
Acta Horticulturae Sinica ›› 2023, Vol. 50 ›› Issue (4): 802-814.doi: 10.16420/j.issn.0513-353x.2022-0023
• Research Papers • Previous Articles Next Articles
XU Yue,*, LI Zixiong,*, CHEN Jie, SUN Liang( )
)
Received:2022-04-21
Revised:2022-12-11
Online:2023-04-25
Published:2023-04-27
Contact:
**(E-mail:liang_sun@cau.edu.cn)
CLC Number:
XU Yue, LI Zixiong, CHEN Jie, SUN Liang. Transcriptional Bases of the Regulation of 2,4-D on Tomato Fruit Shape[J]. Acta Horticulturae Sinica, 2023, 50(4): 802-814.
Add to citation manager EndNote|Ris|BibTeX
URL: https://www.ahs.ac.cn/EN/10.16420/j.issn.0513-353x.2022-0023
| 2,4-D处理/(mmol · L-1) 2,4-D treatment | 最大宽度/cm Maximum width | 最大长度/cm Maximum height | 果形指数 Fruit shape index | 果梗端比例 Proximal end ratio | ||
|---|---|---|---|---|---|---|
| 基因型 Genotype | WT | 0 | 1.20 ± 0.01 a | 1.23 ± 0.01 d | 1.02 ± 0.01 d | 0.13 ± 0.01 d |
| 270 | 0.95 ± 0.03 b | 1.35 ± 0.02 cd | 1.43 ± 0.03 c | 0.31 ± 0.03 bc | ||
| sun | 0 | 1.14 ± 0.04 a | 1.65 ± 0.03 abc | 1.45 ± 0.03 c | 0.15 ± 0.03 d | |
| 270 | 1.65 ± 0.03 b | 1.92 ± 0.03 a | 2.48 ± 0.27 a | 0.36 ± 0.03 b | ||
| ovate | 0 | 1.19 ± 0.03 a | 1.52 ± 0.12 bcd | 1.28 ± 0.13 cd | 0.21 ± 0.03 cd | |
| 270 | 0.92 ± 0.12 b | 1.80 ± 0.25 ab | 1.96 ± 0.05 b | 0.49 ± 0.08 a | ||
| 因素效应 Factorial effects (P-value) | 基因型Genotype | 0.02 | 1.93e-05 | 1.03e-06 | 2.44e-4 | |
| 处理Treatment | 4.56e-07 | 1.28e-03 | 4.24e-08 | 5.36e-08 | ||
| 基因型与处理互作 Genotype × Treatment | 0.31 | 0.45 | 2.94e-03 | 0.17 | ||
Table 1 Morphological analysis of fruits after 2,4-D treatment
| 2,4-D处理/(mmol · L-1) 2,4-D treatment | 最大宽度/cm Maximum width | 最大长度/cm Maximum height | 果形指数 Fruit shape index | 果梗端比例 Proximal end ratio | ||
|---|---|---|---|---|---|---|
| 基因型 Genotype | WT | 0 | 1.20 ± 0.01 a | 1.23 ± 0.01 d | 1.02 ± 0.01 d | 0.13 ± 0.01 d |
| 270 | 0.95 ± 0.03 b | 1.35 ± 0.02 cd | 1.43 ± 0.03 c | 0.31 ± 0.03 bc | ||
| sun | 0 | 1.14 ± 0.04 a | 1.65 ± 0.03 abc | 1.45 ± 0.03 c | 0.15 ± 0.03 d | |
| 270 | 1.65 ± 0.03 b | 1.92 ± 0.03 a | 2.48 ± 0.27 a | 0.36 ± 0.03 b | ||
| ovate | 0 | 1.19 ± 0.03 a | 1.52 ± 0.12 bcd | 1.28 ± 0.13 cd | 0.21 ± 0.03 cd | |
| 270 | 0.92 ± 0.12 b | 1.80 ± 0.25 ab | 1.96 ± 0.05 b | 0.49 ± 0.08 a | ||
| 因素效应 Factorial effects (P-value) | 基因型Genotype | 0.02 | 1.93e-05 | 1.03e-06 | 2.44e-4 | |
| 处理Treatment | 4.56e-07 | 1.28e-03 | 4.24e-08 | 5.36e-08 | ||
| 基因型与处理互作 Genotype × Treatment | 0.31 | 0.45 | 2.94e-03 | 0.17 | ||
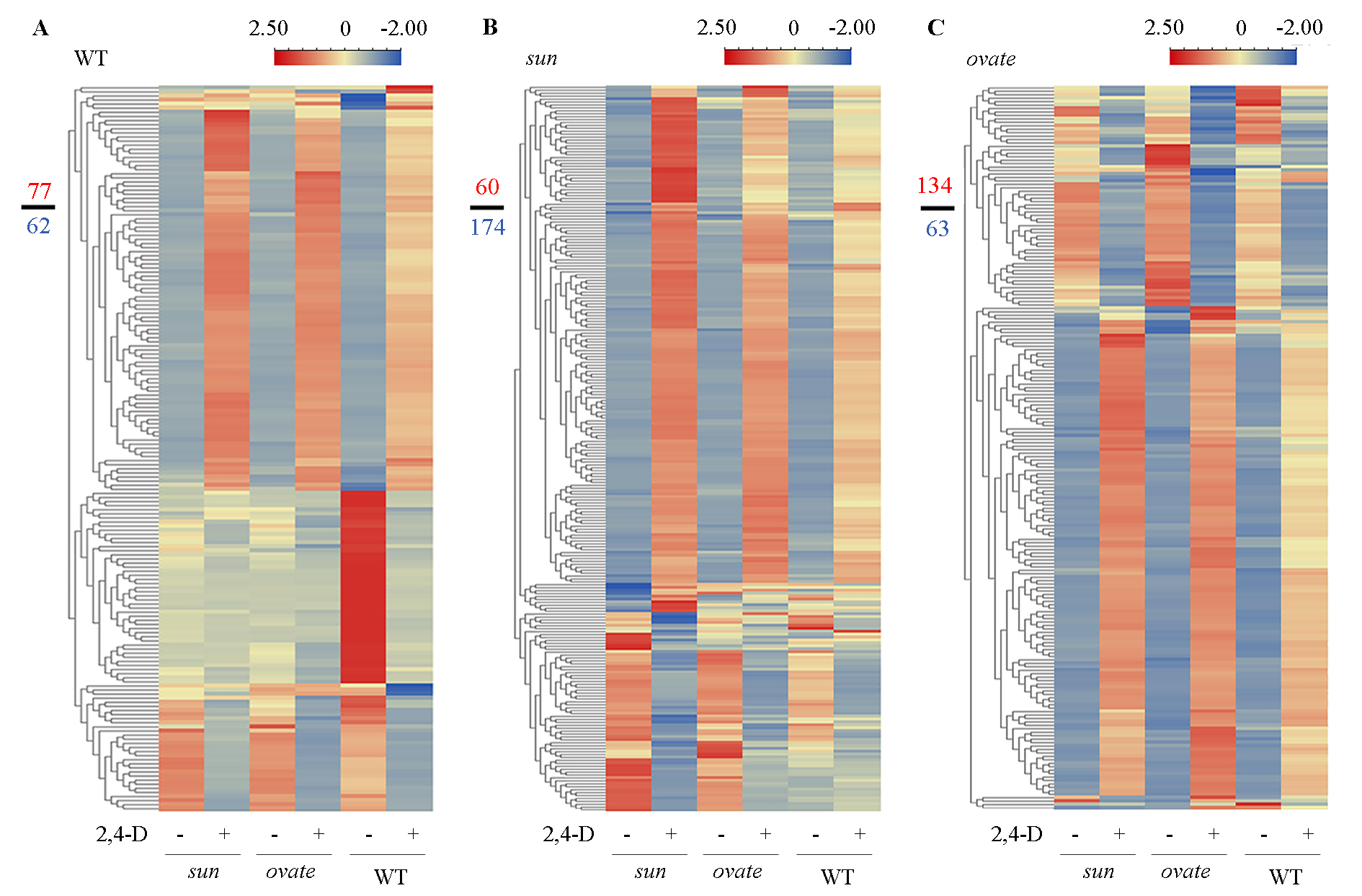
Fig. 3 Heatmaps of the differentially expressed genes(DEGs)in the anthesis ovaries of WT(A),sun(B)and ovate(C)NILs after 2,4-D treatment Numerator and denominator in the left represent the number of the up-regulated and down-regulated genes,respectively.
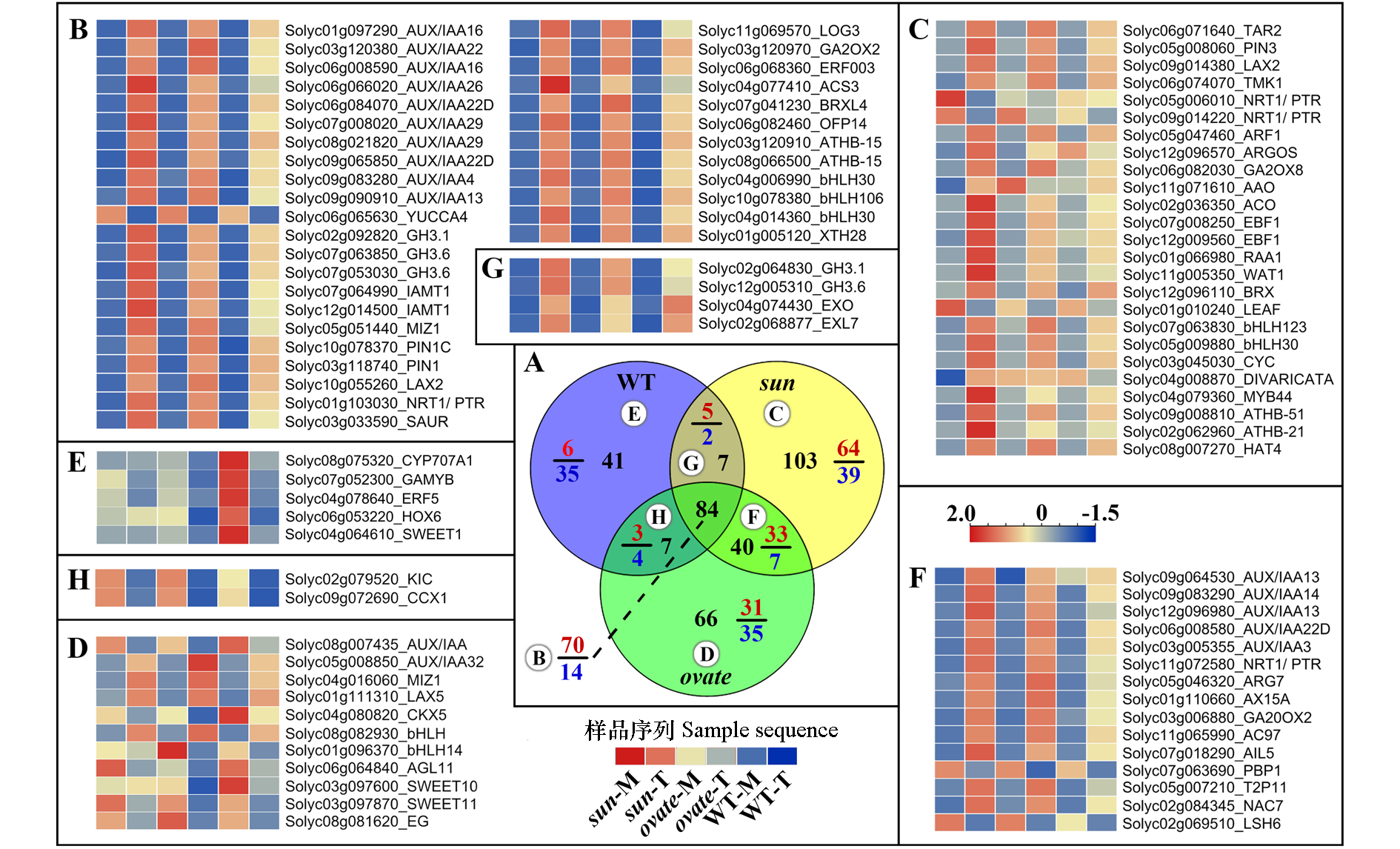
Fig. 4 Venn diagram and heatmap of the interested differentially expressed genes(DEGs)in the anthesis ovaries of WT,sun and ovate NILs after 2,4-D treatment A:Venn diagram of DEGs in WT,sun and ovate NILs after 2,4-D treatment. The black number in each part represents the total number of DEGs,and the numerator and denominator of the fraction represent the number of up-regulated and down-regulated genes,respectively. Letters with circles represent the heatmap corresponding to these genes. B:Heatmap of the interested DEGs shared by the 3 NILs;C-E:heatmaps of the interested genes that only differentially expressed in WT,sun and ovate NILs,respectively. F:Interested DEGs shared by sun and ovate. G:DEGs shared by sun and WT. H:DEGs shared by ovate and WT.

Fig. 5 String analysis of the differentially expressed genes(DEGs)caused by the 2,4-D treatment A:Analysis of DEGs that shared by the three NILs. B:Analysis of DEGs that were only in sun NIL. C:Analysis of DEGs that were only in ovate NIL. D:Analysis of DEGs that were only in WT NIL. E:Analysis of DEGs that were shared by sun and ovate NILs. F:Analysis of DEGs that were shared by WT and ovate NILs.
| [1] |
Azimzadeh J, Nacry P, Christodoulidou A, Drevensek S, Camilleri C, Amiour N, Parcy F, Pastuglia M, Bouchez D. 2008. Arabidopsis TONNEAU1 proteins are essential for preprophase band formation and interact with centrin. Plant Cell, 20 (8):2146-2159.
doi: 10.1105/tpc.107.056812 pmid: 18757558 |
| [2] |
Baima S, Nobili F, Sessa G, Lucchetti S, Ruberti I, Morelli G. 1995. The expression of the Athb-8 homeobox gene is restricted to provascular cells in Arabidopsis thaliana. Development, 121 (12):4171-4182.
doi: 10.1242/dev.121.12.4171 pmid: 8575317 |
| [3] |
Breia R, Conde A, Badim H, Fortes A M, Geros H, Granell A. 2021. Plant SWEETs:from sugar transport to plant-pathogen interaction and more unexpected physiological roles. Plant Physiol, 186 (2):836-852.
doi: 10.1093/plphys/kiab127 URL |
| [4] |
Burstenbinder K, Moller B, Plotner R, Stamm G, Hause G, Mitra D, Abel S. 2017. The IQD family of calmodulin-binding proteins links calcium signaling to microtubules,membrane subdomains,and the nucleus. Plant Physiol, 173 (3):1692-1708.
doi: 10.1104/pp.16.01743 URL |
| [5] |
Burstenbinder K, Savchenko T, Muller J, Adamson A W, Stamm G, Kwong R, Zipp B J, Dinesh D C, Abel S. 2013. Arabidopsis calmodulin-binding protein IQ67-domain 1 localizes to microtubules and interacts with kinesin light chain-related protein-1. Journal of Biological Chemistry, 288 (3):1871-1882.
doi: 10.1074/jbc.M112.396200 URL |
| [6] |
Chen S, Wang X J, Tan G F, Zhou W Q, Wang G L. 2020. Gibberellin and the plant growth retardant Paclobutrazol altered fruit shape and ripening in tomato. Protoplasma, 257 (3):853-861.
doi: 10.1007/s00709-019-01471-2 pmid: 31863170 |
| [7] | Chu Y H, Jang J C, Huang Z, van der Knaap E. 2019. Tomato locule number and fruit size controlled by natural alleles of lc and fas. Plant Direct, 3 (7):e00142. |
| [8] |
Damodharan S, Zhao D, Arazi T. 2016. A common miRNA160-based mechanism regulates ovary patterning,floral organ abscission and lamina outgrowth in tomato. The Plant Journal, 86 (6):458-471.
doi: 10.1111/tpj.13127 pmid: 26800988 |
| [9] |
de Jong M, Wolters-Arts M, Garcia-Martinez J L, Mariani C, Vriezen W H. 2011. The Solanum lycopersicum AUXIN RESPONSE FACTOR 7(SlARF7)mediates cross-talk between auxin and gibberellin signalling during tomato fruit set and development. Journal of Experimental Botany, 62 (2):617-626.
doi: 10.1093/jxb/erq293 URL |
| [10] |
Drevensek S, Goussot M, Duroc Y, Christodoulidou A, Steyaert S, Schaefer E, Duvernois E, Grandjean O, Vantard M, Bouchez D, Pastuglia M. 2012. The Arabidopsis TRM1-TON1 interaction reveals a recruitment network common to plant cortical microtubule arrays and eukaryotic centrosomes. Plant Cell, 24 (1):178-191.
doi: 10.1105/tpc.111.089748 URL |
| [11] | Eyer L, Vain T, Parizkova B, Oklestkova J, Barbez E, Kozubikova H, Pospisil T, Wierzbicka R, Kleine-Vehn J, Franek M, Strnad M, Robert S, Novak O. 2016. 2,4-D and IAA Amino acid conjugates show distinct metabolism in Arabidopsis. PLoS ONE, 11 (7):e0159269. |
| [12] |
Feller A, Machemer K, Braun E L, Grotewold E. 2011. Evolutionary and comparative analysis of MYB and bHLH plant transcription factors. The Plant Journal, 66 (1):94-116.
doi: 10.1111/j.1365-313X.2010.04459.x pmid: 21443626 |
| [13] |
Hao Y, Hu G, Breitel D, Liu M, Mila I, Frasse P, Fu Y, Aharoni A, Bouzayen M, Zouine M. 2015. Auxin response factor SlARF 2 is an essential component of the regulatory mechanism controlling fruit ripening in tomato. PLoS Genetics, 11 (12):e1005649.
doi: 10.1371/journal.pgen.1005649 URL |
| [14] |
Hedden P, Thomas S G. 2012. Gibberellin biosynthesis and its regulation. Biochemical Journal, 444 (1):11-25.
doi: 10.1042/BJ20120245 pmid: 22533671 |
| [15] |
Huang Z, van Houten J, Gonzalez G, Xiao H, van der Knaap E. 2013. Genome-wide identification, phylogeny and expression analysis of SUN,OFP and YABBY gene family in tomato. Molecular Genetics Genomics, 288 (3-4):111-129.
doi: 10.1007/s00438-013-0733-0 URL |
| [16] |
Hur Y S, Um J H, Kim S, Kim K, Park H J, Lim J S, Kim W Y, Jun S E, Yoon E K, Lim J, Ohme-Takagi M, Kim D, Park J, Kim G T, Cheon C I. 2015. Arabidopsis thaliana homeobox 12(ATHB12),a homeodomain-leucine zipper protein,regulates leaf growth by promoting cell expansion and endoreduplication. New Phytologist, 205 (1):316-328.
doi: 10.1111/nph.12998 pmid: 25187356 |
| [17] |
Korasick D A, Enders T A, Strader L C. 2013. Auxin biosynthesis and storage forms. Journal of Experimental Botany, 64 (9):2541-55.
doi: 10.1093/jxb/ert080 pmid: 23580748 |
| [18] |
Kurakawa T, Ueda N, Maekawa M, Kobayashi K, Kojima M, Nagato Y, Sakakibara H, Kyozuka J. 2007. Direct control of shoot meristem activity by a cytokinin-activating enzyme. Nature, 445 (7128):652-655.
doi: 10.1038/nature05504 |
| [19] |
Lazzaro M D, Wu S, Snouffer A, Wang Y, van der Knaap E. 2018. Plant organ shapes are regulated by protein interactions and associations with microtubules. Frontiers of Plant Science, 9:1766.
doi: 10.3389/fpls.2018.01766 URL |
| [20] | Liu J, van Eck J, Cong B, Tanksley S D. 2002. A new class of regulatory genes underlying the cause of pear-shaped tomato fruit. Proceedings of the National Academy of Science of United States of America, 99 (20):13302-13306. |
| [21] |
Mouchel C F, Briggs G C, Hardtke C S. 2004. Natural genetic variation in Arabidopsis identifies BREVIS RADIX,a novel regulator of cell proliferation and elongation in the root. Genes & Development, 18 (6):700-714.
doi: 10.1101/gad.1187704 URL |
| [22] | Perrot-Rechenmann C. 2010. Cellular responses to auxin:division versus expansion. Cold Spring Harb Perspect Biol, 2 (5):a001446. |
| [23] |
Schmulling T, Werner T, Riefler M, Krupkova E, Bartrina Y, Manns I. 2003. Structure and function of cytokinin oxidase/dehydrogenase genes of maize,rice,Arabidopsis and other species. Journal of Plant Research, 116 (3):241-252.
doi: 10.1007/s10265-003-0096-4 URL |
| [24] |
Snouffer A, Kraus C, van der Knaap E. 2020. The shape of things to come:ovate family proteins regulate plant organ shape. Current Opinion in Plant Biology, 53:98-105.
doi: S1369-5266(19)30096-2 pmid: 31837627 |
| [25] |
Spinner L, Pastuglia M, Belcram K, Pegoraro M, Goussot M, Bouchez D, Schaefer D G. 2010. The function of TONNEAU1 in moss reveals ancient mechanisms of division plane specification and cell elongation in land plants. Development, 137 (16):2733-2742.
doi: 10.1242/dev.043810 pmid: 20663817 |
| [26] |
Sun L, Rodriguez G R, Clevenger J P, Illa-Berenguer E, Lin J, Blakeslee J J, Liu W, Fei Z, Wijeratne A, Meulia T, van der Knaap E. 2015. Candidate gene selection and detailed morphological evaluations of fs8.1,a quantitative trait locus controlling tomato fruit shape. Journal of Experimental Botany, 66 (20):6471-6482.
doi: 10.1093/jxb/erv361 URL |
| [27] |
Vaddepalli P, de Zeeuw T, Strauss S, Burstenbinder K, Liao C Y, Ramalho J J, Smith R S, Weijers D. 2021. Auxin-dependent control of cytoskeleton and cell shape regulates division orientation in the Arabidopsis embryo. Current Biology, 31 (22):4946-4955.
doi: 10.1016/j.cub.2021.09.019 URL |
| [28] | van der Knaap E, Chakrabarti M, Chu Y H, Clevenger J P, Illa-Berenguer E, Huang Z, Keyhaninejad N, Mu Q, Sun L, Wang Y, Wu S. 2014. What lies beyond the eye:the molecular mechanisms regulating tomato fruit weight and shape. Frontiers of Plant Science, 5:227. |
| [29] | van der Knaap E, Østergaard L. 2018. Shaping a fruit:developmental pathways that impact growth patterns. Seminars in Cell & Developmental Biology, 79:27-36. |
| [30] |
Wang Y, Clevenger J P, Illa-Berenguer E, Meulia T, van der Knaap E, Sun L. 2019. A comparison of sun,ovate,fs8.1 and auxin application on tomato fruit shape and gene expression. Plant and Cell Physiology, 60 (5):1067-1081.
doi: 10.1093/pcp/pcz024 URL |
| [31] |
Wu S, Clevenger J P, Sun L, Visa S, Kamiya Y, Jikumaru Y, Blakeslee J, van der Knaap E. 2015. The control of tomato fruit elongation orchestrated by sun,ovate and fs8.1 in a wild relative of tomato. Plant Science, 238:95-104.
doi: 10.1016/j.plantsci.2015.05.019 URL |
| [32] |
Wu S, Xiao H, Cabrera A, Meulia T, van der Knaap E. 2011. SUN regulates vegetative and reproductive organ shape by changing cell division patterns. Plant Physiology, 157 (3):1175-1186.
doi: 10.1104/pp.111.181065 URL |
| [33] |
Wu S, Zhang B, Keyhaninejad N, Rodriguez G R, Kim H J, Chakrabarti M, Illa-Berenguer E, Taitano N K, Gonzalo M J, Diaz A, Pan Y, Leisner C P, Halterman D, Buell C R, Weng Y, Jansky S H, van Eck H, Willemsen J, Monforte A J, Meulia T, van der Knaap E. 2018. A common genetic mechanism underlies morphological diversity in fruits and other plant organs. Nature Communications, 9 (1):4734.
doi: 10.1038/s41467-018-07216-8 pmid: 30413711 |
| [34] |
Xiao H, Jiang N, Schaffner E, Stockinger E J, van der Knaap E. 2008. A retrotransposon-mediated gene duplication underlies morphological variation of tomato fruit. Science, 319 (5869):1527-1530.
doi: 10.1126/science.1153040 pmid: 18339939 |
| [35] |
Xiao H, Radovich C, Welty N, Hsu J, Li D, Meulia T, van der Knaap E. 2009. Integration of tomato reproductive developmental landmarks and expression profiles,and the effect of SUN on fruit shape. BMC Plant Biology, 9:49.
doi: 10.1186/1471-2229-9-49 pmid: 19422692 |
| [36] |
Zhang C, Li X Y, Yan H F, Ullah I, Zuo Z Y, Li L L, Yu J J. 2020. Effects of irrigation quantity and biochar on soil physical properties,growth characteristics,yield and quality of greenhouse tomato. Agricultural Water Management, 241:106263.
doi: 10.1016/j.agwat.2020.106263 URL |
| [37] |
Zhang S L, Gu X, Shao J C, Hu Z F, Yang W C, Wang L P, Su H Y, Zhu L Y. 2021. Auxin metabolism is involved in fruit set and early fruit development in the parthenocarpic tomato“R35-P”. Frontiers in Plant Science, 12:671713.
doi: 10.3389/fpls.2021.671713 URL |
| [38] |
Zhao Q, Wu Y, Gao L, Ma J, Li C Y, Xiang C B. 2014. Sulfur nutrient availability regulates root elongation by affecting root indole-3-acetic acid levels and the stem cell niche. Journal of Integrative Plant Biology, 56 (12):1151-1163.
doi: 10.1111/jipb.12217 |
| [1] | LI Ke, SHEN Mengxiao, PAN Weihao, ZHANG Shixuan, MAO Xinye, YIN Yahong, LI Yongqiang, ZHU Youyin, GUO Weidong. Preliminary Investigation of C2H2 Family Genes in Blueberry Flower Bud Dormancy Release [J]. Acta Horticulturae Sinica, 2023, 50(4): 737-753. |
| [2] | ZHENG Jinrong, NIE Jun, LI Yanhong, TAN Delong, XIE Yuming, ZHANG Changyuan. A Cherry Tomato Cultivar‘Yuekeda 101’ [J]. Acta Horticulturae Sinica, 2023, 50(4): 909-910. |
| [3] | RAO Zhixiong, AN Yuyan, CAO Rongxiang, TANG Quan, WANG Liangju. Studies on Mechanisms of ALA Alleviating ABA Inhibiting Root Growth of Strawberry [J]. Acta Horticulturae Sinica, 2023, 50(3): 461-474. |
| [4] | LIU Yuhan, TAO Ning, WANG Qingguo, LI Qingqing. ABC Transporter SlABCG23 Regulates Jasmonic Acid Signaling Pathway in Tomato [J]. Acta Horticulturae Sinica, 2023, 50(3): 559-568. |
| [5] | WANG Zehan, YU Wentao, WANG Pengjie, LIU Caiguo, FAN Xiaojing, GU Mengya, CAI Chunping, WANG Pan, YE Naixing. WGCNA Analysis of Differentially Expressed Genes in Floral Organs of Tea Germplasms with Ovary-glabrous and Ovary-trichome [J]. Acta Horticulturae Sinica, 2023, 50(3): 620-634. |
| [6] | JIANG Yu, TU Xunliang, HE Junrong. Analysis of Differential Expression Genes in Leaves of Leaf Color Mutant of Chinese Orchid [J]. Acta Horticulturae Sinica, 2023, 50(2): 371-381. |
| [7] | YU Tingting, LI Huan, NING Yuansheng, SONG Jianfei, PENG Lulin, JIA Junqi, ZHANG Weiwei, YANG Hongqiang. Genome-wide Identification of GRAS Gene Family in Apple and Expression Analysis of Its Response to Auxin [J]. Acta Horticulturae Sinica, 2023, 50(2): 397-409. |
| [8] | ZHENG Qingbo, BAO Zeyang, LAN Qingqing, ZHOU Yuwen, ZHOU Yufei, ZHENG Caixia, LI Xu. Advances in Studies on Adventitious Root Formation by Juvenile- and Auxin-determined [J]. Acta Horticulturae Sinica, 2023, 50(2): 441-450. |
| [9] | SHI Hongli, LI La, GUO Cuimei, YU Tingting, JIAN Wei, YANG Xingyong. Isolation,Identification and Analysis of Biocontrol Ability of Biocontrol Strain TL1 Against Tomato Botrytis cinerea [J]. Acta Horticulturae Sinica, 2023, 50(1): 79-90. |
| [10] | HU Jingyu, QUE Kaijuan, MIAO Tianli, WU Shaozheng, WANG Tiantian, ZHANG Lei, DONG Xian, JI Pengzhang, DONG Jiahong. Identification of Tomato Spotted Wilt Orthotospovirus Infecting Iris tectorum [J]. Acta Horticulturae Sinica, 2023, 50(1): 170-176. |
| [11] | ZHAO Xueyan, WANG Qi, WANG Li, WANG Fangyuan, WANG Qing, LI Yan. Comparative Transcriptome Analysis of Differential Expression in Different Tissues of Corydalis yanhusuo [J]. Acta Horticulturae Sinica, 2023, 50(1): 177-187. |
| [12] | ZHENG Jirong, WANG Tonglin, and HU Songshen. A New Tomato Cultivar‘Hangza 603’with High Quality [J]. Acta Horticulturae Sinica, 2022, 49(S2): 103-104. |
| [13] | ZHENG Jirong and WANG Tonglin. A New Tomato Cultivar‘Hangza 601’ [J]. Acta Horticulturae Sinica, 2022, 49(S2): 105-106. |
| [14] | ZHENG Jirong and WANG Tonglin. A New Cherry Tomato Cultivar‘Hangza 503’ [J]. Acta Horticulturae Sinica, 2022, 49(S2): 107-108. |
| [15] | HUANG Tingting, LIU Shuqin, ZHANG Yongzhi, LI Ping, ZHANG Zhihuan, and SONG Libo. A New Cherry Tomato Cultivar‘Yingshahong 4’ [J]. Acta Horticulturae Sinica, 2022, 49(S2): 109-110. |
| Viewed | ||||||
|
Full text |
|
|||||
|
Abstract |
|
|||||
Copyright © 2012 Acta Horticulturae Sinica 京ICP备10030308号-2 国际联网备案号 11010802023439
Tel: 010-82109523 E-Mail: yuanyixuebao@126.com
Support by: Beijing Magtech Co.Ltd