuan, CHEN Baihong, MAO Juan. Identification of Apple POD Gene Family and Functional Analysis of MdPOD15 Gene [J]. Acta Horticulturae Sinica, 2022, 49(6): 1181-1199. |
| [10] | ZHANG Kai, MA Mingying, WANG Ping, LI Yi, JIN Yan, SHENG Ling, DENG Ziniu, MA Xianfeng. Identification of HSP20 Family Genes in Citrus and Their Expression in Pathogen Infection Responses Citrus Canker [J]. Acta Horticulturae Sinica, 2022, 49(6): 1213-1232. |
| [11] | LIANG Chen, SUN Ruyi, XIANG Rui, SUN Yimeng, SHI Xiaoxin, DU Guoqiang, WANG Li. Genome-wide Identification of Grape GRF Family and Expression Analysis [J]. Acta Horticulturae Sinica, 2022, 49(5): 995-1007. |
| [12] | XIAO Xuechen, LIU Mengyu, JIANG Mengqi, CHEN Yan, XUE Xiaodong, ZHOU Chengzhe, WU Xingjian, WU Junnan, GUO Yinsheng, YEH Kaiwen, LAI Zhongxiong, LIN Yuling. Whole-genome Identification and Expression Analysis of SNAT,ASMT and COMT Families of Melatonin Synthesis Pathway in Dimocarpus longan [J]. Acta Horticulturae Sinica, 2022, 49(5): 1031-1046. |
| [13] | GAO Weilin, ZHANG Liman, XUE Chaoling, ZHANG Yao, LIU Mengjun, ZHAO Jin. Expression of E-type MADS-box Genes in Flower and Fruits and Protein Interaction Analysis in Chinese Jujube [J]. Acta Horticulturae Sinica, 2022, 49(4): 739-748. |
| [14] | LIU Mengyu, JIANG Mengqi, CHEN Yan, ZHANG Shuting, XUE Xiaodong, XIAO Xuechen, LAI Zhongxiong, LIN Yuling. Genome-wide Identification and Expression Analysis of GDSL Esterase/Lipase Genes in Dimocarpus longan [J]. Acta Horticulturae Sinica, 2022, 49(3): 597-612. |
| [15] | JIANG Cuicui, FANG Zhizhen, ZHOU Danrong, LIN Yanjuan, YE Xinfu. Identification and Expression Analysis of Sugar Transporter Family Genes in‘Furongli’(Prunus salicina) [J]. Acta Horticulturae Sinica, 2022, 49(2): 252-264. |
| Viewed | ||||||
|
Full text |
|
|||||
|
Abstract |
|
|||||
Copyright © 2012 Acta Horticulturae Sinica 京ICP备10030308号-2 国际联网备案号 11010802023439
Tel: 010-82109523 E-Mail: yuanyixuebao@126.com
Support by: Beijing Magtech Co.Ltd