
Acta Horticulturae Sinica ›› 2023, Vol. 50 ›› Issue (4): 737-753.doi: 10.16420/j.issn.0513-353x.2022-0044
• Research Papers • Previous Articles Next Articles
LI Ke1, SHEN Mengxiao1, PAN Weihao1, ZHANG Shixuan1, MAO Xinye1, YIN Yahong1, LI Yongqiang1,2,*( ), ZHU Youyin3,*(
), ZHU Youyin3,*( ), GUO Weidong1,2
), GUO Weidong1,2
Received:2022-10-08
Revised:2023-01-21
Online:2023-04-25
Published:2023-04-27
Contact:
*(E-mail:lyq@zjnu.cn,zhuyouyin@jhc.edu.cn)
E-mail:lyq@zjnu.cn;zhuyouyin@jhc.edu.cn
CLC Number:
LI Ke, SHEN Mengxiao, PAN Weihao, ZHANG Shixuan, MAO Xinye, YIN Yahong, LI Yongqiang, ZHU Youyin, GUO Weidong. Preliminary Investigation of C2H2 Family Genes in Blueberry Flower Bud Dormancy Release[J]. Acta Horticulturae Sinica, 2023, 50(4): 737-753.
Add to citation manager EndNote|Ris|BibTeX
URL: https://www.ahs.ac.cn/EN/10.16420/j.issn.0513-353x.2022-0044
| 目的基因 Gene name | 上游引物(5′-3′) Forward primer | 下游引物(5′-3′) Reverse primer | 扩增长度/bp Amplicon size | 退火温度/℃ Tm |
|---|---|---|---|---|
| Vc38g176.32 | TTATTCGGTGGTTCACGGCA | TTGCGAAGTCACGGATGGAA | 365 | 55 |
| Vc7g51.6 | GCTCGATCTGTCACAAGAGTT | GACGTCGTAAGTCCACTGTTT | 109 | 55 |
| Vc3g10.6 | AACAGAGGATTCGACCTGAAC | CCCAAATCAGGTTGGAAAGATAAA | 155 | 55 |
| Vc31g303.5 | GGAGGAGCAAACAGTGGAATTA | CAAATCCGAATTCCGGCAAAG | 112 | 55 |
| Vc24g106.5 | AAGAAGACGAAGAAGGAGGTAATAAG | GCGACACCGATTTGGTAGAA | 192 | 55 |
| Vc16g355.5 | GGAGGAGCAAACAGTGGAATTA | CCAAATCCATATTCCGGCAAAG | 113 | 55 |
| Vc14g348.9 | GTGTGACGACGACATCTGAA | CCCAAATCAGGTTGGAAAGATAAA | 196 | 55 |
| Vc11g294.5 | CGATATGTGGGTCGGAGTTT | CATCGGTGCTGGTAGATTCA | 183 | 55 |
| Vc21g155.6 | CAAAGGGCCCAACTGCAACAGAT | TACACCCCGAGAAGGGACACGAA | 483 | 55 |
| Vc23g300.2 | CCACAGGTGCTTTCCAACGGGTCA | CGCCGAATCCTTGCCAGAATTCAG | 194 | 60.5 |
| Vc37g104.16 | CTGATAAAATGGAGACTGATGAAA | CCAGACAACAGATAAGAAAACGAA | 244 | 60.5 |
| Vc37g230.20 | TCGACAAAGGTGGTTCAGGT | TGTCCCTGTACCCATACCCA | 213 | 60.5 |
| Vc38g80.29 | GAAGAGGAGAAAGGGAATGCCACT | AACCCATCAAGCAATCCATCAAAA | 152 | 60.5 |
| Vc42g36.8 | GCAAAGGGCCCAACTGCAACAGAT | CACCCCGAGAAGGGACACGAAAAC | 482 | 60.5 |
| Vc6g338.31 | GAAGAGGAGAAAGGGAATGCCACT | AACCCATCAAGCAATCCATCAAAA | 152 | 62 |
| VcGAPDH | TGAGAAAGAATACAAGCCAGAT | CAGGCAACACCTTACCAA | 260 | 55 ~ 62 |
Table 1 Quantitative primers of VcC2H2-ZFP
| 目的基因 Gene name | 上游引物(5′-3′) Forward primer | 下游引物(5′-3′) Reverse primer | 扩增长度/bp Amplicon size | 退火温度/℃ Tm |
|---|---|---|---|---|
| Vc38g176.32 | TTATTCGGTGGTTCACGGCA | TTGCGAAGTCACGGATGGAA | 365 | 55 |
| Vc7g51.6 | GCTCGATCTGTCACAAGAGTT | GACGTCGTAAGTCCACTGTTT | 109 | 55 |
| Vc3g10.6 | AACAGAGGATTCGACCTGAAC | CCCAAATCAGGTTGGAAAGATAAA | 155 | 55 |
| Vc31g303.5 | GGAGGAGCAAACAGTGGAATTA | CAAATCCGAATTCCGGCAAAG | 112 | 55 |
| Vc24g106.5 | AAGAAGACGAAGAAGGAGGTAATAAG | GCGACACCGATTTGGTAGAA | 192 | 55 |
| Vc16g355.5 | GGAGGAGCAAACAGTGGAATTA | CCAAATCCATATTCCGGCAAAG | 113 | 55 |
| Vc14g348.9 | GTGTGACGACGACATCTGAA | CCCAAATCAGGTTGGAAAGATAAA | 196 | 55 |
| Vc11g294.5 | CGATATGTGGGTCGGAGTTT | CATCGGTGCTGGTAGATTCA | 183 | 55 |
| Vc21g155.6 | CAAAGGGCCCAACTGCAACAGAT | TACACCCCGAGAAGGGACACGAA | 483 | 55 |
| Vc23g300.2 | CCACAGGTGCTTTCCAACGGGTCA | CGCCGAATCCTTGCCAGAATTCAG | 194 | 60.5 |
| Vc37g104.16 | CTGATAAAATGGAGACTGATGAAA | CCAGACAACAGATAAGAAAACGAA | 244 | 60.5 |
| Vc37g230.20 | TCGACAAAGGTGGTTCAGGT | TGTCCCTGTACCCATACCCA | 213 | 60.5 |
| Vc38g80.29 | GAAGAGGAGAAAGGGAATGCCACT | AACCCATCAAGCAATCCATCAAAA | 152 | 60.5 |
| Vc42g36.8 | GCAAAGGGCCCAACTGCAACAGAT | CACCCCGAGAAGGGACACGAAAAC | 482 | 60.5 |
| Vc6g338.31 | GAAGAGGAGAAAGGGAATGCCACT | AACCCATCAAGCAATCCATCAAAA | 152 | 62 |
| VcGAPDH | TGAGAAAGAATACAAGCCAGAT | CAGGCAACACCTTACCAA | 260 | 55 ~ 62 |
| 基因名 Gene name | 基因ID Accession No. | 氨基酸数 Amino acid number | 分子量/kD Molecular weight | 等电点 pI | 不稳定系数 Instability index | 亲水值 Hydropathicity | 亚细胞定位 Subcellular location |
|---|---|---|---|---|---|---|---|
| Vc19g325.6 | VaccDscaff19-processed-325.6 | 396 | 43.8 | 8.94 | 49.42 | -0.696 | Nucl,Cyto |
| Vc15g278.5 | VaccDscaff15-processed-278.5 | 394 | 43.7 | 9.00 | 46.09 | -0.704 | Nucl,Cyto |
| Vc11g294.4 | VaccDscaff11-processed-294.4 | 398 | 44.2 | 9.01 | 48.83 | -0.727 | Nucl,Cyto |
| Vc24g106.6 | VaccDscaff24-processed-106.6 | 398 | 44.2 | 8.94 | 48.52 | -0.734 | Nucl,Cyto |
| Vc30g269.43 | VaccDscaff30-augustus-269.43 | 508 | 56.4 | 6.79 | 41.71 | -0.436 | Nucl,Chlo |
| Vc13g13.36 | VaccDscaff13-augustus-13.36 | 456 | 50.7 | 5.87 | 41.61 | -0.398 | Nucl |
| Vc42g47.29 | VaccDscaff42-augustus-47.29 | 456 | 50.6 | 5.87 | 41.10 | -0.397 | Nucl |
| Vc47g65.34 | VaccDscaff47-augustus-65.34 | 388 | 42.9 | 8.51 | 45.21 | -0.701 | Nucl |
| Vc43g136.13 | VaccDscaff43-augustus-136.13 | 379 | 41.9 | 8.61 | 46.09 | -0.635 | Nucl |
| Vc22g249.38 | VaccDscaff22-augustus-249.38 | 378 | 41.7 | 8.61 | 43.32 | -0.599 | Nucl |
| Vc39g85.33 | VaccDscaff39-augustus-85.33 | 381 | 42.3 | 5.60 | 44.46 | -0.452 | Nucl |
| Vc37g230.20 | VaccDscaff37-augustus-230.20 | 392 | 43.3 | 5.58 | 42.06 | -0.421 | Nucl |
| Vc38g80.29 | VaccDscaff38-snap-80.29 | 392 | 43.1 | 5.44 | 43.73 | -0.430 | Nucl |
| Vc6g338.31 | VaccDscaff6-augustus-338.31 | 392 | 43.2 | 5.43 | 42.87 | -0.438 | Nucl,Pero |
| Vc38g176.32 | VaccDscaff38-augustus-176.32 | 478 | 53.4 | 5.84 | 50.44 | -0.775 | Nucl,Pero |
| Vc6g193.34 | VaccDscaff6-augustus-193.34 | 478 | 53.4 | 5.84 | 50.44 | -0.775 | Nucl,Pero |
| Vc37g104.16 | VaccDscaff37-augustus-104.16 | 478 | 53.4 | 5.84 | 50.88 | -0.756 | Nucl,Pero |
| Vc32g62.9 | VaccDscaff32-processed-62.9 | 382 | 41.4 | 6.57 | 53.67 | -0.656 | Nucl |
| Vc32g14.2 | VaccDscaff32-processed-14.2 | 474 | 52.0 | 8.39 | 54.84 | -0.510 | Nucl |
| Vc13g317.8 | VaccDscaff13-processed-317.8 | 477 | 52.1 | 8.09 | 55.80 | -0.521 | Nucl |
| Vc30g79.13 | VaccDscaff30-processed-79.13 | 474 | 52.0 | 8.07 | 54.80 | -0.524 | Nucl |
| Vc19g325.7 | VaccDscaff19-processed-325.7 | 254 | 27.9 | 5.94 | 64.39 | -0.774 | Nucl |
| Vc15g278.6 | VaccDscaff15-processed-278.6 | 254 | 27.9 | 5.94 | 64.05 | -0.784 | Nucl |
| Vc11g294.5 | VaccDscaff11-processed-294.5 | 254 | 28.0 | 5.81 | 65.34 | -0.805 | Nucl |
| Vc24g106.5 | VaccDscaff24-processed-106.5 | 254 | 28.0 | 5.88 | 63.53 | -0.826 | Nucl |
| Vc33g157.2 | VaccDscaff33-processed-157.2 | 244 | 27.5 | 7.11 | 74.79 | -0.976 | Nucl |
| Vc21g197.4 | VaccDscaff21-processed-197.4 | 245 | 27.6 | 7.11 | 73.79 | -0.938 | Nucl |
| Vc29g187.1 | VaccDscaff29-processed-187.1 | 245 | 27.6 | 7.11 | 74.52 | -0.953 | Nucl |
| Vc19g69.12 | VaccDscaff19-processed-69.12 | 355 | 39.0 | 5.51 | 69.83 | -0.699 | Nucl |
| Vc26g194.1 | VaccDscaff26-processed-194.1 | 244 | 27.5 | 6.51 | 76.04 | -0.933 | Nucl |
| Vc20g74.12 | VaccDscaff20-processed-74.12 | 355 | 38.8 | 5.51 | 68.93 | -0.683 | Nucl |
| Vc28g285.19 | VaccDscaff28-processed-285.19 | 355 | 38.9 | 5.49 | 68.69 | -0.692 | Nucl |
| Vc21g236.10 | VaccDscaff21-processed-236.10 | 286 | 31.0 | 7.65 | 56.50 | -0.810 | Nucl |
| Vc26g232.8 | VaccDscaff26-processed-232.8 | 286 | 31.1 | 8.14 | 54.76 | -0.819 | Nucl |
| Vc33g119.6 | VaccDscaff33-processed-119.6 | 287 | 31.1 | 7.65 | 54.81 | -0.808 | Nucl |
| Vc40g163.1 | VaccDscaff40-processed-163.1 | 371 | 41.9 | 5.75 | 64.69 | -1.037 | Nucl |
| Vc41g152.1 | VaccDscaff41-processed-152.1 | 373 | 42.1 | 5.82 | 62.04 | -1.050 | Nucl |
| Vc23g116.9 | VaccDscaff23-processed-116.9 | 373 | 42.1 | 5.68 | 61.92 | -1.053 | Nucl |
| Vc23g130.8 | VaccDscaff23-processed-130.8 | 371 | 41.8 | 5.81 | 64.61 | -1.011 | Nucl |
| Vc29g224.1 | VaccDscaff29-processed-224.1 | 275 | 29.7 | 8.45 | 53.45 | -0.817 | Nucl,Extr |
| Vc47g65.8 | VaccDscaff47-processed-65.8 | 275 | 29.9 | 6.32 | 56.61 | -0.635 | Nucl |
| Vc18g40.7 | VaccDscaff18-processed-40.7 | 236 | 24.6 | 8.93 | 59.17 | -0.567 | Nucl |
| Vc43g135.1 | VaccDscaff43-processed-135.1 | 272 | 29.6 | 6.25 | 55.65 | -0.584 | Nucl |
| Vc16g355.5 | VaccDscaff16-processed-355.5 | 234 | 24.4 | 8.92 | 59.76 | -0.562 | Nucl |
| Vc22g250.7 | VaccDscaff22-processed-250.7 | 272 | 29.6 | 6.25 | 55.53 | -0.606 | Nucl |
| Vc7g51.6 | VaccDscaff7-processed-51.6 | 224 | 23.5 | 9.01 | 62.98 | -0.544 | Nucl |
| Vc31g303.5 | VaccDscaff31-processed-303.5 | 233 | 24.3 | 8.93 | 61.63 | -0.561 | Nucl |
| Vc40g40.5 | VaccDscaff40-processed-40.5 | 240 | 25.5 | 7.72 | 71.73 | -0.707 | Nucl |
| Vc23g300.2 | VaccDscaff23-processed-300.2 | 239 | 25.4 | 8.22 | 71.35 | -0.700 | Nucl |
| Vc12g112.2 | VaccDscaff12-processed-112.2 | 240 | 25.5 | 8.17 | 70.40 | -0.668 | Nucl |
| Vc1g306.32 | VaccDscaff1-processed-306.32 | 239 | 25.8 | 7.87 | 57.87 | -0.782 | Nucl |
| Vc10g124.14 | VaccDscaff10-processed-124.14 | 306 | 33.2 | 7.86 | 64.71 | -0.660 | Nucl |
| Vc3g10.6 | VaccDscaff3-processed-10.6 | 217 | 23.4 | 8.68 | 67.52 | -0.675 | Nucl |
| Vc14g348.9 | VaccDscaff14-processed-348.9 | 233 | 24.7 | 8.67 | 61.00 | -0.624 | Nucl |
| Vc2g402.7 | VaccDscaff2-processed-402.7 | 234 | 24.8 | 8.67 | 59.96 | -0.573 | Nucl |
| Vc22g45.12 | VaccDscaff22-processed-45.12 | 371 | 42.3 | 6.26 | 58.05 | -1.124 | Nucl |
| Vc21g155.6 | VaccDscaff21-processed-155.6 | 182 | 20.4 | 8.84 | 51.53 | -0.348 | Nucl |
| Vc26g158.3 | VaccDscaff26-processed-158.3 | 182 | 20.4 | 8.67 | 52.00 | -0.348 | Nucl |
| Vc33g183.0 | VaccDscaff33-processed-183.0 | 182 | 20.4 | 8.67 | 52.00 | -0.348 | Nucl |
| Vc29g155.8 | VaccDscaff29-processed-155.8 | 182 | 20.4 | 8.66 | 55.59 | -0.415 | Nucl |
| Vc20g57.10 | VaccDscaff20-processed-57.10 | 161 | 17.7 | 9.02 | 53.48 | -0.437 | Nucl,Cyto,Extr |
| Vc48g64.13 | VaccDscaff48-processed-64.13 | 173 | 18.9 | 9.34 | 48.58 | -0.395 | Nucl,Cyto,Extr |
| Vc14g51.11 | VaccDscaff14-processed-51.11 | 228 | 25.1 | 5.19 | 64.33 | -0.483 | Nucl,Pero |
| Vc20g133.0 | VaccDscaff20-processed-133.0 | 228 | 24.2 | 7.08 | 60.52 | -0.638 | Nucl |
| Vc28g213.2 | VaccDscaff28-processed-213.2 | 228 | 24.2 | 7.08 | 60.52 | -0.638 | Nucl |
| Vc2g14.22 | VaccDscaff2-processed-14.22 | 228 | 25.2 | 5.07 | 61.95 | -0.466 | Nucl,Pero |
| Vc42g36.8 | VaccDscaff42-processed-36.8 | 232 | 25.4 | 5.16 | 46.05 | -0.316 | Nucl,Pero |
| Vc19g135.9 | VaccDscaff19-processed-135.9 | 222 | 23.7 | 7.08 | 55.23 | -0.605 | Nucl,Pero |
| Vc30g280.2 | VaccDscaff30-processed-280.2 | 232 | 25.5 | 5.16 | 46.41 | -0.319 | Nucl |
| Vc27g295.0 | VaccDscaff27-processed-295.0 | 263 | 28.8 | 8.80 | 62.35 | -0.655 | Nucl,Pero |
| Vc17g324.14 | VaccDscaff17-processed-324.14 | 263 | 28.8 | 8.59 | 62.70 | -0.664 | Nucl,Pero |
| Vc34g49.12 | VaccDscaff34-processed-49.12 | 263 | 28.8 | 8.59 | 62.70 | -0.664 | Nucl,Pero |
| Vc44g170.19 | VaccDscaff44-augustus-170.19 | 248 | 27.0 | 7.14 | 60.50 | -0.597 | Nucl |
| Vc20g156.15 | VaccDscaff20-augustus-156.15 | 248 | 27.0 | 6.50 | 57.33 | -0.583 | Nucl |
| Vc28g167.32 | VaccDscaff28-augustus-167.32 | 248 | 27.0 | 6.75 | 60.99 | -0.583 | Nucl |
| Vc44g65.1 | VaccDscaff44-processed-65.1 | 247 | 26.6 | 8.38 | 54.41 | -0.681 | Nucl |
| Vc28g73.2 | VaccDscaff28-processed-73.2 | 247 | 26.6 | 8.38 | 53.87 | -0.681 | Nucl |
| Vc20g302.3 | VaccDscaff20-processed-302.3 | 250 | 26.9 | 7.80 | 55.74 | -0.688 | Nucl |
| Vc8g145.9 | VaccDscaff8-processed-145.9 | 237 | 25.8 | 7.87 | 57.57 | -0.784 | Nucl |
Table 2 Basic information of VcC2H2-ZFP
| 基因名 Gene name | 基因ID Accession No. | 氨基酸数 Amino acid number | 分子量/kD Molecular weight | 等电点 pI | 不稳定系数 Instability index | 亲水值 Hydropathicity | 亚细胞定位 Subcellular location |
|---|---|---|---|---|---|---|---|
| Vc19g325.6 | VaccDscaff19-processed-325.6 | 396 | 43.8 | 8.94 | 49.42 | -0.696 | Nucl,Cyto |
| Vc15g278.5 | VaccDscaff15-processed-278.5 | 394 | 43.7 | 9.00 | 46.09 | -0.704 | Nucl,Cyto |
| Vc11g294.4 | VaccDscaff11-processed-294.4 | 398 | 44.2 | 9.01 | 48.83 | -0.727 | Nucl,Cyto |
| Vc24g106.6 | VaccDscaff24-processed-106.6 | 398 | 44.2 | 8.94 | 48.52 | -0.734 | Nucl,Cyto |
| Vc30g269.43 | VaccDscaff30-augustus-269.43 | 508 | 56.4 | 6.79 | 41.71 | -0.436 | Nucl,Chlo |
| Vc13g13.36 | VaccDscaff13-augustus-13.36 | 456 | 50.7 | 5.87 | 41.61 | -0.398 | Nucl |
| Vc42g47.29 | VaccDscaff42-augustus-47.29 | 456 | 50.6 | 5.87 | 41.10 | -0.397 | Nucl |
| Vc47g65.34 | VaccDscaff47-augustus-65.34 | 388 | 42.9 | 8.51 | 45.21 | -0.701 | Nucl |
| Vc43g136.13 | VaccDscaff43-augustus-136.13 | 379 | 41.9 | 8.61 | 46.09 | -0.635 | Nucl |
| Vc22g249.38 | VaccDscaff22-augustus-249.38 | 378 | 41.7 | 8.61 | 43.32 | -0.599 | Nucl |
| Vc39g85.33 | VaccDscaff39-augustus-85.33 | 381 | 42.3 | 5.60 | 44.46 | -0.452 | Nucl |
| Vc37g230.20 | VaccDscaff37-augustus-230.20 | 392 | 43.3 | 5.58 | 42.06 | -0.421 | Nucl |
| Vc38g80.29 | VaccDscaff38-snap-80.29 | 392 | 43.1 | 5.44 | 43.73 | -0.430 | Nucl |
| Vc6g338.31 | VaccDscaff6-augustus-338.31 | 392 | 43.2 | 5.43 | 42.87 | -0.438 | Nucl,Pero |
| Vc38g176.32 | VaccDscaff38-augustus-176.32 | 478 | 53.4 | 5.84 | 50.44 | -0.775 | Nucl,Pero |
| Vc6g193.34 | VaccDscaff6-augustus-193.34 | 478 | 53.4 | 5.84 | 50.44 | -0.775 | Nucl,Pero |
| Vc37g104.16 | VaccDscaff37-augustus-104.16 | 478 | 53.4 | 5.84 | 50.88 | -0.756 | Nucl,Pero |
| Vc32g62.9 | VaccDscaff32-processed-62.9 | 382 | 41.4 | 6.57 | 53.67 | -0.656 | Nucl |
| Vc32g14.2 | VaccDscaff32-processed-14.2 | 474 | 52.0 | 8.39 | 54.84 | -0.510 | Nucl |
| Vc13g317.8 | VaccDscaff13-processed-317.8 | 477 | 52.1 | 8.09 | 55.80 | -0.521 | Nucl |
| Vc30g79.13 | VaccDscaff30-processed-79.13 | 474 | 52.0 | 8.07 | 54.80 | -0.524 | Nucl |
| Vc19g325.7 | VaccDscaff19-processed-325.7 | 254 | 27.9 | 5.94 | 64.39 | -0.774 | Nucl |
| Vc15g278.6 | VaccDscaff15-processed-278.6 | 254 | 27.9 | 5.94 | 64.05 | -0.784 | Nucl |
| Vc11g294.5 | VaccDscaff11-processed-294.5 | 254 | 28.0 | 5.81 | 65.34 | -0.805 | Nucl |
| Vc24g106.5 | VaccDscaff24-processed-106.5 | 254 | 28.0 | 5.88 | 63.53 | -0.826 | Nucl |
| Vc33g157.2 | VaccDscaff33-processed-157.2 | 244 | 27.5 | 7.11 | 74.79 | -0.976 | Nucl |
| Vc21g197.4 | VaccDscaff21-processed-197.4 | 245 | 27.6 | 7.11 | 73.79 | -0.938 | Nucl |
| Vc29g187.1 | VaccDscaff29-processed-187.1 | 245 | 27.6 | 7.11 | 74.52 | -0.953 | Nucl |
| Vc19g69.12 | VaccDscaff19-processed-69.12 | 355 | 39.0 | 5.51 | 69.83 | -0.699 | Nucl |
| Vc26g194.1 | VaccDscaff26-processed-194.1 | 244 | 27.5 | 6.51 | 76.04 | -0.933 | Nucl |
| Vc20g74.12 | VaccDscaff20-processed-74.12 | 355 | 38.8 | 5.51 | 68.93 | -0.683 | Nucl |
| Vc28g285.19 | VaccDscaff28-processed-285.19 | 355 | 38.9 | 5.49 | 68.69 | -0.692 | Nucl |
| Vc21g236.10 | VaccDscaff21-processed-236.10 | 286 | 31.0 | 7.65 | 56.50 | -0.810 | Nucl |
| Vc26g232.8 | VaccDscaff26-processed-232.8 | 286 | 31.1 | 8.14 | 54.76 | -0.819 | Nucl |
| Vc33g119.6 | VaccDscaff33-processed-119.6 | 287 | 31.1 | 7.65 | 54.81 | -0.808 | Nucl |
| Vc40g163.1 | VaccDscaff40-processed-163.1 | 371 | 41.9 | 5.75 | 64.69 | -1.037 | Nucl |
| Vc41g152.1 | VaccDscaff41-processed-152.1 | 373 | 42.1 | 5.82 | 62.04 | -1.050 | Nucl |
| Vc23g116.9 | VaccDscaff23-processed-116.9 | 373 | 42.1 | 5.68 | 61.92 | -1.053 | Nucl |
| Vc23g130.8 | VaccDscaff23-processed-130.8 | 371 | 41.8 | 5.81 | 64.61 | -1.011 | Nucl |
| Vc29g224.1 | VaccDscaff29-processed-224.1 | 275 | 29.7 | 8.45 | 53.45 | -0.817 | Nucl,Extr |
| Vc47g65.8 | VaccDscaff47-processed-65.8 | 275 | 29.9 | 6.32 | 56.61 | -0.635 | Nucl |
| Vc18g40.7 | VaccDscaff18-processed-40.7 | 236 | 24.6 | 8.93 | 59.17 | -0.567 | Nucl |
| Vc43g135.1 | VaccDscaff43-processed-135.1 | 272 | 29.6 | 6.25 | 55.65 | -0.584 | Nucl |
| Vc16g355.5 | VaccDscaff16-processed-355.5 | 234 | 24.4 | 8.92 | 59.76 | -0.562 | Nucl |
| Vc22g250.7 | VaccDscaff22-processed-250.7 | 272 | 29.6 | 6.25 | 55.53 | -0.606 | Nucl |
| Vc7g51.6 | VaccDscaff7-processed-51.6 | 224 | 23.5 | 9.01 | 62.98 | -0.544 | Nucl |
| Vc31g303.5 | VaccDscaff31-processed-303.5 | 233 | 24.3 | 8.93 | 61.63 | -0.561 | Nucl |
| Vc40g40.5 | VaccDscaff40-processed-40.5 | 240 | 25.5 | 7.72 | 71.73 | -0.707 | Nucl |
| Vc23g300.2 | VaccDscaff23-processed-300.2 | 239 | 25.4 | 8.22 | 71.35 | -0.700 | Nucl |
| Vc12g112.2 | VaccDscaff12-processed-112.2 | 240 | 25.5 | 8.17 | 70.40 | -0.668 | Nucl |
| Vc1g306.32 | VaccDscaff1-processed-306.32 | 239 | 25.8 | 7.87 | 57.87 | -0.782 | Nucl |
| Vc10g124.14 | VaccDscaff10-processed-124.14 | 306 | 33.2 | 7.86 | 64.71 | -0.660 | Nucl |
| Vc3g10.6 | VaccDscaff3-processed-10.6 | 217 | 23.4 | 8.68 | 67.52 | -0.675 | Nucl |
| Vc14g348.9 | VaccDscaff14-processed-348.9 | 233 | 24.7 | 8.67 | 61.00 | -0.624 | Nucl |
| Vc2g402.7 | VaccDscaff2-processed-402.7 | 234 | 24.8 | 8.67 | 59.96 | -0.573 | Nucl |
| Vc22g45.12 | VaccDscaff22-processed-45.12 | 371 | 42.3 | 6.26 | 58.05 | -1.124 | Nucl |
| Vc21g155.6 | VaccDscaff21-processed-155.6 | 182 | 20.4 | 8.84 | 51.53 | -0.348 | Nucl |
| Vc26g158.3 | VaccDscaff26-processed-158.3 | 182 | 20.4 | 8.67 | 52.00 | -0.348 | Nucl |
| Vc33g183.0 | VaccDscaff33-processed-183.0 | 182 | 20.4 | 8.67 | 52.00 | -0.348 | Nucl |
| Vc29g155.8 | VaccDscaff29-processed-155.8 | 182 | 20.4 | 8.66 | 55.59 | -0.415 | Nucl |
| Vc20g57.10 | VaccDscaff20-processed-57.10 | 161 | 17.7 | 9.02 | 53.48 | -0.437 | Nucl,Cyto,Extr |
| Vc48g64.13 | VaccDscaff48-processed-64.13 | 173 | 18.9 | 9.34 | 48.58 | -0.395 | Nucl,Cyto,Extr |
| Vc14g51.11 | VaccDscaff14-processed-51.11 | 228 | 25.1 | 5.19 | 64.33 | -0.483 | Nucl,Pero |
| Vc20g133.0 | VaccDscaff20-processed-133.0 | 228 | 24.2 | 7.08 | 60.52 | -0.638 | Nucl |
| Vc28g213.2 | VaccDscaff28-processed-213.2 | 228 | 24.2 | 7.08 | 60.52 | -0.638 | Nucl |
| Vc2g14.22 | VaccDscaff2-processed-14.22 | 228 | 25.2 | 5.07 | 61.95 | -0.466 | Nucl,Pero |
| Vc42g36.8 | VaccDscaff42-processed-36.8 | 232 | 25.4 | 5.16 | 46.05 | -0.316 | Nucl,Pero |
| Vc19g135.9 | VaccDscaff19-processed-135.9 | 222 | 23.7 | 7.08 | 55.23 | -0.605 | Nucl,Pero |
| Vc30g280.2 | VaccDscaff30-processed-280.2 | 232 | 25.5 | 5.16 | 46.41 | -0.319 | Nucl |
| Vc27g295.0 | VaccDscaff27-processed-295.0 | 263 | 28.8 | 8.80 | 62.35 | -0.655 | Nucl,Pero |
| Vc17g324.14 | VaccDscaff17-processed-324.14 | 263 | 28.8 | 8.59 | 62.70 | -0.664 | Nucl,Pero |
| Vc34g49.12 | VaccDscaff34-processed-49.12 | 263 | 28.8 | 8.59 | 62.70 | -0.664 | Nucl,Pero |
| Vc44g170.19 | VaccDscaff44-augustus-170.19 | 248 | 27.0 | 7.14 | 60.50 | -0.597 | Nucl |
| Vc20g156.15 | VaccDscaff20-augustus-156.15 | 248 | 27.0 | 6.50 | 57.33 | -0.583 | Nucl |
| Vc28g167.32 | VaccDscaff28-augustus-167.32 | 248 | 27.0 | 6.75 | 60.99 | -0.583 | Nucl |
| Vc44g65.1 | VaccDscaff44-processed-65.1 | 247 | 26.6 | 8.38 | 54.41 | -0.681 | Nucl |
| Vc28g73.2 | VaccDscaff28-processed-73.2 | 247 | 26.6 | 8.38 | 53.87 | -0.681 | Nucl |
| Vc20g302.3 | VaccDscaff20-processed-302.3 | 250 | 26.9 | 7.80 | 55.74 | -0.688 | Nucl |
| Vc8g145.9 | VaccDscaff8-processed-145.9 | 237 | 25.8 | 7.87 | 57.57 | -0.784 | Nucl |
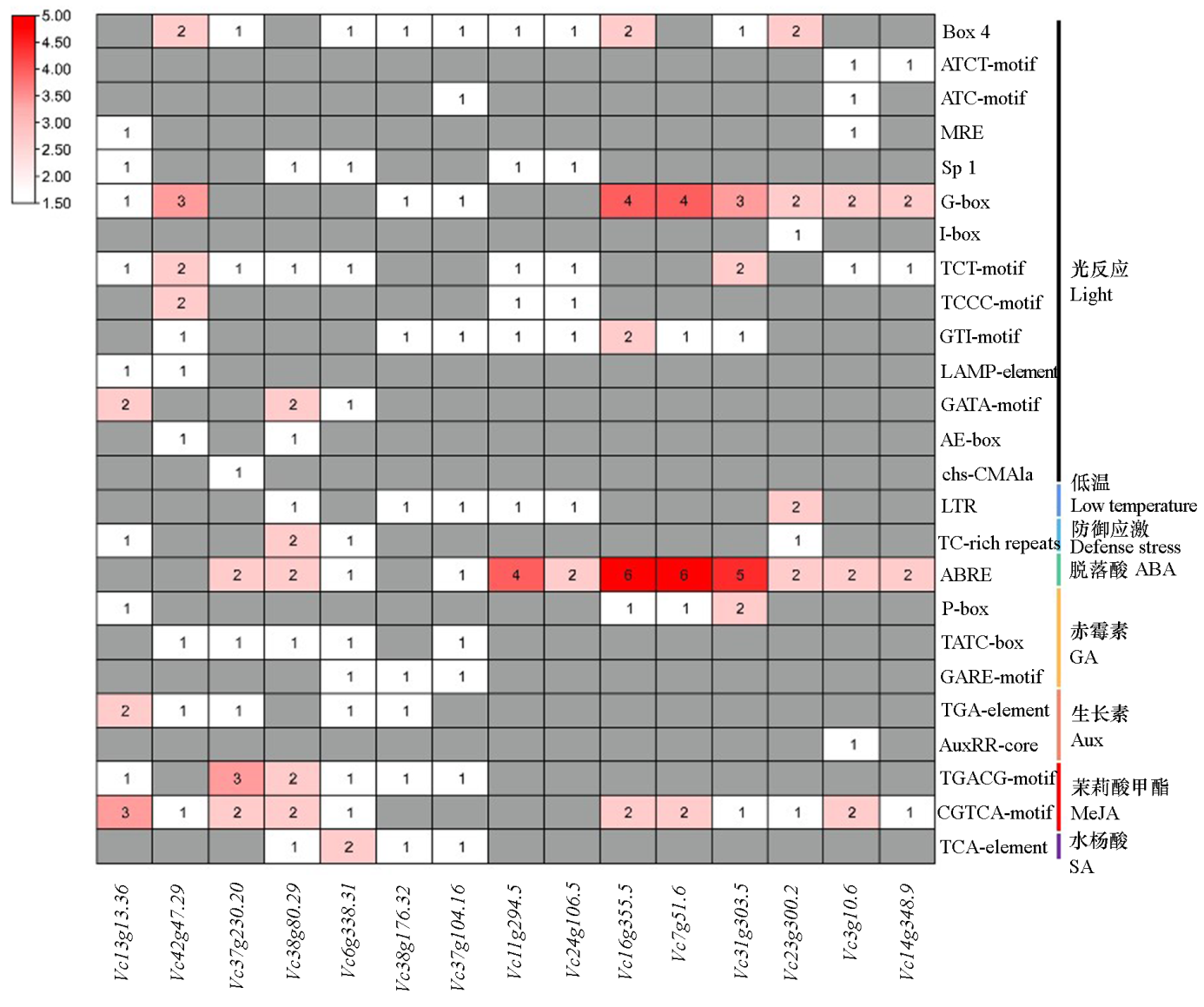
Fig. 7 Analysis of cis-acting elements of VcC2H2-ZFP promoter The numbers in the heat map represent the quantity of corresponding elements contained in the promoter.
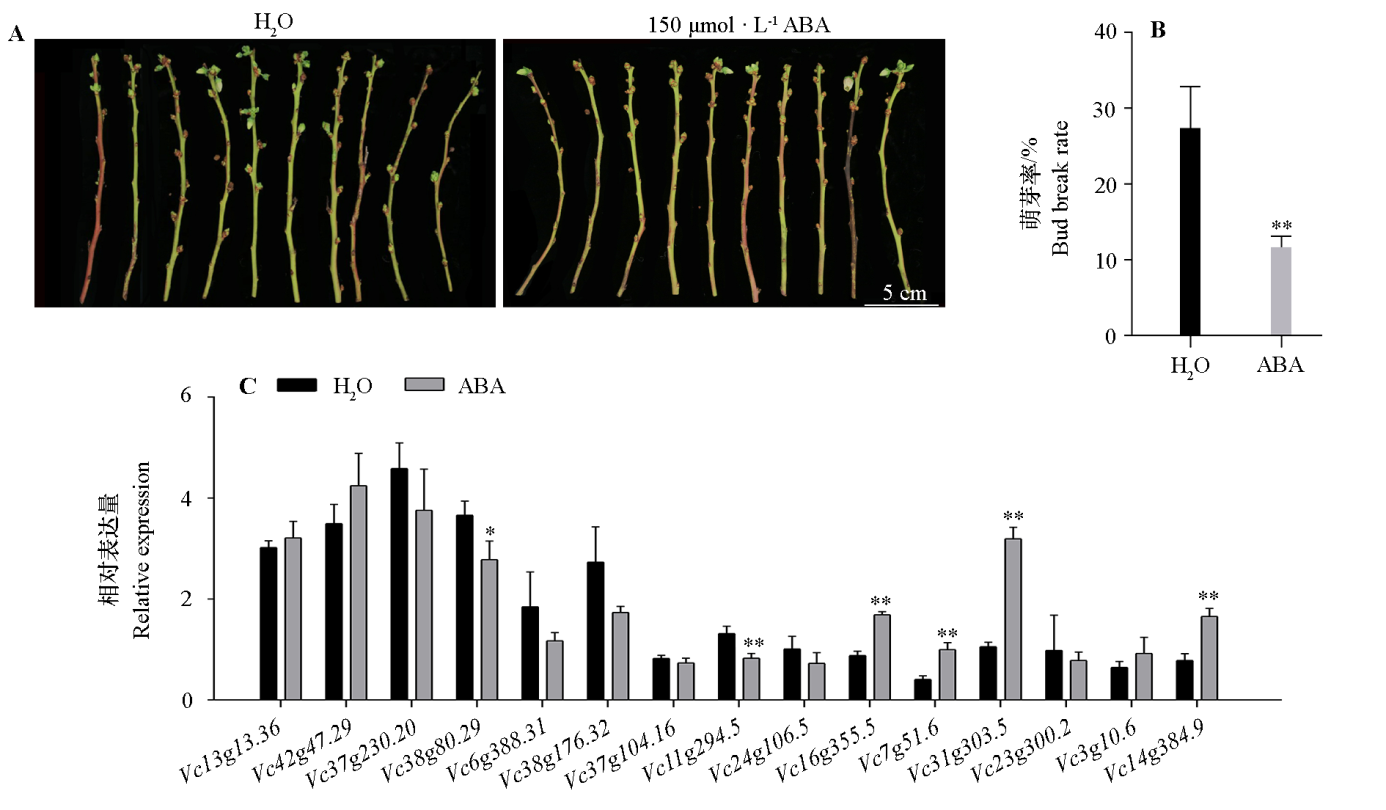
Fig. 8 Break percentage of blueberry flower bud and response of VcC2H2-ZFP after exogenous ABA treatment * indicated the significant difference under P < 0.05,** indicated the significant difference under P < 0.01.
| [1] |
Akhtar A, Becker P B. 2001. The histone H 4 acetyltransferase MOF uses a C2HC zinc finger for substrate recognition. EMBO Reports, 2 (2):113-118.
pmid: 11258702 |
| [2] | Alam I, Batool K, Cui D L, Yang Y Q, Lu Y H. 2019. Comprehensive genomic survey,structural classification and expression analysis of C2H2 zinc finger protein gene family in Brassica rapa L. PLoS ONE, 14 (5):e0216071. |
| [3] | An Shuang, Gao Yu-di, MaidinuerÝusupu, Pan Yi-na, Shao Wan, Zong Yu, Chen Wen-rong, Yang Li, Guo Wei-dong, Li Yong-qiang. 2020. Research on application exogenous abscisic acid in inhibiting blueberry early flowering and associated genes expression characteristics. Journal of Fruit Science, 38 (3):325-334. (in Chinese) |
| 安爽, 高玉迪, 麦迪努尔·玉苏普, 潘益娜, 邵婉, 宗宇, 陈文荣, 杨莉, 郭卫东, 李永强. 2020. 外源脱落酸抑制蓝莓早花及相关基因表达特性研究. 果树学报, 38 (3):325-334. | |
| [4] |
Arora R, Rowland L J, Tanino K J H. 2003. Induction and release of bud dormancy in woody perennials:a science comes of age. HortScience, 38 (5):911-921.
doi: 10.21273/HORTSCI.38.5.911 URL |
| [5] |
Bailey T L, Johnson J, Grant C E, Noble W S. 2015. The MEME Suite. Nucleic Acids Research, 43 (W1):W39-W49.
doi: 10.1093/nar/gkv416 URL |
| [6] |
Berg J M. 1988. Proposed structure for the zinc-binding domains from transcription factor IIIA and related proteins. Proc Natl Acad Sci U S A, 85 (1):99-102.
doi: 10.1073/pnas.85.1.99 URL |
| [7] |
Chen C, Chen H, Zhang Y, Thomas H R, Frank M H, He Y, Xia R. 2020a. TBtools:an integrative toolkit developed for interactive analyses of big biological data. Molecular Plant, 13 (8):1194-1202.
doi: 10.1016/j.molp.2020.06.009 URL |
| [8] |
Chen Y, Wang G, Pan J, Wen H, Du H, Sun J, Zhang K, Lv D, He H, Cai R, Pan J. 2020b. Comprehensive genomic analysis and expression profiling of the C2H2zinc finger protein family under abiotic stresses in cucumber(Cucumis sativus L.). Genes, 11 (2):171.
doi: 10.3390/genes11020171 URL |
| [9] |
Ciftci Y S, Mittler R. 2008. The zinc finger network of plants. Cellular and Molecular Life Sciences, 65:1150-1160.
doi: 10.1007/s00018-007-7473-4 pmid: 18193167 |
| [10] |
Ciftci Y S, Morsy M R, Song L, Coutu A, Krizek B A, Lewis M W, Warren D, Cushman J, Connolly E L, Mittler R. 2007. The EAR-motif of the Cys2/His2-type zinc finger protein Zat 7 plays a key role in the defense response of Arabidopsis to salinity stress. The Journal of Biological Chemistry, 282 (12):9260-9268.
doi: 10.1074/jbc.M611093200 URL |
| [11] | Colle M, Leisner C P, Wai C M, Ou S, Bird K A, Wang J, Wisecaver J H, Yocca A E, Alger E I, Tang H, Xiong Z, Callow P, Ben-Zvi G, Brodt A, Baruch K, Swale T, Shiue L, Song G Q, Childs K L, Schilmiller A, Vorsa N, Buell C R, van Buren R, Jiang N, Edger P P. 2019. Haplotype-phased genome and evolution of phytonutrient pathways of tetraploid blueberry. Gigascience, 8 (3):giz012. |
| [12] |
Davletova S, Schlauc K, Coutu J, Mittler R. 2005. The zinc-finger protein Zat 12 plays a central role in reactive oxygen and abiotic stress signaling in Arabidopsis. Plant Physiology, 139 (2):847-856.
doi: 10.1104/pp.105.068254 pmid: 16183833 |
| [13] |
Englbrecht C C, Schoof H, Bohm S. 2004. Conservation,diversification and expansion of C2H2 zinc finger proteins in the Arabidopsis thaliana genome. BMC Genomics, 5 (1):39.
doi: 10.1186/1471-2164-5-39 pmid: 15236668 |
| [14] |
Gotz K P, Naher J, Fettke J, Chmielewski F M. 2018. Changes of proteins during dormancy and bud development of sweet cherry(Prunus avium L.). Science Horticulturae, 239:41-49.
doi: 10.1016/j.scienta.2018.05.016 URL |
| [15] |
Gourcilleau D, Lenne C, Armenise C, Moulia B, Julien J L, Bronner G, Leblanc-Fournier N. 2011. Phylogenetic study of plant Q-type C2H2 zinc finger proteins and expression analysis of poplar genes in response to osmotic,cold and mechanical stresses. DNA Research, 18 (2):77-92.
doi: 10.1093/dnares/dsr001 pmid: 21367962 |
| [16] |
Han G, Wang M, Yuan F, Sui N, Song J, Wang B. 2014. The CCCH zinc finger protein gene AtZFP1 improves salt resistance in Arabidopsis thaliana. Plant Molecular Biology, 86 (3):237-253.
doi: 10.1007/s11103-014-0226-5 URL |
| [17] | Horton P, Park K J, Obayashi T, Fujita N, Harada H, Adams-Collier C J, Nakai K. 2007. WoLF PSORT:protein localization predictor. Nucleic Acids Research(Web Server Issue):W585-W587. |
| [18] |
Horvath D P, Anderson J V, Chao W S, Foley M E. 2003. Knowing when to grow:signals regulating bud dormancy. Trends in Plant Science, 8 (11):534-540.
doi: 10.1016/j.tplants.2003.09.013 pmid: 14607098 |
| [19] |
Hu X, Zhu L, Zhang Y, Xu L, Li N, Zhang X, Pan Y. 2019. Genome-wide identification of C2H2 zinc-finger genes and their expression patterns under heat stress in tomato(Solanum lycopersicum L.). PeerJ, 7:e7929.
doi: 10.7717/peerj.7929 URL |
| [20] | Jing Jian-ling, Zhang Peng, Wang Zhen-yu, Ma Qiu-xiang. 2020. Genome-wide identification and expression analysis of C2H2-type zinc finger protein transcription factor family in cassava. Plant Physiology Journal, 56 (12):2664-2676. (in Chinese) |
| 井建玲, 张鹏, 王振宇, 马秋香. 2020. 木薯C2H2型锌指蛋白转录因子家族全基因组鉴定及表达分析. 植物生理学报, 56 (12):2664-2676. | |
| [21] |
Kagale S, Links M G, Rozwadowski K. 2010. Genome-wide analysis of ethylene-responsive element binding factor-associated amphiphilic repression motif-containing transcriptional regulators in Arabidopsis. Plant Physiology, 152 (3):1109-1134.
doi: 10.1104/pp.109.151704 URL |
| [22] |
Kazan K. 2006. Negative regulation of defence and stress genes by EAR-motif-containing repressors. Trends in Plant Science, 11 (3):109-112.
doi: 10.1016/j.tplants.2006.01.004 pmid: 16473545 |
| [23] |
Klug A. 2010. The discovery of zinc fingers and their applications in gene regulation and genome manipulation. Annual Review of Biochemistry, 79:213-231.
doi: 10.1146/annurev-biochem-010909-095056 pmid: 20192761 |
| [24] |
Kubo K, Sakamoto A, Kobayashi A, Rybka Z, Kanno Y, Nakagawa H, Takatsuji H. 1998. Cys2/His2 zinc-finger protein family of petunia:evolution and general mechanism of target-sequence recognition. Nucleic Acids Research, 26 (2):608-615.
doi: 10.1093/nar/26.2.608 pmid: 9421523 |
| [25] |
Lang G A, Early J D, Martin G C, Darnell R L. 1987. Endo-,para-,and ecodormancy:physiological terminology and classification for dormancy research. HortScience, 22 (3):371-377.
doi: 10.21273/HORTSCI.22.3.371 URL |
| [26] | Li Ya-dong, Pei Jia-bo, Chen Li, Sun Hai-yue. 2021. China blueberry industry report 2020. Journal of Jilin Agricultural University, 43 (1):1-8. (in Chinese) |
| 李亚东, 裴嘉博, 陈丽, 孙海悦. 2021. 2020中国蓝莓产业年度报告. 吉林农业大学学报, 43 (1):1-8. | |
| [27] |
Li Y Q, Ma R, Li R X, Zhao Q, Zhang Z Z, Zong Y, Yao L B, Chen W R, Yang L, Liao F L, Zhu Y Y, Guo W D. 2022. Comparative transcriptomic analysis provides insight into the key regulatory pathways and differentially expressed genes in blueberry flower bud endo- and ecodormancy release. Horticulturae, 8:176.
doi: 10.3390/horticulturae8020176 URL |
| [28] | Liu Q, Wang Z, Xu X, Zhang H, Li C. 2015. Genome-wide analysis of C2H2zinc-finger family transcription factors and their responses to abiotic stresses in poplar(Populus trichocarpa). PLoS ONE, 10 (8):e0134753. |
| [29] |
Liu Z, Coulter J A, Li Y, Zhang X, Meng J, Zhang J, Liu Y. 2020. Genome-wide identification and analysis of the Q-type C2H2 gene family in potato (Solanum tuberosum L.). International Journal of Biological Macromolecules, 153:327-340.
doi: 10.1016/j.ijbiomac.2020.03.022 URL |
| [30] |
Mackay J P, Crossley M. 1998. Zinc fingers are sticking together. Trends in Biochemical Sciences, 23 (1):1-4.
pmid: 9478126 |
| [31] |
Miller J, McLachlan A D, Klug A. 1985. Repetitive zinc-binding domains in the protein transcription factor Ⅲ A from Xenopus oocytes. The EMBO Journal, 4 (6):1609-1614.
doi: 10.1002/embj.1985.4.issue-6 URL |
| [32] |
Noman A, Aqeel M, Khalid N, Islam W, Sanaullah T, Anwar M, Khan S, Ye W, Lou Y. 2019. Zinc finger protein transcription factors:integrated line of action for plant antimicrobial activity. Microbial Pathogenesis, 132:141-149.
doi: S0882-4010(18)31546-8 pmid: 31051192 |
| [33] |
Ohta M, Matsui K, Hiratsu K, Shinshi H, Ohme-Takagi M. 2001. Repression domains of classⅡERF transcriptional repressors share an essential motif for active repression. The Plant Cell, 13 (8):1959-1968.
doi: 10.1105/TPC.010127 URL |
| [34] |
Saitou N, Nei M. 1987. The neighbor-joining method:a new method for reconstructing phylogenetic trees. Molecular Biology and Evolution, 4 (4):406-425.
doi: 10.1093/oxfordjournals.molbev.a040454 pmid: 3447015 |
| [35] |
Sakamoto H, Maruyama K, Sakuma Y, Meshi T, Iwabuchi M, Shinozaki K, Yamaguchi-Shinozaki K. 2004. Arabidopsis Cys2/His2-type zinc-finger proteins function as transcription repressors under drought,cold,and high-salinity stress conditions. Plant Physiology, 136 (1):2734-2746.
doi: 10.1104/pp.104.046599 pmid: 15333755 |
| [36] |
Samish R M. 1954. Dormancy in woody plants. Annual Review of Plant Physiology, 5 (1):183-204.
doi: 10.1146/arplant.1954.5.issue-1 URL |
| [37] |
Sanchez-Garcia I, Rabbitts T H. 1994. The LIM domain:a new structural motif found in zinc-finger-like proteins. Trends in Genetics, 10 (9):315-320.
doi: 10.1016/0168-9525(94)90034-5 URL |
| [38] | Song Yang, Liu Hongdi, Wang Haibo, Zhang Hongjun, Liu Fengzhi. 2019. Molecular cloning and functional characterization of anthocyanin synthesis related genes VcTTG1of blueberry. Acta Horticulturae Sinica, 46 (7):1270-1278. (in Chinese) |
|
宋杨, 刘红弟, 王海波, 张红军, 刘凤之. 2019. 越橘花青苷合成相关基因VcTTG1的克隆与功能鉴定. 园艺学报, 46 (7):1270-1278.
doi: 10.16420/j.issn.0513-353x.2018-0768 |
|
| [39] | Wu Yue-yan, Li Bo, Zhu Ping, Hu Hua-yong. 2011. Effects of plant growth regulator on flowering and endogenous hormones of Rhododendron hybridum. Acta Horticulturae Sinica, 38 (8):1565-1571. (in Chinese) |
| 吴月燕, 李波, 朱平, 胡华勇. 2011. 植物生长调节剂对西洋杜鹃花期及内源激素的影响. 园艺学报, 38 (4):337-349. | |
| [40] |
Yang Q S, Gao Y H, Wu X Y, Moriguchi T, Bai S L, Teng Y W. 2021. Bud endodormancy in deciduous fruit trees:advances and prospects. Horticulture Research, 8 (1):139.
doi: 10.1038/s41438-021-00575-2 |
| [41] | Yang Yu-chun, Wei Xin, Sun Bin, Zhang Duo, Wang Xing-dong, Liu You-chun, Wei Yong-xiang, Tian Ying, Liu Cheng. 2020. Research analysis of cold requirement of different blueberry varieties at low temperature. Bulletin of Agricultural Science and Technology,(1):178-181. (in Chinese) |
| 杨玉春, 魏鑫, 孙斌, 张舵, 王兴东, 刘有春, 魏永祥, 田颖, 刘成. 2020. 蓝莓不同品种低温需冷量研究分析. 农业科技通讯,(1):178-181. | |
| [42] | Yang Yu-chun, Wei Yong-xiang, Wei Xin, Liu You-chun, Wang Xing-dong, Sun Bin, Zhang Duo, Liu Cheng. 2021. Dynamic changes of antioxidant enzyme activities and endogenous hormone contents during flower bud dormancy of two blueberry cultivars. China Fruits,(8):31-35,42. (in Chinese) |
| 杨玉春, 魏永祥, 魏鑫, 刘有春, 王兴东, 孙斌, 张舵, 刘成. 2021. 2个蓝莓品种花芽休眠期抗氧化酶活性及内源激素含量动态变化分析. 中国果树,(8):31-35,42. | |
| [43] |
Yao Fuwen, Wang Meige, Song Chunhui, Song Shangwei, Jiao Jian, Wang Miaomiao, Wang Kun, Bai Tuanhui, Zheng Xianbo. 2021. Identification and expression analysis of HSP90 gene family under high temperature stress in apple. Acta Horticulturae Sinica, 48 (5):849-859. (in Chinese)
doi: 10.16420/j.issn.0513-353x.2020-0895 |
|
姚富文, 王枚阁, 宋春晖, 宋尚伟, 焦健, 王苗苗, 王昆, 白团辉, 郑先波. 2021. 苹果HSP90家族基因鉴定及高温胁迫下的表达分析. 园艺学报, 48 (5):849-859.
doi: 10.16420/j.issn.0513-353x.2020-0895 |
|
| [44] | Yu Ke-da, Ye Mei-juan, Chen Wen-rong, Zhu Kai-li, Zhang Chang-jing, Guo Wei-dong. 2016. Methods for RNA isolation from blueberry tissues. Journal of Zhejiang Normal University(Nat Sci), 39 (1):60-64. (in Chinese) |
| 余柯达, 叶美娟, 陈文荣, 朱凯丽, 张常晶, 郭卫东. 2016. 蓝莓组织RNA提取方法的研究. 浙江师范大学学报(自然科学版), 39 (1):60-64. | |
| [45] |
Yu Lei, Zhou Ya, Zong Yu, Zhang Ying, Qiu Jiaqi, Li Yongqiang, Yang Li, Guo Weidong. 2021. Characteristic and relative expression pattern analysis of FWL/PLAC 8 family in blueberry. Acta Horticulturae Sinica, 48 (2):336-346. (in Chinese)
doi: 10.16420/j.issn.0513-353x.2020-0151 |
|
俞蕾, 周雅, 宗宇, 张颖, 邱佳琪, 李永强, 杨莉, 郭卫东. 2021. 越橘FWL/PLAC8家族基因特征及表达分析. 园艺学报, 48 (2):336-346.
doi: 10.16420/j.issn.0513-353x.2020-0151 URL |
|
| [46] |
Zhang Huilin, Zhu Wan, Tian Li, Zhang Wei. 2019. Characterization and expression analysis of petunia PhZPT2- 12 transcription factor related to cold response. Acta Horticulturae Sinica, 46 (8):1543-1552. (in Chinese)
doi: 10.16420/j.issn.0513-353x.2018-0891 |
|
张慧琳, 朱婉, 田丽, 张蔚. 2019. 矮牵牛冷响应转录因子PhZPT2-12的特性及表达分析. 园艺学报, 46 (8):1543-1552.
doi: 10.16420/j.issn.0513-353x.2018-0891 URL |
| [1] | XU Yue, LI Zixiong, CHEN Jie, SUN Liang. Transcriptional Bases of the Regulation of 2,4-D on Tomato Fruit Shape [J]. Acta Horticulturae Sinica, 2023, 50(4): 802-814. |
| [2] | WANG Xiaochen, NIE Ziye, LIU Xianju, DUAN Wei, FAN Peige, LIANG Zhenchang. Effects of Abscisic Acid on Monoterpene Synthesis in‘Jingxiangyu’Grape Berries [J]. Acta Horticulturae Sinica, 2023, 50(2): 237-249. |
| [3] | A New Rabbiteye Blueberry Cultivar‘Languan’. A New Rabbiteye Blueberry Cultivar‘Languan’ [J]. Acta Horticulturae Sinica, 2022, 49(S2): 43-44. |
| [4] | LIANG Qin, ZHANG Yanhui, KANG Kaiquan, LIU Jinhang, LI Liang, FENG Yu, WANG Chao, YANG Chao, LI Yongyu. Molecular Evolution of MiR168 Family and Their Expression Profiling During Dormancy of Pyrus pyrifolia [J]. Acta Horticulturae Sinica, 2022, 49(5): 958-972. |
| [5] | XU Tong, SHAO Lingmei, WANG Xiaobin, ZHANG Runlong, ZHANG Kaijing, XIA Yiping, ZHANG Jiaping, LI Danqing. Research Progress on Winter Dormancy of Perennial Monocots [J]. Acta Horticulturae Sinica, 2022, 49(12): 2703-2721. |
| [6] | YU Jianqiang, GU Kaidi, WANG Chuanzeng, HU Dagang. Functional Characterization of An Apple Pyrophosphate-dependent Phosphofructokinase Gene MdPFPβ in Regulating Soluble Sugar Accumulation [J]. Acta Horticulturae Sinica, 2022, 49(10): 2223-2235. |
| [7] | YANG Bo, WEI Jia, LI Kunfeng, WANG Chengliang, NI Junbei, TENG Yuanwen, and BAI Songling. PpyERF060-PpyABF3-PpyMADS71 Regulates Ethylene Signaling Pathway- Mediated Pear Bud Dormancy Process [J]. Acta Horticulturae Sinica, 2022, 49(10): 2249-2262. |
| [8] | XU Guohui, AN Qi, ZHAO Lina, LIU Guoling, LOU Xin, and WANG Hexin, . A New Blueberry Cultivar‘Morning Snow’Suitable for Cluster Harvesting [J]. Acta Horticulturae Sinica, 2021, 48(S2): 2795-2796. |
| [9] | XU Guohui, AN Qi, LIU Guoling, ZHAO Lina, and WANG Hexin, . A New Blueberry Cultivar‘Chasing Dream’ [J]. Acta Horticulturae Sinica, 2021, 48(S2): 2797-2798. |
| [10] | YANG Weihai, ZENG Lizhen, XIAO Qiusheng, SHI Shengyou. Changes of Fruit Abscission and Carbohydrate,ABA and Related Genes Expression in the Pericarp and Fruit Abscission Zone of Longan Under Starvation Stress [J]. Acta Horticulturae Sinica, 2021, 48(8): 1457-1469. |
| [11] | ZENG Zexiang, XIAO Xianmei, TAN Xiaoli, FAN Zhongqi, CHEN Jianye. Characteristics of the Transcription Factor BrWRKY57 and Its Regulation on BrPPH1 and BrNCED3 [J]. Acta Horticulturae Sinica, 2021, 48(3): 518-530. |
| [12] | YU Lei, ZHOU Ya, ZONG Yu, ZHANG Ying, QIU Jiaqi, LI Yongqiang, YANG Li, GUO Weidong. Characteristic and Relative Expression Pattern Analysis of FWL/PLAC8 Family in Blueberry [J]. Acta Horticulturae Sinica, 2021, 48(2): 336-346. |
| [13] | LI Yuzhuo, LIU Ke, YUAN Lu, CAO Liwen, WANG Tingjin, GAN Susheng, CHEN Liping. Cloning and Functional Analyses of BrNAP in Postharvest Leaf Senescence in Chinese Flowering Cabbage [J]. Acta Horticulturae Sinica, 2021, 48(1): 60-72. |
| [14] | QI Qige1,GE Lili1,ZHANG Qichang1,*,JIANG Weiqing2,and WU Banghua1. A New Blueberry Cultivar‘Beihua 1’ [J]. ACTA HORTICULTURAE SINICA, 2020, 47(6): 1217-1218. |
| [15] | DAI Wenshan1,2,WANG Min1,2,and LIU Jihong1,*. Enhanced Dehydration Tolerance in Lemon by Overexpression of CrNCED1(9-cis-epoxycarotenoid dioxygenase gene)from Citrus reshni [J]. ACTA HORTICULTURAE SINICA, 2020, 47(3): 551-561. |
| Viewed | ||||||
|
Full text |
|
|||||
|
Abstract |
|
|||||
Copyright © 2012 Acta Horticulturae Sinica 京ICP备10030308号-2 国际联网备案号 11010802023439
Tel: 010-82109523 E-Mail: yuanyixuebao@126.com
Support by: Beijing Magtech Co.Ltd