
Acta Horticulturae Sinica ›› 2022, Vol. 49 ›› Issue (5): 958-972.doi: 10.16420/j.issn.0513-353x.2021-0103
• Research Papers • Previous Articles Next Articles
LIANG Qin1,2, ZHANG Yanhui3, KANG Kaiquan4, LIU Jinhang1, LI Liang1, FENG Yu1, WANG Chao1, YANG Chao1,*( ), LI Yongyu1,*(
), LI Yongyu1,*( )
)
Received:2022-01-11
Revised:2022-04-01
Online:2022-05-25
Published:2022-05-25
Contact:
YANG Chao,LI Yongyu
E-mail:yycc32@126.com;lilin3182@163.com
CLC Number:
LIANG Qin, ZHANG Yanhui, KANG Kaiquan, LIU Jinhang, LI Liang, FENG Yu, WANG Chao, YANG Chao, LI Yongyu. Molecular Evolution of MiR168 Family and Their Expression Profiling During Dormancy of Pyrus pyrifolia[J]. Acta Horticulturae Sinica, 2022, 49(5): 958-972.
Add to citation manager EndNote|Ris|BibTeX
URL: https://www.ahs.ac.cn/EN/10.16420/j.issn.0513-353x.2021-0103
| 用途Purpose | 引物名称Primer name | 引物序列(5′-3′)Primer sequence |
|---|---|---|
| 克隆Clone | ppy-miR168a-F | ATAGAGGCGTAGGCCTGTGT |
| ppy-miR168a-R | TGATGGTGGGAATTGGGATAT | |
| 克隆Clone | ppy-miR168b-F | TGTCTGTTTGGGCGTTGATT |
| ppy-miR168b-R | AAACTATTGTAGATCAGACAACAACGC | |
| qRT-PCR | Ppy-miR168a | TCGCTTGGTGCAGGTCG |
| AGO1-F | CACCAGCACCACAACAATA | |
| AGO1-R | ACCACCACCATAGCCTCT | |
| CalS3-F | AGAGGCGGATAACTCG | |
| CalS3-R | GCAACACGAAGAATGG | |
| Actin-F | CCATCCAGGCTGTTCTCTC | |
| Actin-R | GCAAGGTCCAGACGAAGG | |
| 5SrRNA-F | TAGGCGTAGAGGAACCACACCAAT | |
| 5SrRNA-R | ACAGTATCGTCACCGCAGTAGAGT |
Table 1 The primers used in this experiment
| 用途Purpose | 引物名称Primer name | 引物序列(5′-3′)Primer sequence |
|---|---|---|
| 克隆Clone | ppy-miR168a-F | ATAGAGGCGTAGGCCTGTGT |
| ppy-miR168a-R | TGATGGTGGGAATTGGGATAT | |
| 克隆Clone | ppy-miR168b-F | TGTCTGTTTGGGCGTTGATT |
| ppy-miR168b-R | AAACTATTGTAGATCAGACAACAACGC | |
| qRT-PCR | Ppy-miR168a | TCGCTTGGTGCAGGTCG |
| AGO1-F | CACCAGCACCACAACAATA | |
| AGO1-R | ACCACCACCATAGCCTCT | |
| CalS3-F | AGAGGCGGATAACTCG | |
| CalS3-R | GCAACACGAAGAATGG | |
| Actin-F | CCATCCAGGCTGTTCTCTC | |
| Actin-R | GCAAGGTCCAGACGAAGG | |
| 5SrRNA-F | TAGGCGTAGAGGAACCACACCAAT | |
| 5SrRNA-R | ACAGTATCGTCACCGCAGTAGAGT |
| 科 Family | 物种 Species | 简写 Logogram | miR168数量 Quantity of miR168 | pre-miR168数量 Quantity of pre-miR168 |
|---|---|---|---|---|
| 十字花科Brassicaceae | 拟南芥Arabidopsis thaliana | ath | 4 | 2 |
| 油菜Brassica napus | bna | 2 | 2 | |
| 琴叶拟南芥Arabidopsis lyrata | aly | 4 | 2 | |
| 芜菁Brassica rapa | bra | 6 | 3 | |
| 亚麻荠Camelina sativa | csa | 1 | 1 | |
| 禾本科Poaceae | 青稞Hordeum vulgare | hvu | 2 | 1 |
| 二穗短柄草Brachypodium distachyon | bdi | 2 | 1 | |
| 甘蔗属Saccharum sp | ssp | 1 | 1 | |
| 甘蔗Saccharum officinarum | sof | 2 | 2 | |
| 水稻Oryza sativa | osa | 3 | 2 | |
| 高粱Sorghum bicolor | sbi | 1 | 1 | |
| 玉米Zea mays | zma | 4 | 2 | |
| 山羊草Aegilops tauschii | ata | 1 | 1 | |
| 豆科Leguminosae | 大豆Glycine max | gma | 2 | 2 |
| 蒺藜苜蓿Medicago truncatula | mtr | 4 | 3 | |
| 大叶相思Acacia auriculiformis | aau | 1 | 1 | |
| 豇豆Vigna unguiculata | vun | 1 | 1 | |
| 百脉根Lotus japonicus | lja | 1 | 1 | |
| 科 Family | 物种 Species | 简写 Logogram | miR168数量 Quantity of miR168 | pre-miR168数量 Quantity of pre-miR168 |
| 杨柳科Salicaceae Mirb | 毛果杨Populus trichocarpa | ptc | 4 | 2 |
| 葡萄科Vitaceae Juss | 葡萄Vitis vinifera | vvi | 1 | 1 |
| 毛茛科Ranunculaceae Juss | 耧斗菜Aquilegia viridiflora | aqc | 1 | 1 |
| 芸香科Rutaceae Juss | 脐橙Citrus sinensis | csi | 2 | 1 |
| 柑橘Citrus reticulata | cre | 1 | 1 | |
| 克萊门柚Citrus clementina | ccl | 1 | 1 | |
| 大戟科Rosaceae Juss | 木薯Manihot esculenta | mes | 1 | 1 |
| 三叶橡胶树 Hevea brasiliensis | hbr | 1 | 1 | |
| 蓖麻 Ricinus communis | rco | 1 | 1 | |
| 葫芦科Cucurbitaceae | 甜瓜Cucumis melo | cme | 1 | 1 |
| 菊科Asteraceae Bercht | 刺苞菜蓟Cynara cardunculus | cca | 1 | 1 |
| 亚麻科Linaceae | 亚麻Linum usitatissimum | lus | 2 | 2 |
| 茄科Solanaceae Juss | 烟草Nicotiana tabacum | nta | 5 | 5 |
| 番茄Lycopersicon esculentum | sly | 4 | 2 | |
| 蔷薇科Rosaceae Juss | 苹果Malus domestica | mdm | 2 | 2 |
| 碧桃Prunus persica | ppe | 1 | 1 | |
| 森林草莓Fragaria vesca | fve | 1 | 1 | |
| 凤梨科Bromeliaceae Juss | 莺歌凤梨Vriesea carinata | vcr | 4 | 2 |
| 松科Pinaceae Lindl | 挪威云杉Picea abies | pab | 2 | 2 |
| 百合科Liliaceae Juss | 芦笋Asparagus officinalis | aof | 2 | 2 |
| 梧桐科Sterculiaceae | 可可Theobroma cacao | tcc | 1 | 1 |
Table 2 Distribution and quantity of plant miR168 family members
| 科 Family | 物种 Species | 简写 Logogram | miR168数量 Quantity of miR168 | pre-miR168数量 Quantity of pre-miR168 |
|---|---|---|---|---|
| 十字花科Brassicaceae | 拟南芥Arabidopsis thaliana | ath | 4 | 2 |
| 油菜Brassica napus | bna | 2 | 2 | |
| 琴叶拟南芥Arabidopsis lyrata | aly | 4 | 2 | |
| 芜菁Brassica rapa | bra | 6 | 3 | |
| 亚麻荠Camelina sativa | csa | 1 | 1 | |
| 禾本科Poaceae | 青稞Hordeum vulgare | hvu | 2 | 1 |
| 二穗短柄草Brachypodium distachyon | bdi | 2 | 1 | |
| 甘蔗属Saccharum sp | ssp | 1 | 1 | |
| 甘蔗Saccharum officinarum | sof | 2 | 2 | |
| 水稻Oryza sativa | osa | 3 | 2 | |
| 高粱Sorghum bicolor | sbi | 1 | 1 | |
| 玉米Zea mays | zma | 4 | 2 | |
| 山羊草Aegilops tauschii | ata | 1 | 1 | |
| 豆科Leguminosae | 大豆Glycine max | gma | 2 | 2 |
| 蒺藜苜蓿Medicago truncatula | mtr | 4 | 3 | |
| 大叶相思Acacia auriculiformis | aau | 1 | 1 | |
| 豇豆Vigna unguiculata | vun | 1 | 1 | |
| 百脉根Lotus japonicus | lja | 1 | 1 | |
| 科 Family | 物种 Species | 简写 Logogram | miR168数量 Quantity of miR168 | pre-miR168数量 Quantity of pre-miR168 |
| 杨柳科Salicaceae Mirb | 毛果杨Populus trichocarpa | ptc | 4 | 2 |
| 葡萄科Vitaceae Juss | 葡萄Vitis vinifera | vvi | 1 | 1 |
| 毛茛科Ranunculaceae Juss | 耧斗菜Aquilegia viridiflora | aqc | 1 | 1 |
| 芸香科Rutaceae Juss | 脐橙Citrus sinensis | csi | 2 | 1 |
| 柑橘Citrus reticulata | cre | 1 | 1 | |
| 克萊门柚Citrus clementina | ccl | 1 | 1 | |
| 大戟科Rosaceae Juss | 木薯Manihot esculenta | mes | 1 | 1 |
| 三叶橡胶树 Hevea brasiliensis | hbr | 1 | 1 | |
| 蓖麻 Ricinus communis | rco | 1 | 1 | |
| 葫芦科Cucurbitaceae | 甜瓜Cucumis melo | cme | 1 | 1 |
| 菊科Asteraceae Bercht | 刺苞菜蓟Cynara cardunculus | cca | 1 | 1 |
| 亚麻科Linaceae | 亚麻Linum usitatissimum | lus | 2 | 2 |
| 茄科Solanaceae Juss | 烟草Nicotiana tabacum | nta | 5 | 5 |
| 番茄Lycopersicon esculentum | sly | 4 | 2 | |
| 蔷薇科Rosaceae Juss | 苹果Malus domestica | mdm | 2 | 2 |
| 碧桃Prunus persica | ppe | 1 | 1 | |
| 森林草莓Fragaria vesca | fve | 1 | 1 | |
| 凤梨科Bromeliaceae Juss | 莺歌凤梨Vriesea carinata | vcr | 4 | 2 |
| 松科Pinaceae Lindl | 挪威云杉Picea abies | pab | 2 | 2 |
| 百合科Liliaceae Juss | 芦笋Asparagus officinalis | aof | 2 | 2 |
| 梧桐科Sterculiaceae | 可可Theobroma cacao | tcc | 1 | 1 |

Fig. 3 Evolutionary tree of miR168(a)and multiple sequences comparison(b) ath. Arabidopsis thaliana;ptc. Populus trichocarpa;vvi. Vitis vinifera;crt. Citrus reticulata;csi. Citrus sinensis;mdm. Malus × domestica;ppe. Prunus persica;fve. Fragaria vesca.
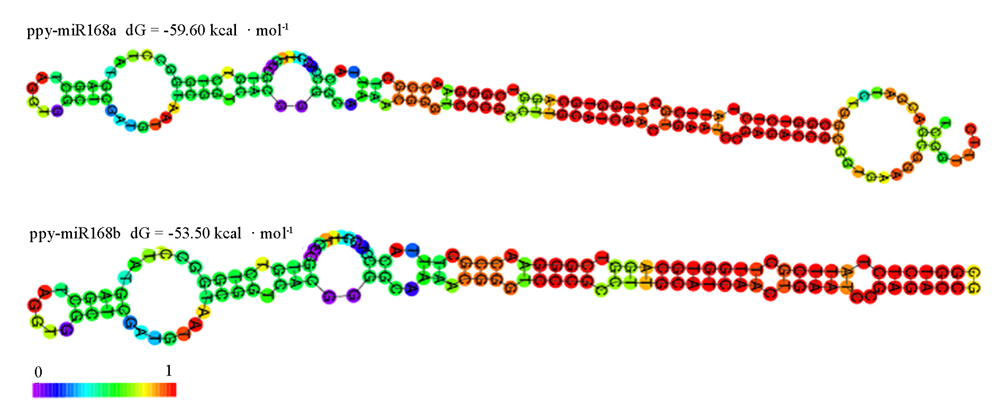
Fig. 5 Analysis of secondary stem-loop structure of Pyrus pyrifolia miR168 precursors. The redder the color,the more likely the base is to be in a pair,and vice versa.
| 靶基因注释 Target gene annotation | 靶基因编号 Target accession | 期望值 Expection |
|---|---|---|
| AGO1蛋白 Protein argonaute 1-like transcript variant | XM_018645622.1 | 0 |
| XM_018647395.1 | 0 | |
| XM_009349800.2 | 0 | |
| XM_018645623.1 | 0 | |
| XM_018647396.1 | 0 | |
| XM_009357682.2 | 0 | |
| 胼胝质合成酶3(CalS3) Callose synthase 3(CalS3)transcript variant | XM_018649103.1 | 2.5 |
| XM_009376827.2 | 2.5 | |
| XM_018651590.1 | 2.5 | |
| XM_018651591.1 | 2.5 | |
| XM_009365800.2 | 2.5 | |
| XM_018649104.1 | 2.5 | |
| XM_009365801.2 | 2.5 | |
| XM_009365802.2 | 2.5 | |
| XM_009365803.2 | 2.5 | |
| AGO1蛋白Protein argonaute 1-like transcript variant | XM_009359066.2 | 2.5 |
| XM_009349798.2 | 2.5 | |
| XM_009359065.2 | 2.5 | |
| XM_009349797.2 | 2.5 | |
| XM_009357684.2 | 2.5 | |
| XM_009357683.2 | 2.5 |
Table 3 Target genes prediction of pear miR168 family(ppy-miR168a-5p,ppy-miR168,ppy-miR168b)
| 靶基因注释 Target gene annotation | 靶基因编号 Target accession | 期望值 Expection |
|---|---|---|
| AGO1蛋白 Protein argonaute 1-like transcript variant | XM_018645622.1 | 0 |
| XM_018647395.1 | 0 | |
| XM_009349800.2 | 0 | |
| XM_018645623.1 | 0 | |
| XM_018647396.1 | 0 | |
| XM_009357682.2 | 0 | |
| 胼胝质合成酶3(CalS3) Callose synthase 3(CalS3)transcript variant | XM_018649103.1 | 2.5 |
| XM_009376827.2 | 2.5 | |
| XM_018651590.1 | 2.5 | |
| XM_018651591.1 | 2.5 | |
| XM_009365800.2 | 2.5 | |
| XM_018649104.1 | 2.5 | |
| XM_009365801.2 | 2.5 | |
| XM_009365802.2 | 2.5 | |
| XM_009365803.2 | 2.5 | |
| AGO1蛋白Protein argonaute 1-like transcript variant | XM_009359066.2 | 2.5 |
| XM_009349798.2 | 2.5 | |
| XM_009359065.2 | 2.5 | |
| XM_009349797.2 | 2.5 | |
| XM_009357684.2 | 2.5 | |
| XM_009357683.2 | 2.5 |
| 处理日期(M-D)Treatment date | ppy-miR168a | AGO1 | CalS3 |
|---|---|---|---|
| 11-01 | 0.22 | 0 | 4.00 |
| 11-25 | 3.37 | 0.05 | 1.03 |
| 12-27 | 1.12 | 0.58 | 0.75 |
| 01-23 | 1.00 | 5.91 | 0.59 |
| 相关系数Correlation coefficient | -0.223 | -0.503 |
Table 4 Correlation analysis of ppy-miR168a and its target genes expression
| 处理日期(M-D)Treatment date | ppy-miR168a | AGO1 | CalS3 |
|---|---|---|---|
| 11-01 | 0.22 | 0 | 4.00 |
| 11-25 | 3.37 | 0.05 | 1.03 |
| 12-27 | 1.12 | 0.58 | 0.75 |
| 01-23 | 1.00 | 5.91 | 0.59 |
| 相关系数Correlation coefficient | -0.223 | -0.503 |
| [1] |
Arora R, Rowland L, Tanino K. 2003. Induction and release of bud dormancy in woody perennials:a science comes of age. HortScience, 38 (5):911-921.
doi: 10.21273/HORTSCI.38.5.911 URL |
| [2] |
Bartel D P. 2004. MicroRNAs:genomics,biogenesis,mechanism,and function. Cell, 116 (2):281-297.
doi: 10.1016/s0092-8674(04)00045-5 pmid: 14744438 |
| [3] | Budak H, Akpinar B A. 2015. Plant miRNAs:biogenesis,organization and origins. Functional & Integrative Genomics, 15 (5):523-531. |
| [4] |
Chen X. 2005. microRNA biogenesis and function in plants. Febs Letters, 579 (26):5923-5931.
doi: 10.1016/j.febslet.2005.07.071 URL |
| [5] | Cui Hai-Fang, Zhang Fan, Yin Jun-long, Guo Ying-qi, Yue Yan-ling. 2017. Callose deposition and pollen development. Journal of Yunnan Agricultural University, 32 (3):551-557. (in Chinese) |
| 崔海芳, 张凡, 尹俊龙, 郭瑛琪, 岳艳玲. 2017. 胼胝质沉积与花粉发育. 云南农业大学学报, 32 (3):551-557. | |
| [6] |
Ding Q, Zeng J, He X-Q. 2016. MiR169 and its target PagHAP2-6 regulated by ABA are involved in poplar cambium dormancy. Journal of Plant Physiology, 198:1-9.
doi: 10.1016/j.jplph.2016.03.017 pmid: 27111502 |
| [7] |
Fazli W, Adeeb S, Taous K, Young K Y. 2010. MicroRNAs:synthesis,mechanism,function,and recent clinical trials. Biochimica et Biophysica Acta, 1803 (11):1231-243.
doi: 10.1016/j.bbamcr.2010.06.013 pmid: 20619301 |
| [8] |
Gao J, Ni X, Li H, Hayat F, Shi T, Gao Z. 2021. miR169 and PmRGL2 synergistically regulate the NF-Y complex to activate dormancy release in Japanese apricot(Prunus mume Siebet Zucc.). Plant Molecular Biology, 105:83-87.
doi: 10.1007/s11103-020-01070-3 URL |
| [9] | Hammond S M, Bernstein E, Beach D, Hannon G J. 2000. An RNA-directed nuclease mediates post-transcriptional gene silencing in Drosophila cells of science. Nature:International Weekly Journal of Science, 404 (6775):293-296. |
| [10] |
Hemery G E, Savill P S, Pyror S N. 2005. Applications of the crown diameter-stem diameter relationship for different species of broadleaved trees. Forest Ecology Management, 215:285-294.
doi: 10.1016/j.foreco.2005.05.016 URL |
| [11] | Jian Ling-cheng, Wang Hong. 2004. Plasmodesmatal dynamics in both woody poplar and herbaceous winter wheat under controlled short day and in field winter period. Acta Botanica Sinica, 46 (2):230-235. (in Chinese) |
| 简令成, 王红. 2004. 杨树和冬小麦在短日照和寒冬中的胞间连丝动态. 植物学报, 46 (2):230-235. | |
| [12] | Jiang Weibing, Han Haozhang, Wang Liangju, Ma Kai. 2003. Advance in research of chilling requirement and the mechanism of deciduous fruit crops. Journal of Fruit Science, 20 (5):364-368. (in Chinese) |
| 姜卫兵, 韩浩章, 汪良驹, 马凯. 2003. 落叶果树需冷量及其机理研究进展. 果树学报, 20 (5):364-368. | |
| [13] |
Mathieu J, Levi J, Yant, Felix Mürdter, Frank Küttner, Markus Schmid. 2009. Repression of flowering by the miR172 target SMZ. PLoS Biology, 7 (7):e1000148.
doi: 10.1371/journal.pbio.1000148 URL |
| [14] |
Ko J H, Prassinos C, Han K H. 2006. Developmental and seasonal expression of PtaHB1,a Populus gene encoding a class III HD-Zip protein,is closely associated with secondary growth and inversely correlated with the level of microRNA(miR166). New Phytologist, 169 (3):469-478.
doi: 10.1111/j.1469-8137.2005.01623.x URL |
| [15] | Lang G A, Early J D, Martin G C, Darnell R L. 1987. Endo-,para-,and ecodormancy:physiological terminology and classification for dormancy research. HortScience, 22 (3):371-377. |
| [16] | Li Su-fen, Zhao Yan, Ji Lu-sha. 2014. Influences of AGOl to the development of the Arabidopsis leaves with the serration margin. Journal of Hebei University of Science and Technology, 35 (1):51-57. (in Chinese) |
| 李素芬, 赵燕, 冀芦沙. 2014. AGOl基因对拟南芥叶边缘锯齿状发育的影响. 河北科技大学学报, 35 (1):51-57. | |
| [17] |
Li W, Cui X, Meng Z, Huang X, Xie Q, Wu H, Jin H, Zhang D, Liang W. 2012. Transcriptional regulation of Arabidopsis MIR168a and ARGONAUTE1 homeostasis in abscisic acid and abiotic stress responses. Plant Physiology, 158 (3):1279-1292.
doi: 10.1104/pp.111.188789 URL |
| [18] | Li Xiao-dong, Lu Guo-yi, Yu Yan 1996. Effect of endogenous hormones on the dormancy of garlic(Allium sativum L.). Acta Horticulturae Sinica. 23 (2):150-154 (in Chinese) |
| 李晓东, 陆帼一, 于燕. 1996. 休眠‘蔡家坡’蒜内源激素水平的变化规律. 园艺学报, 23 (2):150-154. | |
| [19] |
Liu C, Xin Y, Xu L, Cai Z, Xue Y, Liu Y, Xie D, Liu Y, Qi Y. 2018. Arabidopsis ARGONAUTE 1 binds chromatin to promote gene transcription in response to hormones and stresses. Developmental Cell, 44 (3):348-361.
doi: 10.1016/j.devcel.2017.12.002 URL |
| [20] | Liu Chang. 2017. Function of nuclear Argonaute 1 in response to various stimuli in Arabidopsis[Ph. D. Dissertation]. Beijing: China Agricultural University. |
| 刘畅. 2017. 拟南芥核内Argonaute 1在响应各类刺激中的功能研究[博士论文]. 北京: 中国农业大学. (in Chinese) | |
| [21] |
Liu G, Li W, Zheng P, Xu T, Chen L, Liu D, Hussain S, Teng Y. 2012. Transcriptomic analysis of‘Suli’pear(Pyrus pyrifolia white pear group) buds during the dormancy by RNA-Seq. BMC Genomics, 13 (1): 10.1186/1471-2164-13-700.
doi: 10.1186/1471-2164-13-700 |
| [22] | Liu Guo-qin, Zheng Peng-hua, Sayed Hussain, Teng Yuan-wen. 2013. Characteristics of two dormancy-associated MADS-box genes in pear and their expression analysis during the dormancy. Acta Horticulturae Sinica, 40 (4):724-732. (in Chinese) |
| 刘国琴, 郑鹏华, Hussain S, 滕元文. 2013. 梨两个休眠相关MADS-box基因特征及在其休眠过程中的表达分析. 园艺学报, 40 (4):724-732. | |
| [23] |
Liu Lin, Quan Xian-qing, Zhao Xiao-mei, Huang Li-hua, Feng Shang-cai, Huang Kun-yan, Zhou Xiao-yan, Su Wen-ting. 2013. Arabidopsis callose synthase gene GSL8 is required for cell wall formation and establishment and maintenance of quiescent center. Chinese Bulletin of Botany, 48 (4):389-397. (in Chinese)
doi: 10.3724/SP.J.1259.2013.00389 URL |
|
刘林, 全先庆, 赵小梅, 黄力华, 冯尚彩, 黄坤艳, 周晓燕, 粟文婷. 2013. 拟南芥胼胝质合酶基因GSL8参与细胞壁形成和根端静止中心建立与维持. 植物学报, 48 (4):389-397.
doi: 10.3724/SP.J.1259.2013.00389 |
|
| [24] | Ma Xin-rui, Li Liang, Liu Jin-hang, Yang Meng-jie, Chen Jie, Liang Qin, Wu Shao-hua, LI Yong-yu. 2018. Identification and differentially expressed analysis of microRNA associated with dormancy of pear flower buds. Acta Horticulturae Sinica, 45 (11):2089-2105. (in Chinese) |
| 马鑫瑞, 李亮, 刘瑾航, 杨梦洁, 陈洁, 梁沁, 吴少华, 李永裕. 2018. 梨花芽休眠相关miRNA的鉴定和差异表达分析. 园艺学报, 45 (11):2089-2105. | |
| [25] |
Miao C, Wang Z, Zhang L, Yao J, Zhu J K. 2019. The grain yield modulator miR156 regulates seed dormancy through the gibberellin pathway in rice. Nature Communications, 10 (1):3822.
doi: 10.1038/s41467-019-11830-5 URL |
| [26] | Muthamilarasan M, Manoj Prasad. 2013. Cutting-edge research on plant miRNAs. Current Science, 104 (3):287-289. |
| [27] |
Niu Q, Li J, Cai D, Qian M, Jia H, Bai S, Sayed H, Liu G, Teng Y, Zheng X. 2016. Dormancy-associated MADS-box genes and microRNAs jointly control dormancy transition in pear(Pyrus pyrifolia white pear group)flower bud. Journal of Experimental Botany, 67 (1):239-257.
doi: 10.1093/jxb/erv454 URL |
| [28] | Niu Qing-feng. 2016. The molecular mechanism and molecular network of pear flower bud dormancy transition[Ph. D. Dissertation]. Hangzhou: Zhejiang University. (in Chinese) |
| 牛庆丰. 2016. 梨花芽休眠转换的分子网络及调控机制[博士论文]. 杭州: 浙江大学. | |
| [29] |
Olsen J E. 2010. Light and temperature sensing and signaling in induction of bud dormancy in woody plants. Plant Molecular Biology, 73 (1-2):37-47.
doi: 10.1007/s11103-010-9620-9 URL |
| [30] |
Pacey-Miller T, Scott K, Ablett E, Tingey S, Ching A, Henry R. 2003. Genes associated with the end of dormancy in grapes. Functional Integrative Genomics, 3 (4):144-152.
doi: 10.1007/s10142-003-0094-6 URL |
| [31] |
Peng S, Liang D, Zhang Z, Yin W, Xia X. 2013. Identification of drought-responsive and novel Populus trichocarpa microRNAs by high-throughput sequencing and their targets using degradome analysis. BMC Genomics, 14 (1):233.
doi: 10.1186/1471-2164-14-233 URL |
| [32] |
Rajesh Kumar Singh, Pal Miskolczi, Jay P Maurya, Rishikesh P Bhalerao. 2019. A tree ortholog of SHORT VEGETATIVE PHASE floral repressor mediates photoperiodic control of bud dormancy. Current Biology, 29 (1):128-133.
doi: S0960-9822(18)31472-6 pmid: 30554900 |
| [33] |
Rhoades M, Reinhart B, Lim L, Burge C, Bartel B, Bartel D. 2002. Prediction of plant microRNA targets. Cell, 110 (4):513-520.
doi: 10.1016/S0092-8674(02)00863-2 URL |
| [34] |
Rinne P L, Kaikuranta P M, Schoot C V D. 2001. The shoot apical meristem restores its symplasmic organization during chilling-induced release from dormancy. Plant Journal, 26 (3):249-264.
pmid: 11439114 |
| [35] | Rinne P L, Schoot C V D. 2003. Plasmodesmata at the crossroads between development,dormancy,and defense. Canadian Journal of Botany, 276 (1-2):133-137. |
| [36] |
Rohde A, Bhalerao R. 2007. Plant dormancy in the perennial context. Trends in Plant Science, 12 (5):217-223.
doi: 10.1016/j.tplants.2007.03.012 URL |
| [37] |
Rui Shi, Vincent L Ching. 2005. Facile means for quantifying microRNA expression by real-time PCR. Biotechniques, 39 (4):519-524.
doi: 10.2144/000112010 pmid: 16235564 |
| [38] | Ruttink T, Arend M, Morreel K, Storme V, Rombauts S, Fromm J, Bhalerao R P, Boerjan W, Rohde A. 2007. A molecular timetable for apical bud formation and dormancy induction in poplar. Plant Cell Reports, 19 (8):2370-2390. |
| [39] | Shi Ting, Gao Zhi-hong, Zhang Zhen, Zhuang Wei-bing. 2010. Advance of research on microRNA in flower development regulation. Chinese Agricultural Science Bulletin, 26 (13):267-271. (in Chinese) |
| 侍婷, 高志红, 章镇, 庄维兵. 2010. MicroRNA参与植物花发育调控的研究进展. 中国农学通报, 26 (13):267-271. | |
| [40] |
Siré C, Moreno A B, Garcia-Chapa M, López-Moya J J, Segundo B S. 2009. Diurnal oscillation in the accumulation of Arabidopsis microRNAs,miR167,miR168,miR171 and miR398. Febs Letters, 583 (6):1039-1044.
doi: 10.1016/j.febslet.2009.02.024 URL |
| [41] | Su Li-Yao, Jiang Meng-Qi, Huang Shu-Qi, Xu Xiao-Ping, Li Xue, Chen Xu, Lai Zhong-Xiong, Lin Yu-Ling. 2019. Analysis of the molecular evolution characteristics of miR403 and its expression pattern during analysis of the molecular evolution characteristics of miR403 and its expression pattern during early somatic embryogenesis in Dimocarpus longan. Journal of Fruit Science, 36 (7):846-856. (in Chinese) |
| 苏立遥, 蒋梦琦, 黄倏祺, 徐小萍, 厉雪, 陈旭, 赖钟雄, 林玉玲. 2019. miR403分子进化特性及其在龙眼体胚发生早期的表达模式分析. 果树学报, 36 (7):846-856. | |
| [42] |
Sun L, Ai X, Li W, Guo W, Deng X, Hu C, Zhang J, Wu K. 2012. Identification and comparative profiling of miRNAs in an early flowering mutant of trifoliate orange and its wild type by genome-wide deep sequencing. PLoS ONE, 7 (8):doi: 10.1371/journal.pone.0043760.
doi: 10.1371/journal.pone.0043760 URL |
| [43] | Várallyay Éva, Válóczi Anna, Ágyi Ákos, Burgyán József, Havelda Zoltán. 2017. Plant virus-mediated induction of miR168 is associated with repression of ARGONAUTE1 accumulation. The EMBO Journal, 36 (7):846-856. |
| [44] |
Wang J W, Park M, Wang L J, Koo Y, Chen X Y, Weigel D, Poethig R, Sederoff R. 2011. MiRNA control of vegetative phase change in trees. PLoS Genetics, 7 (2):doi: 10.1371/journal.pgen.1002012.
doi: 10.1371/journal.pgen.1002012 URL |
| [45] | Wang Xin-chao, Ma Chun-lei, Yang Ya-jun. 2011. Progress in research on bud dormancy and its regulation mechanisms in perennial plants. Chinese Journal of Applied & Environmental Biology, 17 (4):589-595. (in Chinese) |
| 王新超, 马春雷, 杨亚军. 2011. 多年生植物的芽休眠及调控机理研究进展. 应用与环境生物学报, 17 (4):589-595. | |
| [46] | Wang Yan-fang, Zhou Rui-lian, Zhao Yan-hong. 2015. Molecular evolution of miR-171 gene family and prediction of their target genes. Life Science Research, 19 (6):479-483. (in Chinese) |
| 王艳芳, 周瑞莲, 赵彦宏. 2015. miR-171基因家族进化分析及靶基因预测. 生命科学研究, 19 (6):479-483. | |
| [47] |
Welling A, Palva E T. 2010. Molecular control of cold acclimation in trees. Physiologia Plantarum, 127 (2):167-181.
doi: 10.1111/j.1399-3054.2006.00672.x URL |
| [48] |
Wu G, Park M Y, Conway R. S, Wang J-W, Weigel D, Poethig R S. 2009. The sequential action of miR156 and miR172 regulates developmental timing in Arabidopsis. Cell, 138 (4):750-759.
doi: 10.1016/j.cell.2009.06.031 URL |
| [49] | Xian Zhi-qiang. 2014. Isolation and characterisation of SlAGO genes and study on biological function of miR168 and AGO1 in tomato[Ph. D. Dissertation]. Chongqing: Chongqing University. (in Chinese) |
| 先志强. 2014. 番茄AGO基因的分离鉴定及SlmiR168,AGO1在番茄生长发育过程中的功能研究[博士论文]. 重庆: 重庆大学. | |
| [50] |
Yadav S R, Yan D, Sevilem I, Ykä Helariutta. 2014. Plasmodesmata-mediated intercellular signaling during plant growth and development. Frontiers in Plant Science, 5,doi: 10.3389/fpls.2014.00044.
doi: 10.3389/fpls.2014.00044 URL |
| [51] |
Yamane H, Ooka T, Jotatsu H, Hosaka Y, Sasaki R, Tao R. 2011. Expressional regulation of PpDAM5 and PpDAM6,peach(Prunus persica) dormancy-associated MADS-box genes,by low temperature and dormancy-breaking reagent treatment. Journal of Experimental Botany, 62 (10):3481-3488.
doi: 10.1093/jxb/err028 URL |
| [52] |
Zhang Y, Wang Y, Gao X, Liu C, Gai S. 2018. Identification and characterization of microRNAs in tree peony during chilling induced dormancy release by high-throughput sequencing. Scientific Reports, 8 (1):4537.
doi: 10.1038/s41598-018-22415-5 pmid: 29540706 |
| [53] | Zhao X, Chen S, Zhao L, Zhang X, Ma P, Gou Z. 2014. Evolution of MIR166 gene family in land plants. Plant Diversity and Resources, 36 (3):331-341. |
| [1] | SONG Jiankun, YANG Yingjie, LI Dingli, MA Chunhui, WANG Caihong, and WANG Ran. A New Pear Cultivar‘Luxiu’ [J]. Acta Horticulturae Sinica, 2022, 49(S2): 3-4. |
| [2] | DONG Xingguang, CAO Yufen, ZHANG Ying, TIAN Luming, HUO Hongliang, QI Dan, XU Jiayu, LIU Chao, and WANG Lidong. A New Cold-resistant Crispy Pear Cultivar‘Yucuixiang’ [J]. Acta Horticulturae Sinica, 2022, 49(S2): 5-6. |
| [3] | OU Chunqing, JIANG Shuling, WANG Fei, MA Li, ZHANG Yanjie, and LIU Zhenjie. A New Early-ripening Pear Cultivar‘Xingli Mishui’ [J]. Acta Horticulturae Sinica, 2022, 49(S2): 7-8. |
| [4] | ZHANG Yanjie, WANG Fei, OU Chunqing, MA Li, JIANG Shuling, and LIU Zhenjie. A New Pear Cultivar‘Zhongli Yucui 3’ [J]. Acta Horticulturae Sinica, 2022, 49(S2): 9-10. |
| [5] | WANG Suke, LI Xiugen, YANG Jian, WANG Long, SU Yanli, ZHANG Xiangzhan, and XUE Huabai. A New Red Pear Cultivar‘Danxiahong’ [J]. Acta Horticulturae Sinica, 2022, 49(S2): 13-14. |
| [6] | WANG Fei, OU Chunqing, ZHANG Yanjie, MA Li, and JIANG Shuling. A New Late Ripening Pear Cultivar‘Huaqiu’with Long Storage Period [J]. Acta Horticulturae Sinica, 2022, 49(S1): 9-10. |
| [7] | SONG Jiankun, LI Dingli, YANG Yingjie, MA Chunhui, WANG Caihong, and WANG Ran. A New Pear Cultivar‘Qindaohong’ [J]. Acta Horticulturae Sinica, 2022, 49(S1): 11-12. |
| [8] | GUO Weizhen, ZHAO Jingxian, LI Ying. A New Mid-early Ripening Pear Cultivar‘Meiyu’ [J]. Acta Horticulturae Sinica, 2022, 49(9): 2051-2052. |
| [9] | LIU Jinming, GUO Caihua, YUAN Xing, KANG Chao, QUAN Shaowen, NIU Jianxin. Genome-wide Identification of Dof Family Genes and Expression Analysis Sepal Persistent and Abscission in Pear [J]. Acta Horticulturae Sinica, 2022, 49(8): 1637-1649. |
| [10] | TAO Xin, ZHU Rongxiang, GONG Xin, WU Lei, ZHANG Shaoling, ZHAO Jianrong, ZHANG Huping. Fructokinase Gene PpyFRK5 Plays an Important Role in Sucrose Accumulation of Pear Fruit [J]. Acta Horticulturae Sinica, 2022, 49(7): 1429-1440. |
| [11] | XIANG Miaolian, WU Fan, LI Shucheng, MA Qiaoli, WANG Yinbao, XIAO Liuhua, CHEN Jinyin, CHEN Ming. Exogenous Melatonin Regulates Reactive Oxygen Metabolism to Induce Resistance of Postharvest Pear Fruit to Black Spot [J]. Acta Horticulturae Sinica, 2022, 49(5): 1102-1110. |
| [12] | XU Tong, SHAO Lingmei, WANG Xiaobin, ZHANG Runlong, ZHANG Kaijing, XIA Yiping, ZHANG Jiaping, LI Danqing. Research Progress on Winter Dormancy of Perennial Monocots [J]. Acta Horticulturae Sinica, 2022, 49(12): 2703-2721. |
| [13] | SU Yanli, GAO Xiaoming, YANG Jian, WANG Long, WANG Suke, ZHANG Xiangzhan, XUE Huabai. Dynamic Changes of Browning Degree,Phenolics Contents and Related Enzyme Activities During Pear Fruit Development [J]. Acta Horticulturae Sinica, 2022, 49(11): 2304-2312. |
| [14] | YANG Bo, WEI Jia, LI Kunfeng, WANG Chengliang, NI Junbei, TENG Yuanwen, and BAI Songling. PpyERF060-PpyABF3-PpyMADS71 Regulates Ethylene Signaling Pathway- Mediated Pear Bud Dormancy Process [J]. Acta Horticulturae Sinica, 2022, 49(10): 2249-2262. |
| [15] | WANG Yaru, LI Yong, WANG Jin, WANG Yingtao, LI Xiao, and WANG Yongbo. A New Mid-ripening Pear Cultivar‘Jijin’ [J]. Acta Horticulturae Sinica, 2021, 48(S2): 2771-2772. |
| Viewed | ||||||
|
Full text |
|
|||||
|
Abstract |
|
|||||
Copyright © 2012 Acta Horticulturae Sinica 京ICP备10030308号-2 国际联网备案号 11010802023439
Tel: 010-82109523 E-Mail: yuanyixuebao@126.com
Support by: Beijing Magtech Co.Ltd