
Acta Horticulturae Sinica ›› 2022, Vol. 49 ›› Issue (5): 995-1007.doi: 10.16420/j.issn.0513-353x.2021-0533
• Research Papers • Previous Articles Next Articles
LIANG Chen, SUN Ruyi, XIANG Rui, SUN Yimeng, SHI Xiaoxin, DU Guoqiang( ), WANG Li(
), WANG Li( )
)
Received:2022-01-27
Revised:2022-03-28
Online:2022-05-25
Published:2022-05-25
Contact:
DU Guoqiang,WANG Li
E-mail:gdu@hebau.edu.cn;vivi@hebau.edu.cn
CLC Number:
LIANG Chen, SUN Ruyi, XIANG Rui, SUN Yimeng, SHI Xiaoxin, DU Guoqiang, WANG Li. Genome-wide Identification of Grape GRF Family and Expression Analysis[J]. Acta Horticulturae Sinica, 2022, 49(5): 995-1007.
Add to citation manager EndNote|Ris|BibTeX
URL: https://www.ahs.ac.cn/EN/10.16420/j.issn.0513-353x.2021-0533
| 基因Gene | 上游引物(5'-3')Forward primer | 下游引物(5'-3')Reverse primer |
|---|---|---|
| VvGRF1 | GGGCTTCCAGTGCCTTATC | TTCCCATCCGTTCTTCTACAT |
| VvGRF2 | CTGGTGGGACTCATTTCGC | AGGTTGCCTGGCTGTTATTG |
| VvGRF3 | GCCTCAATGACTTCTGAAACCT | CGATGATGCCTGTAATCCCTAT |
| VvGRF4 | CAGAAAGCCTGTGGAAGTTATG | TGTAGCAAGAAGGGTGACGAT |
| VvGRF5 | CGCCTTTGGACAGCACAT | CGAGACTGACCCTCCTGTTTC |
| VvGRF6 | TCGTTCTATCTTCAACCTCCGT | AGTACTTCTGATTGAGTGCCACAT |
| VvGRF7 | GAAGCCCTTGATTCCGTTG | TCTCACAATACTTCTGATCGGC |
| VvGRF8 | CGCCACTCAGGCTATCTCG | AAGCAGGCTCCCATGTTGT |
| EF1-α | AGGAGGCAGCCAACTTCACC | CAAACCCTGCATCACCATTC |
Table 1 The primer sequences used for qRT-PCR amplification
| 基因Gene | 上游引物(5'-3')Forward primer | 下游引物(5'-3')Reverse primer |
|---|---|---|
| VvGRF1 | GGGCTTCCAGTGCCTTATC | TTCCCATCCGTTCTTCTACAT |
| VvGRF2 | CTGGTGGGACTCATTTCGC | AGGTTGCCTGGCTGTTATTG |
| VvGRF3 | GCCTCAATGACTTCTGAAACCT | CGATGATGCCTGTAATCCCTAT |
| VvGRF4 | CAGAAAGCCTGTGGAAGTTATG | TGTAGCAAGAAGGGTGACGAT |
| VvGRF5 | CGCCTTTGGACAGCACAT | CGAGACTGACCCTCCTGTTTC |
| VvGRF6 | TCGTTCTATCTTCAACCTCCGT | AGTACTTCTGATTGAGTGCCACAT |
| VvGRF7 | GAAGCCCTTGATTCCGTTG | TCTCACAATACTTCTGATCGGC |
| VvGRF8 | CGCCACTCAGGCTATCTCG | AAGCAGGCTCCCATGTTGT |
| EF1-α | AGGAGGCAGCCAACTTCACC | CAAACCCTGCATCACCATTC |
| 基因 Gene | 登录号 Accession No. | 基因ID Gene locus ID | VCost.v3 ID | 起始与终止位点/bp Start and end position | CDS/bp | ORF/ aa | 亚细胞定位 Subcellular localization |
|---|---|---|---|---|---|---|---|
| VvGRF1 | XP_002281639.1 | GSVIVT01019913001 | Vitvi02g00449 | 4 421 518-4 424 601 | 639 | 213 | 细胞核Nucleus |
| VvGRF2 | XP_002277233.3 | GSVIVT01033800001 | Vitvi08g01498 | 17 725 858-17 728 519 | 1 176 | 392 | 细胞核Nucleus |
| VvGRF3 | XP_010654438.1 | GSVIVT01016762001 | Vitvi09g00107 | 1 135 208-1 137 036 | 1 092 | 364 | 细胞核Nucleus |
| VvGRF4 | XP_010656126.2 | GSVIVT01015095001 | Vitvi11g00092 | 1 000 359-1 001 747 | 1 164 | 388 | 细胞核Nucleus |
| VvGRF5 | XP_002270427.2 | GSVIVT01024326001 | Vitvi16g00073 | 975 755-979 958 | 1 122 | 374 | 细胞核Nucleus |
| VvGRF6 | XP_002271355.3 | GSVIVT01038629001 | Vitvi16g01354 | 21 346 554-21 349 482 | 1 812 | 604 | 细胞核Nucleus |
| VvGRF7 | CBI19365.3 | GSVIVT01009299001 | Vitvi18g00623 | 7 086 633-7 089 964 | 1 428 | 476 | 细胞核Nucleus |
| VvGRF8 | CBI33069.3 | GSVIVT01007165001 | Vitvi07g01551 | 30 596 800-30 600 327 | 1 674 | 558 | 细胞核Nucleus |
Table 2 Information of GRF family gene in grape genome
| 基因 Gene | 登录号 Accession No. | 基因ID Gene locus ID | VCost.v3 ID | 起始与终止位点/bp Start and end position | CDS/bp | ORF/ aa | 亚细胞定位 Subcellular localization |
|---|---|---|---|---|---|---|---|
| VvGRF1 | XP_002281639.1 | GSVIVT01019913001 | Vitvi02g00449 | 4 421 518-4 424 601 | 639 | 213 | 细胞核Nucleus |
| VvGRF2 | XP_002277233.3 | GSVIVT01033800001 | Vitvi08g01498 | 17 725 858-17 728 519 | 1 176 | 392 | 细胞核Nucleus |
| VvGRF3 | XP_010654438.1 | GSVIVT01016762001 | Vitvi09g00107 | 1 135 208-1 137 036 | 1 092 | 364 | 细胞核Nucleus |
| VvGRF4 | XP_010656126.2 | GSVIVT01015095001 | Vitvi11g00092 | 1 000 359-1 001 747 | 1 164 | 388 | 细胞核Nucleus |
| VvGRF5 | XP_002270427.2 | GSVIVT01024326001 | Vitvi16g00073 | 975 755-979 958 | 1 122 | 374 | 细胞核Nucleus |
| VvGRF6 | XP_002271355.3 | GSVIVT01038629001 | Vitvi16g01354 | 21 346 554-21 349 482 | 1 812 | 604 | 细胞核Nucleus |
| VvGRF7 | CBI19365.3 | GSVIVT01009299001 | Vitvi18g00623 | 7 086 633-7 089 964 | 1 428 | 476 | 细胞核Nucleus |
| VvGRF8 | CBI33069.3 | GSVIVT01007165001 | Vitvi07g01551 | 30 596 800-30 600 327 | 1 674 | 558 | 细胞核Nucleus |
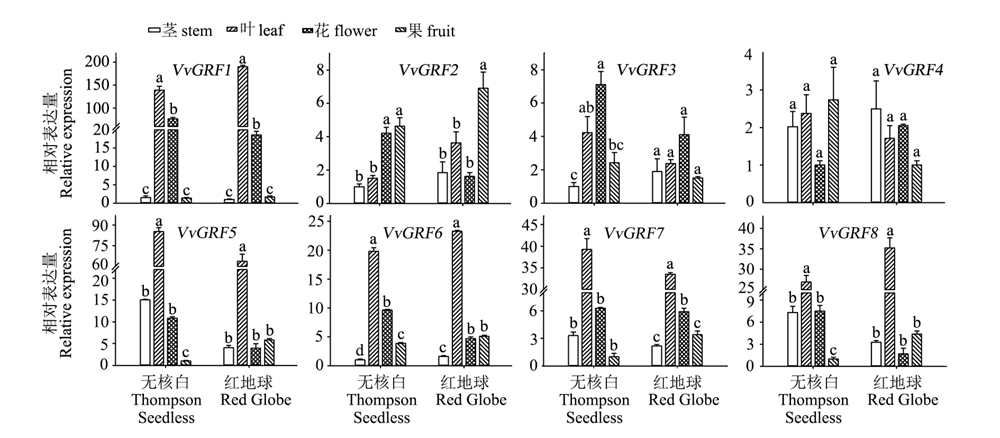
Fig. 5 Expression of GRF family genes in‘Thompson Seedless’and‘Red Globe’grape in different organs(qRT-PCR) Different letters indicate statistically significant differences among different organs in same cultivar. ANOVA with a Tukey post-hoc analysis,P < 0.05.
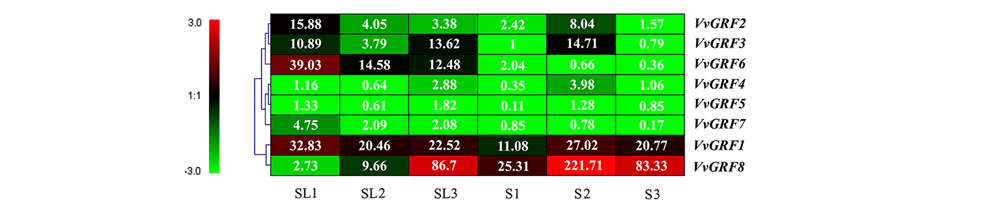
Fig. 6 Expression of GRF genes in the‘Red Globe’and‘Centennial Seedless’hybridization separation seeded(S)and seedless(SL) progenies during seed development

Fig. 7 Expression of GRF genes in seeded and seedless grapes during seed development Different letters indicate significant differences among different cultivars in the same day. ANOVA with a Tukey post-hoc analysis,P < 0.05.
| [1] |
Ai G, Zhang D, Huang R, Zhang S, Li W, Ahiakpa J K, Zhang J. 2020. Genome-wide identification and molecular characterization of the growth-regulating factors-interacting factor gene family in tomato. Genes, 11 (12):1435.
doi: 10.3390/genes11121435 URL |
| [2] | Bailey T L, Boden M, Buske F A, Frith M, Grant C E, Clementi L, Ren J, Li W W, Noble W S. 2009. MEME SUITE:tools for motif discovery and searching. Nucleic Acids Research, 37 (s2):W202-W208. |
| [3] | Cao Y, Han Y, Jin Q, Lin Y, Cai Y. 2016. Comparative genomic analysis of the GRF genes in Chinese pear(Pyrus bretschneideri Rehd),poplar(Populous),grape(Vitis vinifera),Arabidopsis and rice(Oryza sativa). Frontiers in Plant Science, 7:1750. |
| [4] |
Chen C, Chen H, Zhang Y, Thomas H R, Frank M H, He Y, Xia R. 2020. TBtools:an integrative toolkit developed for interactive analyses of big biological data. Molecular Plant, 13 (8):1194-1202.
doi: 10.1016/j.molp.2020.06.009 URL |
| [5] |
Choi D, Kim J H, Kende H. 2004. Whole genome analysis of the OsGRF gene family encoding plant-specific putative transcription activators in rice(Oryza sativa L.). Plant and Cell Physiology, 45 (7):897-904.
doi: 10.1093/pcp/pch098 URL |
| [6] |
Horiguchi G, Kim G T, Tsukaya H. 2005. The transcription factor AtGRF5 and the transcription coactivator AN3 regulate cell proliferation in leaf primordia of Arabidopsis thaliana. Plant Journal, 43 (1):68-78.
pmid: 15960617 |
| [7] |
Howe E A, Sinha R, Schlauch D, Quackenbush J. 2011. RNA-Seq analysis in MeV. Bioinformatics, 27 (22):3209-3210.
doi: 10.1093/bioinformatics/btr490 URL |
| [8] |
Hu B, Jin J, Guo A Y, Zhang H, Luo J, Gao G. 2015. GSDS 2.0:an upgraded gene feature visualization server. Bioinformatics, 31 (8):1296-1297.
doi: 10.1093/bioinformatics/btu817 URL |
| [9] |
Khatun K, Robin A H K, Park J I, Nath U K, Kim C K, Lim K B, Nou I S, Chung M Y. 2017. Molecular characterization and expression profiling of tomato GRF transcription factor family genes in response to abiotic stresses and phytohormones. International Journal of Molecular Sciences, 18 (5):1056.
doi: 10.3390/ijms18051056 URL |
| [10] |
Kim J H, Choi D, Kende H. 2003. The AtGRF family of putative transcription factors is involved in leaf and cotyledon growth in Arabidopsis. Plant Journal, 36 (1):94-104.
doi: 10.1046/j.1365-313X.2003.01862.x URL |
| [11] | Kim J H, Kende H. 2004. A transcriptional coactivator,AtGIF1,is involved in regulating leaf growth and morphology in Arabidopsis. Proceedings of the National Academy of Sciences, 101 (36):13374-13379. |
| [12] |
Kim J H, Lee B H. 2006. Growth-regulating factor 4 of Arabidopsis thaliana is required for development of leaves,cotyledons,and shoot apical meristem. Journal of Plant Biology, 49 (6):463-468.
doi: 10.1007/BF03031127 URL |
| [13] |
Kim J S, Mizoi J, Kidokoro S, Maruyama K, Nakajima J, Nakashima K, Mitsuda N, Takiguchi Y, Ohme-Takagi M, Kondou Y, Yoshizumi T, Matsui M, Shinozaki K, Yamaguchi-Shinozaki K. 2012. Arabidopsis GROWTH-REGULATING FACTOR7 functions as a transcriptional repressor of abscisic acid- and osmotic stress-responsive genes,including DREB2A. Plant Cell, 24 (8):3393-3405.
doi: 10.1105/tpc.112.100933 URL |
| [14] |
Krzywinski M, Schein J, Birol I, Connors J, Gascoyne R, Horsman D, Jones S J, Marra M A. 2009. Circos:an information aesthetic for comparative genomics. Genome Research, 19 (9):1639-1645.
doi: 10.1101/gr.092759.109 pmid: 19541911 |
| [15] |
Kumar S, Stecher G, Li M, Knyaz C, Tamura K. 2018. MEGA X:molecular evolutionary genetics analysis across computing platforms. Molecular Biology and Evolution, 35 (6):1547-1549.
doi: 10.1093/molbev/msy096 URL |
| [16] | Lee T H, Tang H, Wang X, Paterson A H. 2013. PGDD:a database of gene and genome duplication in plants. Nucleic Acids Research, 41:D1152-1158. |
| [17] | Lescot M, Déhais P, Thijs G, Marchal K, Moreau Y, Van de Peer Y, Rouzé P, Rombauts S. 2002. PlantCARE,a database of plant cis-acting regulatory elements and a portal to tools for in silico analysis of promoter sequences. Nucleic Acids Research, 30 (1):325-327. |
| [18] |
Li S, Gao F, Xie K, Zeng X, Cao Y, Zeng J, He Z, Ren Y, Li W, Deng Q, Wang S, Zheng A, Zhu J, Liu H, Wang L, Li P. 2016. The OsmiR396c-OsGRF4-OsGIF 1 regulatory module determines grain size and yield in rice. Plant Biotechnology Journal, 14 (11):2134-2146.
doi: 10.1111/pbi.12569 URL |
| [19] | Liu Pudong, Zhang Shuting, Chen Xiaohui, Su Liyao, Yao Deheng, Li Hansheng, Zhang Zihao. 2020. Genomic identification and expression patterns of the longan GRF family. Chinese Journal of Applied and Environmental Biology, 26 (2):236-245. (in Chinese) |
| 刘蒲东, 张舒婷, 陈晓慧, 苏立遥, 姚德恒, 李汉生, 张梓浩, 陈裕坤, 林玉玲, 赖钟雄. 2020. 龙眼GRF家族全基因组鉴定及表达模式. 应用与环境生物学报, 26 (2):236-245. | |
| [20] |
Liu X, Guo L X, Jin L F, Liu Y Z, Liu T, Fan Y H, Peng S A. 2016. Identification and transcript profiles of citrus growth-regulating factor genes involved in the regulation of leaf and fruit development. Molecular Biology Reports, 43 (10):1059-1067.
doi: 10.1007/s11033-016-4048-1 URL |
| [21] |
Livak K J, Schmittgen T D. 2001. Analysis of relative gene expression data using real-time quantitative PCR and the 2-ΔΔCT method. Methods, 25 (4):402-408.
doi: 10.1006/meth.2001.1262 pmid: 11846609 |
| [22] |
Omidbakhshfard M A, Proost S, Fujikura U, Mueller-Roeber B. 2015. Growth-regulating factors(GRFs):a small transcription factor family with important functions in plant biology. Molecular Plant, 8 (7):998-1010.
doi: 10.1016/j.molp.2015.01.013 pmid: 25620770 |
| [23] |
Sun P, Zhang W, Wang Y, He Q, Shu F, Liu H, Wang J, Wang J, Yuan L, Deng H. 2016. OsGRF4 controls grain shape,panicle length and seed shattering in rice. Journal of Integrative Plant Biology, 58 (10):836-847.
doi: 10.1111/jipb.12473 URL |
| [24] |
van der Knaap E, Kim J H, Kende H. 2000. A novel gibberellin-induced gene from rice and its potential regulatory role in stem growth. Plant Physiology, 122 (3):695-704.
pmid: 10712532 |
| [25] |
Wang F, Qiu N, Ding Q, Li J, Zhang Y, Li H, Gao J. 2014. Genome-wide identification and analysis of the growth-regulating factor family in Chinese cabbage(Brassica rapa L. ssp. pekinensis). BMC Genomics, 15:807.
doi: 10.1186/1471-2164-15-807 URL |
| [26] |
Wang L, Ahmad B, Liang C, Shi X X, Sun R Y, Zhang S L, Du G Q. 2020a. Bioinformatics and expression analysis of histone modification genes in grapevine predict their involvement in seed development,powdery mildew resistance,and hormonal signaling. BMC Plant Biology, 20 (1):412.
doi: 10.1186/s12870-020-02618-7 URL |
| [27] |
Wang L, Hu X, Jiao C, Li Z, Fei Z, Yan X, Liu C, Wang Y, Wang X. 2016. Transcriptome analyses of seed development in grape hybrids reveals a possible mechanism influencing seed size. BMC Genomics, 17 (1):898.
pmid: 27829355 |
| [28] |
Wang L, Yin X, Cheng C, Wang H, Guo R, Xu X, Zhao J, Zheng Y, Wang X. 2015. Evolutionary and expression analysis of a MADS-box gene superfamily involved in ovule development of seeded and seedless grapevines. Molecular Genetics and Genomics, 290 (3):825-846.
doi: 10.1007/s00438-014-0961-y pmid: 25429734 |
| [29] |
Wang L, Zhang S L, Zhang X M, Hu X Y, Guo C L, Wang X P, Song J Y. 2018. Evolutionary and expression analysis of Vitis vinifera OFP gene family. Plant Systematics and Evolution, 304 (8):995-1008.
doi: 10.1007/s00606-018-1528-x URL |
| [30] |
Wang J N, Zhou H J, Zhao Y Q, Sun P B, Tang F, Song X Q, Lu M Z. 2020b. Characterization of poplar growth regulating factors and analysis of their function in leaf size control. BMC Plant Biology, 20:509.
doi: 10.1186/s12870-020-02699-4 URL |
| [31] | Wang Xiaofang, Chang Qinxiang, Wang Shufang, Yan Zhao, Li Xuyan, Gary Gao, Ji Wei. 2021. Bioinformatics analysis of GRF gene family in grape. Molecular Plant Breeding, 19 (18):5975-5983. (in Chinese) |
| 王晓芳, 昌秦湘, 王淑芳, 闫钊, 李戌彦, Gary Gao, 纪薇. 2021. 葡萄GRF基因家族的生物信息学分析. 分子植物育种, 19 (18):5975-5983. | |
| [32] | Wei Yanhong, Liu Zhen, Li Ke, Meng Yuan, Wang Hui, Mao Jiangping, Ma Doudou, Li Shaohuan, Ma Juanjuan, Lu Xian, Zhang Dong. 2020. Genome-wide identification and expression analysis of miR396 family during adventitious root development in apple. Acta Horticulturae Sinica, 47 (7):1237-1252. (in Chinese) |
| 韦燕红, 刘桢, 李珂, 孟媛, 汪蕙, 毛江萍, 马豆豆, 李少欢, 马娟娟, 卢显, 张东. 2020. 苹果miR396 家族鉴定及在不定根发育过程中的表达分析. 园艺学报, 47 (7):1237-1252. | |
| [33] |
Wu L, Zhang D, Xue M, Qian J, He Y, Wang S. 2014. Overexpression of the maize GRF10,an endogenous truncated growth-regulating factor protein,leads to reduction in leaf size and plant height. Journal of Integrative Plant Biology, 56 (11):1053-1063.
doi: 10.1111/jipb.12220 URL |
| [34] | Wu Z J, Wang W L, Zhuang J. 2017. Developmental processes and responses to hormonal stimuli in tea plant(Camellia sinensis)leaves are controlled by GRF and GIF gene families. Functional & Integrative Genomics, 17 (5):503-512. |
| [35] |
Zhang D F, Li B, Jia G Q, Zhang T F, Dai J R, Li J S, Wang S C. 2008. Isolation and characterization of genes encoding GRF transcription factors and GIF transcriptional coactivators in maize(Zea mays L.). Plant Science, 175 (6):809-817.
doi: 10.1016/j.plantsci.2008.08.002 URL |
| [36] |
Zhang J, Li Z, Jin J, Xie X, Zhang H, Chen Q, Luo Z, Yang J. 2018. Genome-wide identification and analysis of the growth-regulating factor family in tobacco(Nicotiana tabacum). Gene, 639:117-127.
doi: 10.1016/j.gene.2017.09.070 URL |
| [37] |
Zhang S, Wang L, Sun X, Li Y, Yao J, van Nocker S, Wang X. 2019. Genome-wide analysis of the YABBY gene family in grapevine and functional characterization of VvYABBY4. Frontiers in Plant Science, 10:1207.
doi: 10.3389/fpls.2019.01207 URL |
| [38] | Zhao Huixia, Li Fengxia, Guo Rui, Chen Chanyou. 2020. Genome-wide characterization and expression analysis of GRF gene family in Capsicum. Acta Horticulturae Sinica, 47 (11):2145-2160. (in Chinese) |
| 赵慧霞, 李凤霞, 郭瑞, 陈禅友. 2020. 辣椒属GRF基因家族全基因组鉴定和表达分析. 园艺学报, 47 (11):2145-2160. | |
| [39] | Zheng L W, Ma J J, Song C H, Zhang L Z, Gao C, Zhang D, An N, Mao J P, Han M Y. 2018. Genome-wide identification and expression analysis of GRF genes regulating apple tree architecture. Tree Genetics & Genomes, 14 (4):54. |
| [40] |
Zhou H J, Song X Q, Wei K L, Zhao Y Q, Jiang C, Wang J N, Tang F, Lu M Z. 2019. Growth-regulating factor 15 is required for leaf size control in Populus. Tree Physiology, 39 (3):381-390.
doi: 10.1093/treephys/tpy107 URL |
| [41] |
Zhu Y X, Li Y D, Zhang S L, Zhang X M, Yao J, Luo Q W, Sun F, Wang X P. 2019. Genome-wide identification and expression analysis reveal the potential function of ethylene responsive factor gene family in response to Botrytis cinerea infection and ovule development in grapes(Vitis vinifera L.). Plant Biology, 21 (4):571-584.
doi: 10.1111/plb.12943 pmid: 30468551 |
| [1] | WANG Xiaochen, NIE Ziye, LIU Xianju, DUAN Wei, FAN Peige, and LIANG Zhenchang, . Effects of Abscisic Acid on Monoterpene Synthesis in‘Jingxiangyu’Grape Berries [J]. Acta Horticulturae Sinica, 2023, 50(2): 237-249. |
| [2] | YUAN Xin, XU Yunhe, ZHANG Yupei, SHAN Nan, CHEN Chuying, WAN Chunpeng, KAI Wenbin, ZHAI Xiawan, CHEN Jinyin, GAN Zengyu. Studies on AcAREB1 Regulating the Expression of AcGH3.1 During Postharvest Ripening of Kiwifruit [J]. Acta Horticulturae Sinica, 2023, 50(1): 53-64. |
| [3] | ZHAO Xueyan, WANG Qi, WANG Li, WANG Fangyuan, WANG Qing, LI Yan. Comparative Transcriptome Analysis of Differential Expression in Different Tissues of Corydalis yanhusuo [J]. Acta Horticulturae Sinica, 2023, 50(1): 177-187. |
| [4] | WANG Baoliang, LIU Fengzhi, JI Xiaohao, WANG Xiaodi, SHI Xiangbin, ZHANG Yican, LI Peng, and WANG Haibo. A New Early Ripening Grape Cultivar‘Huapu Zaoyu’for Table [J]. Acta Horticulturae Sinica, 2022, 49(S2): 33-34. |
| [5] | WANG Baoliang, WANG Haibo, JI Xiaohao, WANG Xiaodi, SHI Xiangbin, WANG Zhiqiang, WANG Xiaolong, and LIU Fengzhi. A New Middle Ripening Grape Cultivar‘Huapu Huangyu’for Table [J]. Acta Horticulturae Sinica, 2022, 49(S2): 35-36. |
| [6] | A Late-maturing Seedless Grape Cultivar‘Zilongzhu’. A Late-maturing Seedless Grape Cultivar‘Zilongzhu’ [J]. Acta Horticulturae Sinica, 2022, 49(S2): 37-38. |
| [7] | SHI Xiaoxin, DU Guoqiang, YANG Lili, QIAO Yuelian, HUANG Chengli, WANG Suyue, ZHAO Yuexin, WEI Xiaohui, WANG Li, and QI Xiangli. A Late-ripening Seedless Grape Cultivar‘Hongfeng Wuhe’ [J]. Acta Horticulturae Sinica, 2022, 49(S2): 39-40. |
| [8] | WU Yueyan, CHEN Tianchi, WANG Liru, HAN Shanqi, and FU Tao. A New Table Grape Cultivar‘Yongzaohong’ [J]. Acta Horticulturae Sinica, 2022, 49(S2): 41-42. |
| [9] | WANG Xiaoyue, YAN Ailing, ZHANG Guojun, WANG Huiling, REN Jiancheng, LIU Zhenhua, SUN Lei, and XU Haiying, . A New Grape Cultivar‘Ruidu Wanhong’ [J]. Acta Horticulturae Sinica, 2022, 49(S1): 29-30. |
| [10] | GAO Yanlong, WU Yuxia, ZHANG Zhongxing, WANG Shuangcheng, ZHANG Rui, ZHANG De, WANG Yanxiu. Bioinformatics Analysis of Apple ELO Gene Family and Its Expression Analysis Under Low Temperature Stress [J]. Acta Horticulturae Sinica, 2022, 49(8): 1621-1636. |
| [11] | QIU Ziwen, LIU Linmin, LIN Yongsheng, LIN Xiaojie, LI Yongyu, WU Shaohua, YANG Chao. Cloning and Functional Analysis of the MbEGS Gene from Melaleuca bracteata [J]. Acta Horticulturae Sinica, 2022, 49(8): 1747-1760. |
| [12] | WANG Yongjian, KONG Junhua, FAN Peige, LIANG Zhenchang, JIN Xiuliang, LIU Buchun, DAI Zhanwu. Grape Phenome High-throughput Acquisition and Analysis Methods:A Review [J]. Acta Horticulturae Sinica, 2022, 49(8): 1815-1832. |
| [13] | ZHENG Lin, WANG Shuai, LIU Yunuo, DU Meixia, PENG Aihong, HE Yongrui, CHEN Shanchun, ZOU Xiuping. Gene Cloning and Expression Analysis of NAC Gene in Citrus in Response to Huanglongbing [J]. Acta Horticulturae Sinica, 2022, 49(7): 1441-1457. |
| [14] | MA Weifeng, LI Yanmei, MA Zonghuan, CHEN Baihong, MAO Juan. Identification of Apple POD Gene Family and Functional Analysis of MdPOD15 Gene [J]. Acta Horticulturae Sinica, 2022, 49(6): 1181-1199. |
| [15] | WEI Xiaoyu, WANG Yuejin. Correlation Between Anatomical Structure and Resistance to Powdery Mildew in Chinese Wild Vitis Species [J]. Acta Horticulturae Sinica, 2022, 49(6): 1200-1212. |
| Viewed | ||||||
|
Full text |
|
|||||
|
Abstract |
|
|||||
Copyright © 2012 Acta Horticulturae Sinica 京ICP备10030308号-2 国际联网备案号 11010802023439
Tel: 010-82109523 E-Mail: yuanyixuebao@126.com
Support by: Beijing Magtech Co.Ltd