
Acta Horticulturae Sinica ›› 2023, Vol. 50 ›› Issue (6): 1187-1202.doi: 10.16420/j.issn.0513-353x.2022-0297
• Genetic & Breeding·Germplasm Resources·Molecular Biology • Previous Articles Next Articles
RUAN Ruoxin1, LUO Huifeng1, ZHANG Chen1, HUANG Kangkang1, XI Dujun1, PEI Jiabo1, XING Mengyun1,2, LIU Hui1,*( )
)
Received:2023-01-28
Revised:2023-05-12
Online:2023-06-25
Published:2023-06-27
Contact:
* (E-mail:CLC Number:
RUAN Ruoxin, LUO Huifeng, ZHANG Chen, HUANG Kangkang, XI Dujun, PEI Jiabo, XING Mengyun, LIU Hui. Transcriptome Analysis of Flower Buds of Sweet Cherry Cultivars with Different Chilling Requirements During Dormancy Stages in Hangzhou[J]. Acta Horticulturae Sinica, 2023, 50(6): 1187-1202.
Add to citation manager EndNote|Ris|BibTeX
URL: https://www.ahs.ac.cn/EN/10.16420/j.issn.0513-353x.2022-0297
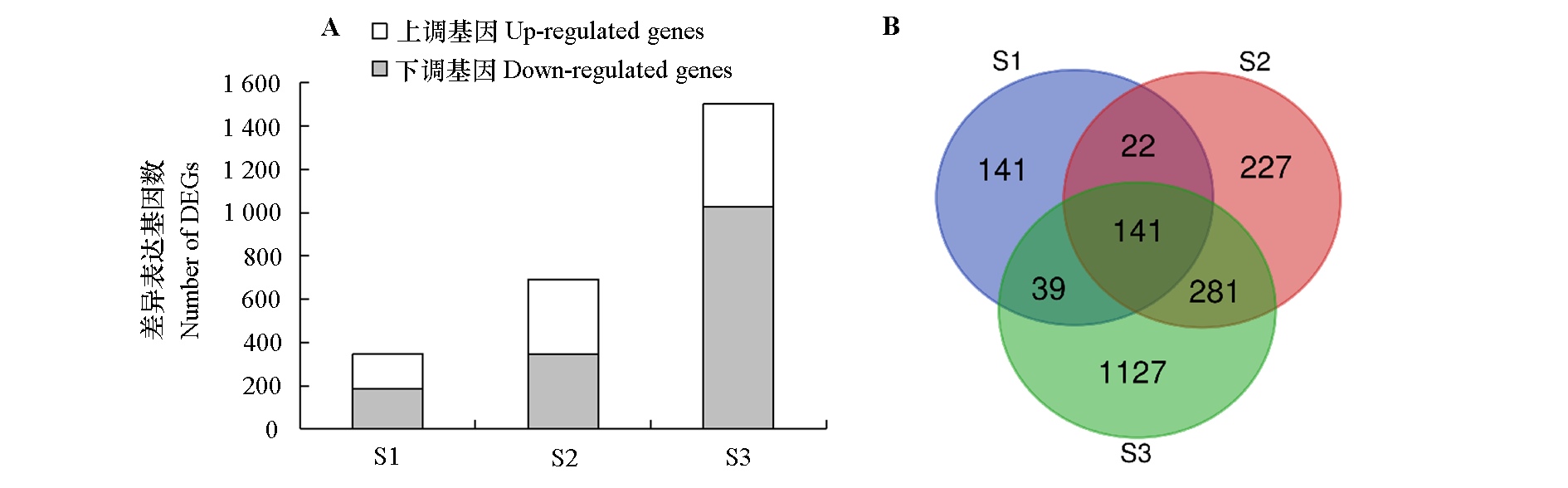
Fig. 2 Number(A)and Venn diagram(B)of DEGs in flower buds of sweet cherry‘Brooks’and‘Summit’at different dormancy stages S1:Pre-dormancy stage;S2:Endormancy stage;S3:Pre-budbreak stage. The same below.
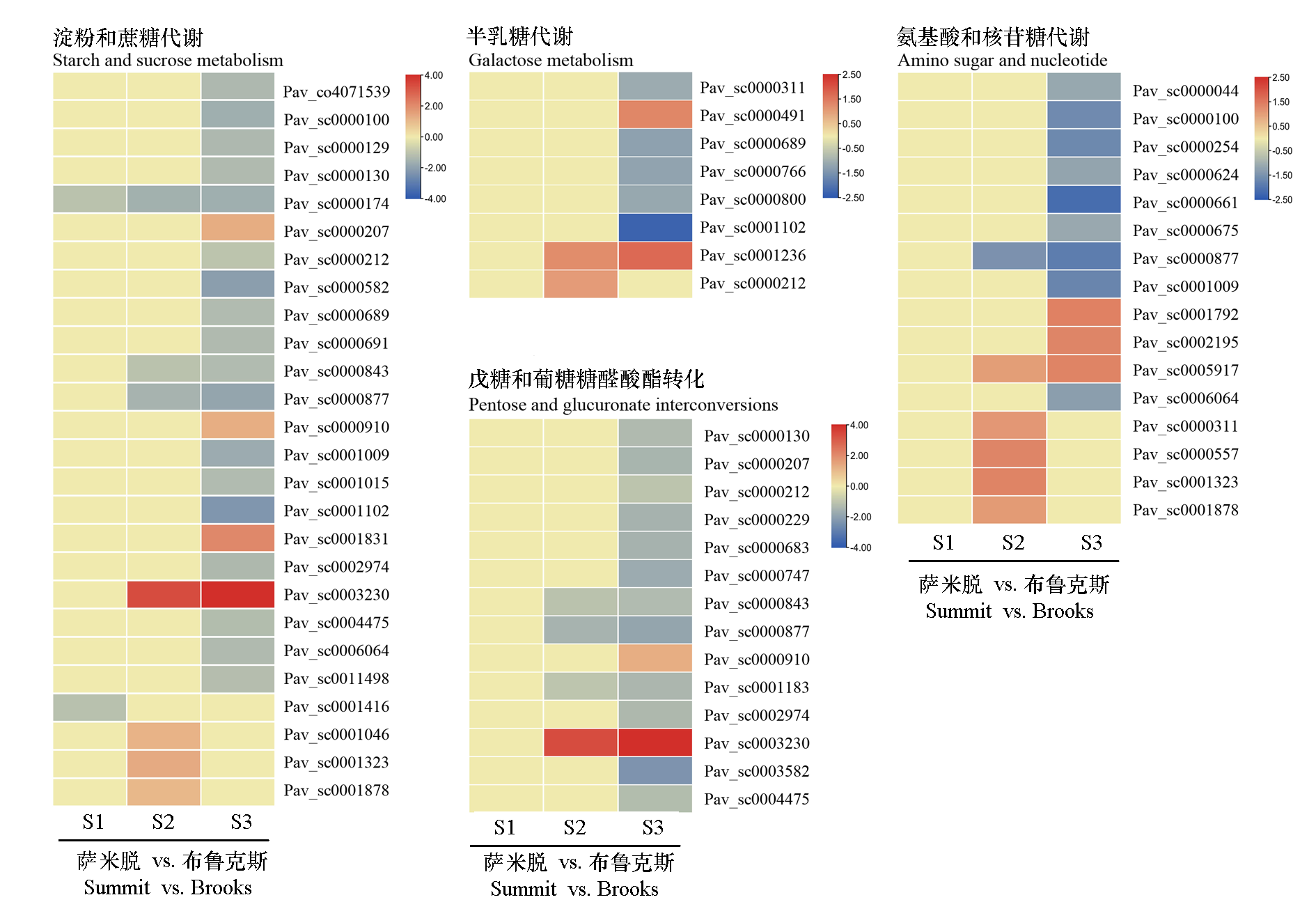
Fig. 6 Expression profiles of DEGs involved in carbohydrate metabolism related pathways between flower buds of sweet cherry‘Brooks’and‘Summit’at different dormancy stages The values shown in the heatmap represent the average relative gene expression levels(log2FC)of DEGs in‘Summit’versus‘Brooks’at the same stage.
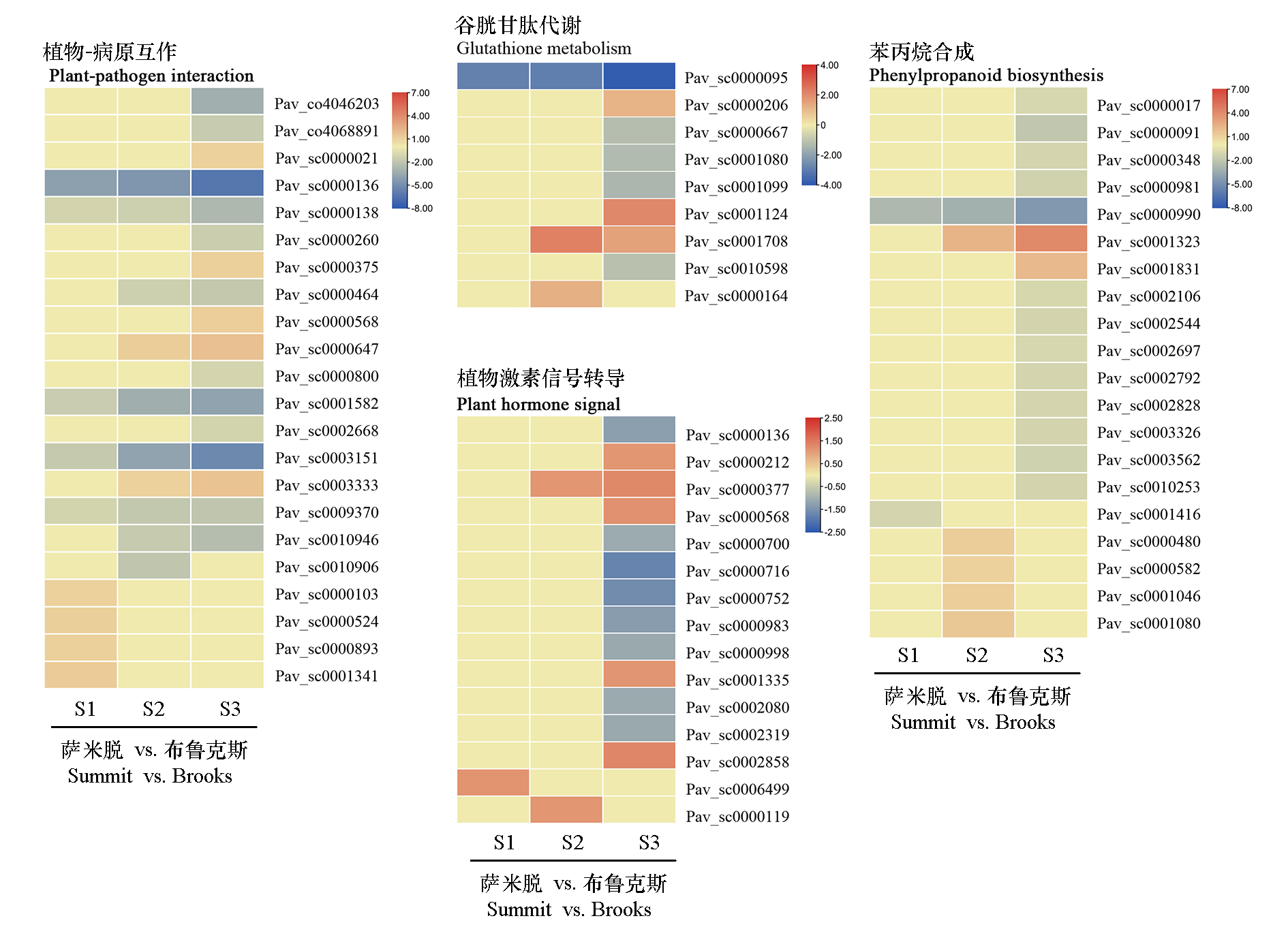
Fig. 7 Expression profiles of DEGs involved in stress response-related pathways between flower buds of sweet cherry‘Brooks’and‘Summit’at different dormancy stages The values shown in the heatmap represent the average relative gene expression levels(log2FC)of DEGs in‘Summit’versus‘Brooks’at the same stage.
| [1] |
Beauvieux R, Wenden B, Dirlewanger E. 2018. Bud dormancy in perennial fruit tree species :a pivotal role for oxidative cues. Frontiers in Plant Science, 9:1-13.
doi: 10.3389/fpls.2018.00001 URL |
| [2] |
Casanova-Sáez R, Mateo-Bonmatí E, Ljung K. 2021. Auxin Metabolism in Plants. Cold Spring Harbor Perspectives in Biology, 13 (3):a039867. doi:10.1101/cshperspect.a039867.
doi: 10.1101/cshperspect.a039867 URL |
| [3] | Chen Xiao-dan, Wang Lei, Huang Jia-bin, Liao Zhang-hua, Ma Chao, Wang Shi-ping, Zhang Cai-xi. 2017. Preliminary study on evaluation of cooling requirements of different varieties of sweet cherry. South China Fruits, 46 (3):109-112. (in Chinese) |
| 陈晓丹, 王磊, 黄嘉赟, 廖樟华, 马超, 王世平, 张才喜. 2017. 甜樱桃不同品种需冷量评估初探. 中国南方果树, 46 (3):109-112. | |
| [4] | Cui Dong-dong, Li Bo, Wang Shi-ping. 2014. Cultivation techniques of sweet cherry in Yangtze River Delta region. South China Fruits, 43 (6):141-144. (in Chinese) |
| 崔冬冬, 李勃, 王世平. 2014. 长三角地区甜樱桃栽培技术. 中国南方果树, 43 (6):141-144. | |
| [5] | Cui Jian-chao, Wang Wen-hui, Jia Xiao-hui, Wang Zhi-hua, Tong Wei. 2017. The domestic industry problems from the sweet cherry import situation its development countermeasure for the future. Journal of Fruit Science, 34 (5):620-631. (in Chinese) |
| 崔建潮, 王文辉, 贾晓辉, 王志华, 佟伟. 2017. 从国内外甜樱桃生产现状看国内甜樱桃产业存在的问题及发展对策. 果树学报, 34 (5): 620-631. | |
| [6] |
Fennell A, Schlauch K, Gouthu S, Deluc L, Khadka V, Sreekantan L, Grimplet J, Cramer G, Mathiason K. 2015. Short day transcriptomic programming during induction of dormancy in grapevine. Frontiers in Plant Science, 6:834.
doi: 10.3389/fpls.2015.00834 pmid: 26582400 |
| [7] | Gao Hua-jun, Yu Yun-zheng, Wang Cai-bo, Wang Jia-xi. 2012. Causes research progress of abnormal fruit-setting of sweet cherry in the middle lower reaches of Yangtze River. Deciduous Fruits, 44 (2):17-21. (in Chinese) |
| 高华君, 于云政, 王彩波, 王家喜. 2012. 长江中下游地区甜樱桃坐果异常的原因及研究进展. 落叶果树, 44 (2):17-21. | |
| [8] |
Götz S, García-Gómez J, Terol J, Williams T, Nagaraj S, Nueda M, Robles M, Talón M, Dopazo J, Conesa A. 2008. High-throughput functional annotation data mining with the Blast2GO suite. Nucleic Acids Research, 36 (10):3420-3435.
doi: 10.1093/nar/gkn176 URL |
| [9] |
Kim D, Langmead B, Salzberg S. 2015. HISAT:a fast spliced aligner with low memory requirements. Nature Methods, 12 (4):357-360.
doi: 10.1038/NMETH.3317 |
| [10] | Li Bo, Li Shu-ping, Xie Zhao-sen, Xu Wen-ping, Zhang Cai-xi, Liu Qing-zhong, Liu Cheng-lian, Wang Shi-ping. 2010. Research on biological characteristics flower bud differentiation in sweet cherry in Shanghai Yantai. Journal of Fruit Science, 27 (3):349-354. (in Chinese) |
| 李勃, 李淑平, 谢兆森, 许文平, 张才喜, 刘庆忠, 刘成连, 王世平. 2010. 上海和烟台地区甜樱桃生物学特性及花芽分化进程研究. 果树学报, 27 (3):349-354. | |
| [11] |
Li Bo, Xia Xiu-ying, Liu Si. 2015. Changes in physiological biochemical properties variation in DNA methylation patterns during dormancy dormancy release in blueberry(Vaccinium corymbosum L.). Plant Physiology Journal, 51 (7):1133-1141. (in Chinese)
doi: 10.1104/pp.51.6.1133 URL |
| 李波, 夏秀英, 刘思. 2015. 蓝莓花芽休眠与解除过程中生理生化变化及DNA甲基化差异分析. 植物生理学报, 51 (7):1133-1141. | |
| [12] |
Linsley-Noakes G C, Allan P. 1994. Comparison of 2 models for the prediction of rest completion in peaches. Scientia Horticulturae, 59:107-113.
doi: 10.1016/0304-4238(94)90077-9 URL |
| [13] | Liu Cong-li, Zhao Gai-rong, Li Ming, Li Yu-hong, Qi Xi-liang. 2017. Determination of chilling requirements and cluster analysis of 66 sweet cherry cultivars. Journal of Fruit Science, 34 (4):464-472. (in Chinese) |
| 刘聪利, 赵改荣, 李明, 李玉红, 齐希梁. 2017. 66个甜樱桃品种需冷量的评价与聚类分析. 果树学报, 34 (4):464-472. | |
| [14] | Liu Guo-qin, Zheng Peng-hua, Teng Yuan-wen. 2012. Research progresses on genes associated with bud dormancy in deciduous fruit trees. Journal of Fruit Science, 29 (5):911-917. (in Chinese) |
| 刘国琴, 郑鹏华, 滕元文. 2012. 落叶果树芽休眠相关基因的研究进展. 果树学报, 29 (5):911-917. | |
| [15] | Liu Ren-dao, Liu Jian-jun. 2009. The study on the chilling requirement of sweet cherry cultivars. Northern Horticulture,(2):84-85. (in Chinese) |
| 刘仁道, 刘建军. 2009. 甜樱桃不同品种需冷量研究. 北方园艺,(2):84-85. | |
| [16] | Liu X, Zhang H, Zhao Y, Feng Z, Li Q, Yang H Q, Luan S, Li J, He Z H. 2013. Auxin controls seed dormancy through stimulation of abscisic acid signaling by inducing ARF-mediated ABI3 activation in Arabidopsis. Proceedings of the National Academy of Sciences of the United States of America, 110 (38):15485-15490. |
| [17] |
Love M I, Huber W, Anders S. 2014. Moderated estimation of fold change dispersion for RNA-seq data with DESeq2. Genome Biology, 15 (12):0550.
doi: 10.1186/s13059-014-0550-8 URL |
| [18] | Luo Hui-feng, Liao Yi-min, Shen Guo-zheng, Huang Kang-kang, Pei Jia-bo, Xi Dun-jun, Zhang Chen, Ruan Ruo-xin, Liu Hui. 2021a. Study on fruit cracking and quality of sweet cherry based on IoT technology. South China Fruits, 50 (5):124-129. (in Chinese) |
| 骆慧枫, 廖益民, 寿国忠, 黄康康, 裴嘉博, 郗笃隽, 张琛, 阮若昕, 刘辉. 2021a. 基于物联网技术的甜樱桃裂果及其品质研究. 中国南方果树, 50 (5):124-129. | |
| [19] | Luo Hui-feng, Shen Guo-zheng, Huang Kang-kang, Pei Jia-bo, Xi Dun-jun, Zhang Chen, Ruan Ruo-xin, Liu Hui. 2021b. Impact of hydrogen cyanamide on germination and flowering of sweet cherry in Hangzhou. Journal of Zhengjiang Agricultural Sciences, 62 (7):1331-1333. (in Chinese) |
| 骆慧枫, 沈国正, 黄康康, 裴嘉博, 郗笃隽, 张琛, 阮若昕, 刘辉. 2021b. 单氰胺处理对杭州地区甜樱桃萌芽和开花的影响. 浙江农业科学, 62 (7):1331-1333. | |
| [20] |
Mao X, Cai T, Olyarchuk J G, Wei L. 2005. Automated genome annotation pathway identification using the KEGG Orthology(KO)as a controlled vocabulary. Bioinformatics, 21 (19):3787-3793.
doi: 10.1093/bioinformatics/bti430 URL |
| [21] | Mohr T J, Mammarella N D, Hoff T, Woffenden B J, Jelesko J G, McDowell J M. 2010. The Arabidopsis downy mildew resistance gene RPP8 is induced by pathogens and salicylic acid and is regulated by W box cis elements. Molecular Plant-Microbe Iinteractions, 23 (10):1303-1315. |
| [22] |
Peng Fangfang, Long Zhijian, Wei Zhaoxin, Li Xunlan, Luo Youjin, Han Guohui. 2021. Association analysis of SCoT markers and leaf phenotypic traits in cherry germplasm. Acta Horticulturae Sinica, 48 (2):325-335. (in Chinese)
doi: 10.16420/j.issn.0513-353x.2020-0262 URL |
|
彭芳芳, 龙治坚, 魏召新, 李勋兰, 罗友进, 韩国辉. 2021. 樱桃种质SCoT分子标记与叶片表型性状关联分析. 园艺学报, 48 (2):325-335.
doi: 10.16420/j.issn.0513-353x.2020-0262 URL |
|
| [23] |
Pertea M, Pertea G M, Antonescu C M, Chang T C, Mendell J T, Salzberg S L. 2015. StringTie enables improved reconstruction of a transcriptome from RNA-seq reads. Nature Biotechnology, 33 (3):290-295.
doi: 10.1038/nbt.3122 pmid: 25690850 |
| [24] |
Prudencio Á S, Hoeberichts F A, Dicenta F, Martínez-Gómez P, Sánchez-Pérez R. 2021. Identification of early late flowering time candidate genes in endodormant ecodormant almond flower buds. Tree Physiology, 41 (4):589-605.
doi: 10.1093/treephys/tpaa151 pmid: 33200186 |
| [25] |
Robin A H K, Saha G, Laila R, Park J I, Kim H T, Nou I S. 2020. Expression and role of biosynthetic,transporter,receptor,and responsive genes for auxin signaling during clubroot disease development. International Journal of Molecular Sciences, 21 (15):5554.
doi: 10.3390/ijms21155554 URL |
| [26] | Rose L, Atwell S, Grant M, Holub E B. 2012. Parallel loss-of-function at the RPM1 bacterial resistance locus in Arabidopsis thaliana. Frontiers in Plant Science, 3:287. |
| [27] |
Rothkegel K, Sandoval P, Soto E, Ulloa L, Riveros A, Lillo-Carmona V, Cáceres-Molina J, Almeida A M, Meneses C. 2020. Dormant but active:chilling accumulation modulates the epigenome and transcriptome of Prunus avium during bud dormancy. Frontiers in Plant Science, 11:1115.
doi: 10.3389/fpls.2020.01115 pmid: 32765576 |
| [28] | Shen Guo-zheng, Liu Hui, Zhang Chen, Xi Du-jun, Pei Jia-bo, Huang Kang-kang. 2020. Comparison analyzation on the floral organ development quality of three sweet cherry cultivars in Hangzhou and Tai’an areas. Journal of Zhejiang University(Agriculture and Life Sciences), 46 (2):168-176. (in Chinese) |
| 沈国正, 刘辉, 张琛, 郗笃隽, 裴嘉博, 黄康康. 2020. 杭州和泰安地区3个甜樱桃品种花器官发育质量比较分析. 浙江大学学报(农业与生命科学版), 46 (2):168-176. | |
| [29] |
Shirasawa K, Isuzugawa K, Ikenaga M, Saito Y, Yamamoto T, Hirakawa H, Isobe S. 2017. The genome sequence of sweet cherry(Prunus avium)for use in genomics-assisted breeding. DNA Research, 24 (5):499-508.
doi: 10.1093/dnares/dsx020 pmid: 28541388 |
| [30] | Sun Li-xin, Zhang Ya-hong. 2016. Research progress on lifting of dormancy of deciduous fruit trees. China Fruits,(2):55-59. (in Chinese) |
| 孙利鑫, 张亚红. 2016. 落叶果树休眠期解除界定的研究进展. 中国果树,(2):55-59. | |
| [31] | Sun Ling-jun, Ma Li, Gao Sheng-hua, Zhao Hai-liang, Zhu Shao-kun. 2019. Research achievement on sugar metabolism in deciduous fruit trees during dormancy period. Northern Fruits,(2):1-4. (in Chinese) |
| 孙凌俊, 马丽, 高圣华, 赵海亮, 朱绍坤. 2019. 落叶果树休眠期糖代谢研究进展. 北方果树,(2):1-4. | |
| [32] | Sun Wan-xia, Liu Xun-ju, Wang Li, Wang Ji-yuan, Jiu Song-tao, Wang Lei, Wang Shi-ping, Zhang Cai-xi. 2021. Characteristics of low temperature and its impact on chilling accumulation of sweet cherries in the Yangtze River Delta. Journal of Fruit Science, 38 (11):1900-1910. (in Chinese) |
| 孙菀霞, 刘勋菊, 王丽, 王继源, 纠松涛, 王磊, 王世平, 张才喜. 2021. 长三角地区低温特征及其对甜樱桃蓄冷量的影响. 果树学报, 38 (11):1900-1910. | |
| [33] |
Tarancón C, González-Grandío E, Oliveros J C, Nicolas M, Cubas P. 2017. A conserved carbon starvation response underlies bud dormancy in woody herbaceous species. Frontiers in Plant Science, 8:788.
doi: 10.3389/fpls.2017.00788 pmid: 28588590 |
| [34] |
Vimont N, Fouché M, Campoy J A, Tong M, Arkoun M, Yvin J C, Wigge P A, Dirlewanger E, Cortijo S, Wenden B. 2019. From bud formation to flowering:transcriptomic state defines the cherry developmental phases of sweet cherry bud dormancy. BMC Genomics, 20:974.
doi: 10.1186/s12864-019-6348-z |
| [35] |
Vimont N, Schwarzenberg A, Domijan M, Donkpegan A S L, Beauvieux R le Dantec L, Arkoun M, Jamois F, Yvin J C, Wigge P A, Dirlewanger E, Cortijo S, Wenden B. 2021. Fine tuning of hormonal signaling is linked to dormancy status in sweet cherry flower buds. Tree Physiology, 41:544-561.
doi: 10.1093/treephys/tpaa122 pmid: 32975290 |
| [36] |
Wang Hui, Li Ling, Tan Yue, Li Dong-mei, Tan Qiu-ping, Chen Xiu-de, Gao Dong-sheng. 2011. Changes in carbohydrate metabolism related gene expression in nectarine floral buds during dormancy. Plant Physiology Journal, 47 (6):595-600. (in Chinese)
doi: 10.1104/pp.47.5.595 URL |
| 王慧, 李玲, 谭钺, 李冬梅, 谭秋平, 陈修德, 高东升. 2011. 休眠期间油桃花芽碳水化合物代谢及其相关基因的表达变化. 植物生理学报, 47 (6):595-600. | |
| [37] | Weiberger J H. 1950. Chilling requirements of peach varieties. Proceedings. American Society for Horticultural Science, 56:122-128. |
| [38] | Wen Luhua, Gao Zhihong, Huo Ximei, Zhuang Weibing, Lü Lin. 2017. Signal effect of H2O 2 on seasonal dormancy release induced by exogenous GA4 in Japanese apricots. Acta Horticulturae Sinica, 44 (2):255-267. (in Chinese) |
|
温璐华, 高志红, 霍希美, 庄维兵, 吕林. 2017. H2O2在外源GA4解除果梅季节性休眠中的信号作用. 园艺学报, 44 (2):255-267.
doi: 10.16420/j.issn.0513-353x.2016-0501 |
|
| [39] |
Yamane H, Ooka T, Jotatsu H, Hosaka Y, Sasaki R, Tao R. 2011. Expressional regulation of PpDAM5 and PpDAM6,peach(Prunus persica)dormancy-associated MADS-box genes,by low temperature and dormancy-breaking reagent treatment. Journal of Experimental Botany, 62 (10):3481-3488.
doi: 10.1093/jxb/err028 URL |
| [40] | Yang Bo, Wei Jia, Li Kunfeng, Wang Chengliang, Ni Junbei, Teng Yuanwen, Bai Songling. 2022. PpyERF060-PpyABF3-PpyMADS71 regulates ethylene signaling pathway- mediated pear bud dormancy process. Acta Horticulturae Sinica, 49 (10):2249-2262. (in Chinese) |
|
杨博, 魏佳, 李坤峰, 王程亮, 倪隽蓓, 滕元文, 白松龄. 2022. PpyERF060-PpyABF3-PpyMADS71调控乙烯信号通路介导的梨芽休眠进程. 园艺学报, 49 (10):2249-2262.
doi: 10.16420/j.issn.0513-353x.2022-0585 URL |
|
| [41] |
Yu J, Conrad A O, Decroocq V, Zhebentyayeva T, Williams D E, Bennett D, Roch G, Audergon J M, Dardick C, Liu Z, Abbott A G, Staton M E. 2020. Distinctive gene expression patterns define endodormancy to ecodormancy transition in apricot and peach. Frontiers in Plant Science, 11:180.
doi: 10.3389/fpls.2020.00180 pmid: 32180783 |
| [42] | Zhang Chen, Liu Hui, Xi Du-jun, Pei Jia-bo, Huang Kang-kang, Luo Hui-feng, Shen Guo-zheng. 2021a. Comparative study on flower bud inclusion quality in different development stages of sweet cherry from Hangzhou and Tai’an. Journal of Fruit Science, 38 (8):1308-1318. (in Chinese) |
| 张琛, 刘辉, 郗笃隽, 裴嘉博, 黄康康, 骆慧枫, 沈国正. 2021a. 杭州和泰安甜樱桃不同发育阶段花芽内含物及花芽质量的比较研究. 果树学报, 38 (8):1308-1318. | |
| [43] | Zhang Chen, Xi Dun-jun, Luo Hui-feng, Pei Jia-bo, Huang Kang-kang, Liu Hui. 2021b. The effect of foliar spray of urea in autumn on the defoliation of sweet cherry in Hangzhou. Chinese Agricultural Science Bulletin, 37 (33):64-68. (in Chinese) |
| 张琛, 郗笃隽, 骆慧枫, 裴嘉博, 黄康康, 刘辉. 2021b. 杭州地区秋施尿素对甜樱桃落叶效应的研究. 中国农学通报, 37 (33):64-68. | |
| [44] | Zhang Qing, Shen Guo-zheng, Liu Hui, Huang Kang-kang, Sun Chun-guang, Pei Jia-bo, Xi Du-jun, Zhang Chen, Pan Feng-rong. 2017. Introduction and cultivation of sweet cherry in high altitude mountainous area of Zhejiang Province. China Fruits,(1):26-29. (in Chinese) |
| 张青, 沈国正, 刘辉, 黄康康, 孙春光, 裴嘉博, 郗笃隽, 张琛, 潘凤荣. 2017. 浙江高海拔山区甜樱桃引种栽培初报. 中国果树,(1):26-29. | |
| [45] |
Zhang Z, Zhuo X, Zhao K, Zheng T, Han Y, Yuan C, Zhang Q. 2018. Transcriptome profiles reveal the crucial roles of hormone sugar in the bud dormancy of Prunus mume. Scientific Reports, 8 (1):5090.
doi: 10.1038/s41598-018-23108-9 |
| [46] |
Zhu You-yin, Li Yong-qiang, Xin De-dong, Chen Wen-rong, Shao Xu, Wang Yue, Guo Weidong. 2015. RNA-Seq-based transcriptome analysis of dormant flower buds of Chinese cherry ( Prunus pseudocerasus ). Gene, 555 (2):362-376.
doi: 10.1016/j.gene.2014.11.032 pmid: 25447903 |
| [1] | JIN Zhou, LU Shan, JIANG Junhao, LI Shouren, ZHANG Nan, JIANG Xiaoyu, WU Fan. Research Progress on Influencing Factors and Mechanisms of Flower Bud Differentiation in Horticultural Plants [J]. Acta Horticulturae Sinica, 2023, 50(5): 1151-1164. |
| [2] | LI Ke, SHEN Mengxiao, PAN Weihao, ZHANG Shixuan, MAO Xinye, YIN Yahong, LI Yongqiang, ZHU Youyin, GUO Weidong. Preliminary Investigation of C2H2 Family Genes in Blueberry Flower Bud Dormancy Release [J]. Acta Horticulturae Sinica, 2023, 50(4): 737-753. |
| [3] | XU Yue, LI Zixiong, CHEN Jie, SUN Liang. Transcriptional Bases of the Regulation of 2,4-D on Tomato Fruit Shape [J]. Acta Horticulturae Sinica, 2023, 50(4): 802-814. |
| [4] | WANG Zehan, YU Wentao, WANG Pengjie, LIU Caiguo, FAN Xiaojing, GU Mengya, CAI Chunping, WANG Pan, YE Naixing. WGCNA Analysis of Differentially Expressed Genes in Floral Organs of Tea Germplasms with Ovary-glabrous and Ovary-trichome [J]. Acta Horticulturae Sinica, 2023, 50(3): 620-634. |
| [5] | JIANG Yu, TU Xunliang, HE Junrong. Analysis of Differential Expression Genes in Leaves of Leaf Color Mutant of Chinese Orchid [J]. Acta Horticulturae Sinica, 2023, 50(2): 371-381. |
| [6] | ZHAO Xueyan, WANG Qi, WANG Li, WANG Fangyuan, WANG Qing, LI Yan. Comparative Transcriptome Analysis of Differential Expression in Different Tissues of Corydalis yanhusuo [J]. Acta Horticulturae Sinica, 2023, 50(1): 177-187. |
| [7] | ZHANG Xiaoming, YAN Guohua, ZHOU Yu, WANG Jing, DUAN Xuwei, WU Chuanbao, and ZHANG Kaichun. A New Sweet Cherry Rootstock Cultivar‘Jingchun 2’ [J]. Acta Horticulturae Sinica, 2022, 49(S2): 31-32. |
| [8] | LIANG Qin, ZHANG Yanhui, KANG Kaiquan, LIU Jinhang, LI Liang, FENG Yu, WANG Chao, YANG Chao, LI Yongyu. Molecular Evolution of MiR168 Family and Their Expression Profiling During Dormancy of Pyrus pyrifolia [J]. Acta Horticulturae Sinica, 2022, 49(5): 958-972. |
| [9] | LI Lixian, WANG Shuo, CHEN Ying, WU Yingtao, WANG Yaqian, FANG Yue, CHEN Xuesen, TIAN Changping, FENG Shouqian. PavMYB10.1 Promotes Anthocyanin Accumulation by Positively Regulating PavRiant in Sweet Cherry [J]. Acta Horticulturae Sinica, 2022, 49(5): 1023-1030. |
| [10] | ZHOU Xuzixin, YANG Wei, MAO Meiqin, XUE Yanbin, MA Jun. Identification of Pigment Components and Key Genes in Carotenoid Pathway in Mutants of Chimeric Ananas comosus var. bracteatus [J]. Acta Horticulturae Sinica, 2022, 49(5): 1081-1091. |
| [11] | SHEN Nan, ZHANG Jingcheng, WANG Chengchen, BIAN Yinbing, XIAO Yang. Studies on Transcriptome During Fruiting Body Development of Lentinula edodes [J]. Acta Horticulturae Sinica, 2022, 49(4): 801-815. |
| [12] | XIA Ming, LI Jingwei, LUO Zhangrui, ZU Guidong, WANG Ya, ZHANG Wanping. Research on Mechanism of Exogenous Melatonin Effects on Radish Growth and Resistence to Alternaria brassicae [J]. Acta Horticulturae Sinica, 2022, 49(3): 548-560. |
| [13] | DENG Jiao, SU Mengyue, LIU Xuelian, OU Kefang, HU Zhengrong, YANG Pingfang. Transcriptome Analysis Revealed the Formation Mechanism of Floral Color of Lotus‘Dasajin’with Bicolor Petal [J]. Acta Horticulturae Sinica, 2022, 49(2): 365-377. |
| [14] | XU Tong, SHAO Lingmei, WANG Xiaobin, ZHANG Runlong, ZHANG Kaijing, XIA Yiping, ZHANG Jiaping, LI Danqing. Research Progress on Winter Dormancy of Perennial Monocots [J]. Acta Horticulturae Sinica, 2022, 49(12): 2703-2721. |
| [15] | QIAO Jun, WANG Liying, LIU Jing, LI Suweng. Expression Analysis of Genes Related to Photosensitive Color Under the Caylx in Eggplant Based on Transcriptome Sequencing [J]. Acta Horticulturae Sinica, 2022, 49(11): 2347-2356. |
| Viewed | ||||||
|
Full text |
|
|||||
|
Abstract |
|
|||||
Copyright © 2012 Acta Horticulturae Sinica 京ICP备10030308号-2 国际联网备案号 11010802023439
Tel: 010-82109523 E-Mail: yuanyixuebao@126.com
Support by: Beijing Magtech Co.Ltd