
Acta Horticulturae Sinica ›› 2024, Vol. 51 ›› Issue (8): 1803-1822.doi: 10.16420/j.issn.0513-353x.2023-0384
• Genetic & Breeding·Germplasm Resources·Molecular Biology • Previous Articles Next Articles
WANG Yanhong1, XI Keyong1, TIAN Ye1, LIU Deqi1, ZHOU Kegui1, YIN Junliang2, LIU Yiqing1, ZHU Yongxing1,*( )
)
Received:2024-03-22
Revised:2024-05-17
Online:2024-08-25
Published:2024-08-21
Contact:
ZHU Yongxing
WANG Yanhong, XI Keyong, TIAN Ye, LIU Deqi, ZHOU Kegui, YIN Junliang, LIU Yiqing, ZHU Yongxing. Identification and Expression Pattern Analysis of GST in Zingiber officinale[J]. Acta Horticulturae Sinica, 2024, 51(8): 1803-1822.
Add to citation manager EndNote|Ris|BibTeX
URL: https://www.ahs.ac.cn/EN/10.16420/j.issn.0513-353x.2023-0384
| 登录号 Accession ID | 处理样品描述 Description | 数据大小 Data size | 参考文献 Reference |
|---|---|---|---|
| PRJNA788194 | 不同发育阶段根茎、不同组织 Different developmental stages of rhizomes,different tissues | 未知 Unknown | Li et al., |
| PRJNA592215 | 红茎内部组织、绿茎、黄根茎、红茎表皮 Internal tissue of red stem,green stem,yellow rhizome,red stem epidermis | 254 Mb | Li et al., |
| PRJNA380847 | 姜根茎低/高土壤湿度下用无菌水和青枯菌接种 Inoculate ginger rhizomes with sterile water and Ralstonia solanacearum under low/high soil moisture | 未知 Unknown | Jiang et al., |
| PRJNA380972 | 高土壤湿度增加姜中青枯菌感染率 High-soil-moisture-elevated susceptibility to R. solanacearum infection in ginger | 31.5 Gb | Huang et al., |
| PRJNA911443 | 姜采收后26、10和2 ℃贮藏条件下的根茎 Rhizome of ginger harvested under storage conditions of 26,10,and 2 ℃ | 16.8 Gb | Zhang et al., |
Table 1 Source of data sets of transcriptome of ginger
| 登录号 Accession ID | 处理样品描述 Description | 数据大小 Data size | 参考文献 Reference |
|---|---|---|---|
| PRJNA788194 | 不同发育阶段根茎、不同组织 Different developmental stages of rhizomes,different tissues | 未知 Unknown | Li et al., |
| PRJNA592215 | 红茎内部组织、绿茎、黄根茎、红茎表皮 Internal tissue of red stem,green stem,yellow rhizome,red stem epidermis | 254 Mb | Li et al., |
| PRJNA380847 | 姜根茎低/高土壤湿度下用无菌水和青枯菌接种 Inoculate ginger rhizomes with sterile water and Ralstonia solanacearum under low/high soil moisture | 未知 Unknown | Jiang et al., |
| PRJNA380972 | 高土壤湿度增加姜中青枯菌感染率 High-soil-moisture-elevated susceptibility to R. solanacearum infection in ginger | 31.5 Gb | Huang et al., |
| PRJNA911443 | 姜采收后26、10和2 ℃贮藏条件下的根茎 Rhizome of ginger harvested under storage conditions of 26,10,and 2 ℃ | 16.8 Gb | Zhang et al., |
| 基因名 Gene name | 正向引物(5′-3′) Forward primer | 反向引物(5′-3′) Reverse primer | 长度/bp Length |
|---|---|---|---|
| ZoGSTDHAR2 | CTCCTCCTGCCATTTCCTCG | GATTCACCTCCACGACTTTGTATG | 228 |
| ZoGSTEF1G2 | AAGGTGTTAGGCGAGGTCAAG | TGGGTGCTTCTTCCTCTTCTTC | 169 |
| ZoGSTCHQ1 | GGTCACAGAATGGGTGGAGAG | TGGACCATCCGAGCAATCAC | 129 |
| ZoGSTT1 | AGAGTCATGCCATATTGAGCTACC | ATTGAACCACGGCGTAGATTG | 140 |
| ZoGSTL1 | CTTCCTCCGTCCCTCGATTC | GTTATCCAGGTGCGTTGTGC | 107 |
| ZoGSTL2 | TCCCGCTTTGTGAAGGAGG | CGCAAATGGGCAGGTGTATG | 111 |
| ZoGSTF9 | GGTGGTACGACGGACGAGAAG | TGACGGGATAGTGGCAGAGG | 145 |
| ZoGSTU13 | TTGCTGGATCTGTGGGTGAGTC | GTATTGCACGATGATGAGCGACT | 201 |
| ZoGSTU16 | TGGATCTGTGGGTGAGTCCTTTC | TGTTGTCCAAGTCCTGCTCCTG | 96 |
| ZoGSTU27 | GGTGTTACTGGATCTGTGGGTGAG | GCAACGGGCTCTTGTTGTCC | 116 |
| RBP | CCTATGAAGCGTAGAAACACAAG | GGAAGGACAACATCCCAAATC | 123 |
Table 2 Primer sequences for fluorescence quantitative PCR of ZoGST
| 基因名 Gene name | 正向引物(5′-3′) Forward primer | 反向引物(5′-3′) Reverse primer | 长度/bp Length |
|---|---|---|---|
| ZoGSTDHAR2 | CTCCTCCTGCCATTTCCTCG | GATTCACCTCCACGACTTTGTATG | 228 |
| ZoGSTEF1G2 | AAGGTGTTAGGCGAGGTCAAG | TGGGTGCTTCTTCCTCTTCTTC | 169 |
| ZoGSTCHQ1 | GGTCACAGAATGGGTGGAGAG | TGGACCATCCGAGCAATCAC | 129 |
| ZoGSTT1 | AGAGTCATGCCATATTGAGCTACC | ATTGAACCACGGCGTAGATTG | 140 |
| ZoGSTL1 | CTTCCTCCGTCCCTCGATTC | GTTATCCAGGTGCGTTGTGC | 107 |
| ZoGSTL2 | TCCCGCTTTGTGAAGGAGG | CGCAAATGGGCAGGTGTATG | 111 |
| ZoGSTF9 | GGTGGTACGACGGACGAGAAG | TGACGGGATAGTGGCAGAGG | 145 |
| ZoGSTU13 | TTGCTGGATCTGTGGGTGAGTC | GTATTGCACGATGATGAGCGACT | 201 |
| ZoGSTU16 | TGGATCTGTGGGTGAGTCCTTTC | TGTTGTCCAAGTCCTGCTCCTG | 96 |
| ZoGSTU27 | GGTGTTACTGGATCTGTGGGTGAG | GCAACGGGCTCTTGTTGTCC | 116 |
| RBP | CCTATGAAGCGTAGAAACACAAG | GGAAGGACAACATCCCAAATC | 123 |
| 基因1 | 基因2 | 异义替换频率(Ka) | 同义替换频率(Ks) | 异义替换/同义替换 |
|---|---|---|---|---|
| Gene 1 | Gene 2 | Non-synonymous | Synonymous | Ka/Ks |
| ZoGSTL1 | ZoGSTL3 | 0.141203891 | 0.527940985 | 0.267461505 |
| ZoGSTL2 | ZoGSTL3 | 0.096538384 | 0.315771474 | 0.305722310 |
| ZoGSTDHAR2 | ZoGSTDHAR3 | 0.059875159 | 0.365643255 | 0.163752943 |
| ZoGSTF2 | ZoGSTF4 | 0.389135669 | 0.971421814 | 0.400583622 |
| ZoGSTU2 | ZoGSTU3 | 0.113145323 | 0.259868329 | 0.435394813 |
| ZoGSTU11 | ZoGSTU14 | 0.073775974 | 0.133473189 | 0.552740031 |
Table 3 Ka/Ks of ZoGST genes
| 基因1 | 基因2 | 异义替换频率(Ka) | 同义替换频率(Ks) | 异义替换/同义替换 |
|---|---|---|---|---|
| Gene 1 | Gene 2 | Non-synonymous | Synonymous | Ka/Ks |
| ZoGSTL1 | ZoGSTL3 | 0.141203891 | 0.527940985 | 0.267461505 |
| ZoGSTL2 | ZoGSTL3 | 0.096538384 | 0.315771474 | 0.305722310 |
| ZoGSTDHAR2 | ZoGSTDHAR3 | 0.059875159 | 0.365643255 | 0.163752943 |
| ZoGSTF2 | ZoGSTF4 | 0.389135669 | 0.971421814 | 0.400583622 |
| ZoGSTU2 | ZoGSTU3 | 0.113145323 | 0.259868329 | 0.435394813 |
| ZoGSTU11 | ZoGSTU14 | 0.073775974 | 0.133473189 | 0.552740031 |
| 名称 Name | 基因ID Gene ID | 长度/aa Length | 分子量/kD Mw | 等电点 pI | 不稳定指数 Instability index | 亲水性 Hydrophilicity | 亚细胞定位 Subcellular localization |
|---|---|---|---|---|---|---|---|
| ZoGSTDHAR1 | Maker00035113 | 330 | 37.20 | 5.06 | 51.08 | -0.278 | Cp,Cy,Mt |
| ZoGSTDHAR2 | Maker00052306 | 328 | 36.71 | 9.10 | 47.47 | -0.285 | Cp |
| ZoGSTDHAR3 | Maker00046027 | 350 | 39.34 | 8.79 | 44.65 | -0.139 | Mt |
| ZoGSTDHAR5 | Maker00035024 | 255 | 28.17 | 6.25 | 43.08 | -0.232 | Cp,Cy |
| ZoGSTDHAR4 | Maker00022334 | 267 | 29.40 | 6.91 | 48.36 | -0.098 | Cp,Cy |
| ZoGSTEF1G1 | Maker00033503 | 418 | 47.80 | 5.62 | 40.78 | -0.387 | Cy |
| ZoGSTEF1G2 | Maker00031875 | 419 | 47.96 | 5.68 | 39.85 | -0.346 | Cy |
| ZoGSTCHQ1 | Maker00046252 | 265 | 31.38 | 9.20 | 36.80 | -0.314 | Cy |
| ZoGSTT1 | Maker00029139 | 263 | 29.87 | 8.99 | 41.86 | -0.200 | Cy |
| ZoGSTL1 | Maker00033284 | 218 | 24.45 | 4.57 | 49.88 | -0.138 | Nu |
| ZoGSTL2 | Maker00059206 | 262 | 29.82 | 5.32 | 41.03 | -0.344 | Cp |
| ZoGSTL3 | Maker00009568 | 241 | 27.61 | 5.28 | 42.39 | -0.194 | Cy |
| ZoGSTZ1 | Maker00037272 | 346 | 39.86 | 6.47 | 34.33 | -0.469 | Mt |
| ZoGSTZ2 | Maker00013992 | 420 | 46.61 | 8.85 | 57.36 | -0.186 | Cy |
| ZoGSTZ3 | Maker00051190 | 490 | 55.36 | 4.71 | 53.11 | -0.735 | Cp |
| ZoGSTZ4 | Maker00021183 | 334 | 37.27 | 8.20 | 52.82 | -0.159 | Cp |
| ZoGSTZ5 | Maker00078047 | 402 | 44.92 | 5.87 | 44.01 | -0.161 | Cy |
| ZoGSTF1 | Maker00077732 | 675 | 72.51 | 5.62 | 60.97 | -0.818 | Nu |
| ZoGSTF2 | Maker00076791 | 220 | 24.34 | 6.91 | 55.02 | -0.311 | Cy |
| ZoGSTF3 | Maker00068242 | 318 | 35.47 | 6.56 | 54.65 | -0.470 | Cp,Cy,Nu |
| ZoGSTF4 | Maker00047494 | 221 | 25.42 | 7.76 | 50.63 | -0.422 | Cy |
| ZoGSTF5 | Maker00076803 | 221 | 25.31 | 6.97 | 55.37 | -0.513 | Cy |
| ZoGSTF6 | Maker00050934 | 243 | 27.74 | 6.34 | 39.92 | -0.216 | Cy |
| ZoGSTF7 | Maker00023234 | 224 | 25.10 | 5.35 | 41.26 | -0.144 | Cy |
| ZoGSTF8 | Maker00077970 | 251 | 28.38 | 6.72 | 38.38 | 0.051 | Cy |
| ZoGSTF9 | Maker00078922 | 217 | 24.39 | 5.52 | 49.34 | -0.089 | Cy |
| ZoGSTF10 | Maker00078286 | 217 | 24.47 | 5.71 | 37.72 | 0.043 | Cy |
| ZoGSTF11 | Maker00078094 | 255 | 28.60 | 6.31 | 39.33 | 0.025 | Cy |
| ZoGSTF12 | Maker00077516 | 219 | 24.50 | 5.53 | 36.69 | 0.050 | Cy |
| ZoGSTF13 | Maker00040344 | 217 | 23.76 | 5.97 | 18.75 | 0.107 | Cy |
| ZoGSTU1 | Maker00008855 | 222 | 24.81 | 5.39 | 39.79 | -0.027 | Cy |
| ZoGSTU2 | Maker00015777 | 244 | 27.20 | 5.02 | 38.10 | -0.195 | Cy |
| ZoGSTU3 | Maker00049005 | 247 | 27.51 | 5.40 | 34.94 | -0.146 | Cy |
| ZoGSTU4 | Maker00045495 | 232 | 26.80 | 6.20 | 55.74 | -0.338 | Cy |
| ZoGSTU5 | Maker00045023 | 228 | 26.25 | 5.47 | 48.11 | -0.321 | Cy |
| ZoGSTU6 | Maker00061695 | 227 | 25.65 | 5.49 | 55.78 | 0.027 | Cy |
| ZoGSTU7 | Maker00045310 | 222 | 24.36 | 5.43 | 24.27 | 0.064 | Cy |
| ZoGSTU8 | Maker00042846 | 222 | 24.37 | 5.43 | 23.19 | 0.060 | Cy |
| ZoGSTU9 | Maker00044620 | 219 | 24.93 | 5.29 | 34.73 | -0.096 | Cy |
| ZoGSTU10 | Maker00042772 | 219 | 24.91 | 5.39 | 34.62 | -0.086 | Cy |
| ZoGSTU11 | Maker00042767 | 117 | 13.53 | 5.26 | 31.22 | -0.222 | Cp |
| ZoGSTU12 | Maker00042721 | 252 | 28.66 | 5.40 | 37.67 | -0.124 | Cy |
| ZoGSTU13 | Maker00042760 | 216 | 24.66 | 5.15 | 34.00 | -0.111 | Cy |
| ZoGSTU14 | Maker00045562 | 252 | 28.79 | 5.28 | 38.35 | -0.142 | Cy |
| ZoGSTU15 | Maker00045539 | 216 | 24.59 | 5.25 | 34.89 | -0.143 | Cy |
| ZoGSTU16 | Maker00042725 | 216 | 24.58 | 5.14 | 35.78 | -0.118 | Cy |
| ZoGSTU17 | Maker00052435 | 237 | 25.65 | 5.67 | 23.52 | 0.175 | Cy |
| ZoGSTU18 | Maker00052130 | 202 | 22.36 | 7.78 | 39.56 | 0.117 | Cp |
| ZoGSTU19 | Maker00052070 | 236 | 25.51 | 5.30 | 30.69 | 0.262 | Cy |
| ZoGSTU20 | Maker00075976 | 313 | 35.78 | 4.55 | 48.14 | -0.332 | Cy |
| ZoGSTU21 | Maker00075948 | 237 | 26.04 | 4.86 | 32.24 | 0.078 | Cy |
| ZoGSTU22 | Maker00008319 | 223 | 25.38 | 7.71 | 40.26 | -0.146 | Cy |
| ZoGSTU23 | Maker00008163 | 110 | 12.21 | 9.51 | 46.97 | -0.215 | Cp |
| ZoGSTU24 | Maker00071927 | 233 | 26.10 | 5.37 | 36.13 | 0.203 | Cy |
| ZoGSTU25 | Maker00045210 | 233 | 26.42 | 5.34 | 24.14 | 0.042 | Cy |
| ZoGSTU26 | Maker00045476 | 232 | 25.95 | 5.35 | 32.96 | -0.001 | Cy |
| ZoGSTU27 | Maker00045093 | 233 | 26.43 | 5.92 | 25.92 | 0.012 | Cy |
| ZoGSTU28 | Maker00044883 | 233 | 26.32 | 6.11 | 27.64 | 0.045 | Cy |
Table 4 Protein characterization of ZoGSTs
| 名称 Name | 基因ID Gene ID | 长度/aa Length | 分子量/kD Mw | 等电点 pI | 不稳定指数 Instability index | 亲水性 Hydrophilicity | 亚细胞定位 Subcellular localization |
|---|---|---|---|---|---|---|---|
| ZoGSTDHAR1 | Maker00035113 | 330 | 37.20 | 5.06 | 51.08 | -0.278 | Cp,Cy,Mt |
| ZoGSTDHAR2 | Maker00052306 | 328 | 36.71 | 9.10 | 47.47 | -0.285 | Cp |
| ZoGSTDHAR3 | Maker00046027 | 350 | 39.34 | 8.79 | 44.65 | -0.139 | Mt |
| ZoGSTDHAR5 | Maker00035024 | 255 | 28.17 | 6.25 | 43.08 | -0.232 | Cp,Cy |
| ZoGSTDHAR4 | Maker00022334 | 267 | 29.40 | 6.91 | 48.36 | -0.098 | Cp,Cy |
| ZoGSTEF1G1 | Maker00033503 | 418 | 47.80 | 5.62 | 40.78 | -0.387 | Cy |
| ZoGSTEF1G2 | Maker00031875 | 419 | 47.96 | 5.68 | 39.85 | -0.346 | Cy |
| ZoGSTCHQ1 | Maker00046252 | 265 | 31.38 | 9.20 | 36.80 | -0.314 | Cy |
| ZoGSTT1 | Maker00029139 | 263 | 29.87 | 8.99 | 41.86 | -0.200 | Cy |
| ZoGSTL1 | Maker00033284 | 218 | 24.45 | 4.57 | 49.88 | -0.138 | Nu |
| ZoGSTL2 | Maker00059206 | 262 | 29.82 | 5.32 | 41.03 | -0.344 | Cp |
| ZoGSTL3 | Maker00009568 | 241 | 27.61 | 5.28 | 42.39 | -0.194 | Cy |
| ZoGSTZ1 | Maker00037272 | 346 | 39.86 | 6.47 | 34.33 | -0.469 | Mt |
| ZoGSTZ2 | Maker00013992 | 420 | 46.61 | 8.85 | 57.36 | -0.186 | Cy |
| ZoGSTZ3 | Maker00051190 | 490 | 55.36 | 4.71 | 53.11 | -0.735 | Cp |
| ZoGSTZ4 | Maker00021183 | 334 | 37.27 | 8.20 | 52.82 | -0.159 | Cp |
| ZoGSTZ5 | Maker00078047 | 402 | 44.92 | 5.87 | 44.01 | -0.161 | Cy |
| ZoGSTF1 | Maker00077732 | 675 | 72.51 | 5.62 | 60.97 | -0.818 | Nu |
| ZoGSTF2 | Maker00076791 | 220 | 24.34 | 6.91 | 55.02 | -0.311 | Cy |
| ZoGSTF3 | Maker00068242 | 318 | 35.47 | 6.56 | 54.65 | -0.470 | Cp,Cy,Nu |
| ZoGSTF4 | Maker00047494 | 221 | 25.42 | 7.76 | 50.63 | -0.422 | Cy |
| ZoGSTF5 | Maker00076803 | 221 | 25.31 | 6.97 | 55.37 | -0.513 | Cy |
| ZoGSTF6 | Maker00050934 | 243 | 27.74 | 6.34 | 39.92 | -0.216 | Cy |
| ZoGSTF7 | Maker00023234 | 224 | 25.10 | 5.35 | 41.26 | -0.144 | Cy |
| ZoGSTF8 | Maker00077970 | 251 | 28.38 | 6.72 | 38.38 | 0.051 | Cy |
| ZoGSTF9 | Maker00078922 | 217 | 24.39 | 5.52 | 49.34 | -0.089 | Cy |
| ZoGSTF10 | Maker00078286 | 217 | 24.47 | 5.71 | 37.72 | 0.043 | Cy |
| ZoGSTF11 | Maker00078094 | 255 | 28.60 | 6.31 | 39.33 | 0.025 | Cy |
| ZoGSTF12 | Maker00077516 | 219 | 24.50 | 5.53 | 36.69 | 0.050 | Cy |
| ZoGSTF13 | Maker00040344 | 217 | 23.76 | 5.97 | 18.75 | 0.107 | Cy |
| ZoGSTU1 | Maker00008855 | 222 | 24.81 | 5.39 | 39.79 | -0.027 | Cy |
| ZoGSTU2 | Maker00015777 | 244 | 27.20 | 5.02 | 38.10 | -0.195 | Cy |
| ZoGSTU3 | Maker00049005 | 247 | 27.51 | 5.40 | 34.94 | -0.146 | Cy |
| ZoGSTU4 | Maker00045495 | 232 | 26.80 | 6.20 | 55.74 | -0.338 | Cy |
| ZoGSTU5 | Maker00045023 | 228 | 26.25 | 5.47 | 48.11 | -0.321 | Cy |
| ZoGSTU6 | Maker00061695 | 227 | 25.65 | 5.49 | 55.78 | 0.027 | Cy |
| ZoGSTU7 | Maker00045310 | 222 | 24.36 | 5.43 | 24.27 | 0.064 | Cy |
| ZoGSTU8 | Maker00042846 | 222 | 24.37 | 5.43 | 23.19 | 0.060 | Cy |
| ZoGSTU9 | Maker00044620 | 219 | 24.93 | 5.29 | 34.73 | -0.096 | Cy |
| ZoGSTU10 | Maker00042772 | 219 | 24.91 | 5.39 | 34.62 | -0.086 | Cy |
| ZoGSTU11 | Maker00042767 | 117 | 13.53 | 5.26 | 31.22 | -0.222 | Cp |
| ZoGSTU12 | Maker00042721 | 252 | 28.66 | 5.40 | 37.67 | -0.124 | Cy |
| ZoGSTU13 | Maker00042760 | 216 | 24.66 | 5.15 | 34.00 | -0.111 | Cy |
| ZoGSTU14 | Maker00045562 | 252 | 28.79 | 5.28 | 38.35 | -0.142 | Cy |
| ZoGSTU15 | Maker00045539 | 216 | 24.59 | 5.25 | 34.89 | -0.143 | Cy |
| ZoGSTU16 | Maker00042725 | 216 | 24.58 | 5.14 | 35.78 | -0.118 | Cy |
| ZoGSTU17 | Maker00052435 | 237 | 25.65 | 5.67 | 23.52 | 0.175 | Cy |
| ZoGSTU18 | Maker00052130 | 202 | 22.36 | 7.78 | 39.56 | 0.117 | Cp |
| ZoGSTU19 | Maker00052070 | 236 | 25.51 | 5.30 | 30.69 | 0.262 | Cy |
| ZoGSTU20 | Maker00075976 | 313 | 35.78 | 4.55 | 48.14 | -0.332 | Cy |
| ZoGSTU21 | Maker00075948 | 237 | 26.04 | 4.86 | 32.24 | 0.078 | Cy |
| ZoGSTU22 | Maker00008319 | 223 | 25.38 | 7.71 | 40.26 | -0.146 | Cy |
| ZoGSTU23 | Maker00008163 | 110 | 12.21 | 9.51 | 46.97 | -0.215 | Cp |
| ZoGSTU24 | Maker00071927 | 233 | 26.10 | 5.37 | 36.13 | 0.203 | Cy |
| ZoGSTU25 | Maker00045210 | 233 | 26.42 | 5.34 | 24.14 | 0.042 | Cy |
| ZoGSTU26 | Maker00045476 | 232 | 25.95 | 5.35 | 32.96 | -0.001 | Cy |
| ZoGSTU27 | Maker00045093 | 233 | 26.43 | 5.92 | 25.92 | 0.012 | Cy |
| ZoGSTU28 | Maker00044883 | 233 | 26.32 | 6.11 | 27.64 | 0.045 | Cy |

Fig. 6 Expression of ZoGST in ginger tissues of different colors(A),rhizomes at different storage temperatures(B)and leaves of ginger seedlings under melatonin treatment(C)

Fig. 7 Expression of ZoGST under different soil pore moisture content of ginger inoculated with Ralstonia solanacearum(A)and rhizome inoculated with Fusarium solani + chitosan treatment(B)
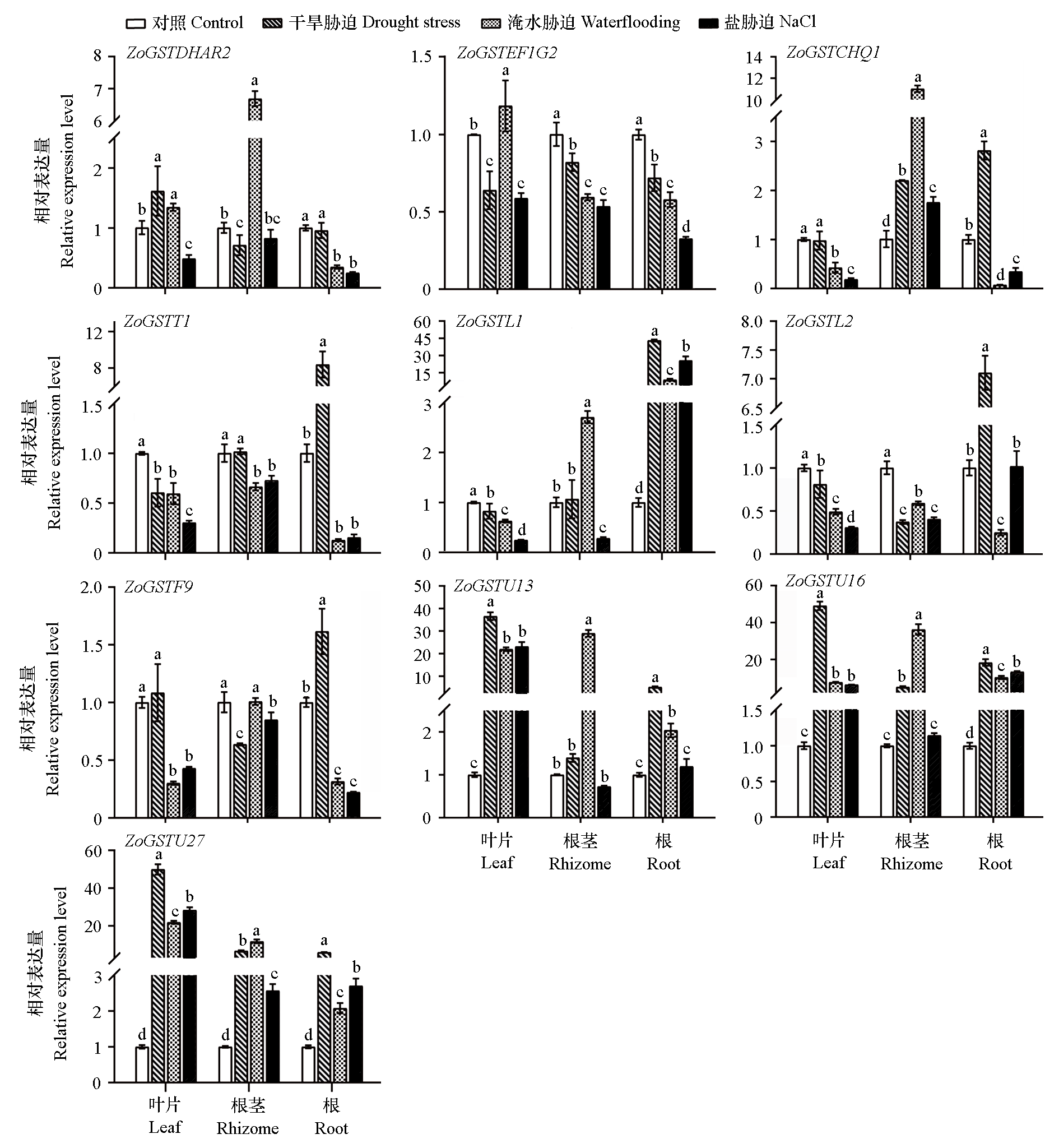
Fig. 8 Expression of ZoGST gene members under different stresses The different letters on the graph bar represent significant differences between different treatments of the same organization(P < 0.05).
| [1] |
|
| [2] |
|
| [3] |
|
|
毕艳, 郑伟, 周涛, 江维克, 杨昌贵, 肖承鸿, 周太敏. 2021. 太子参谷胱甘肽-S-转移酶基因家族生物信息学及表达分析. 广西植物, 41 (4):535-554.
|
|
| [4] |
doi: 10.16288/j.yczz.15-069 pmid: 26351171 |
|
曹运鹏, 方志, 李姝妹, 闫冲冲, 丁庆庆, 程曦, 林毅, 郭宁, 蔡永萍. 2015. 砀山酥梨4CL基因家族的全基因组鉴定与分析. 遗传, 37 (7):711-719.
pmid: 26351171 |
|
| [5] |
doi: 10.1093/dnares/dsq031 pmid: 21169340 |
| [6] |
|
| [7] |
doi: 10.1186/s12870-017-1179-z pmid: 29179697 |
| [8] |
|
| [9] |
|
| [10] |
doi: 10.1016/s1360-1385(00)01601-0 pmid: 10785664 |
| [11] |
|
| [12] |
|
| [13] |
|
| [14] |
|
| [15] |
|
| [16] |
|
| [17] |
|
| [18] |
|
|
姜栋柱, 赵小琴, 刘燃, 李洪雷, 刘奕清, 蔡小东. 2022. 姜种质资源农艺性状的遗传多样性分析. 中国蔬菜,(11):86-91.
|
|
| [19] |
|
| [20] |
|
| [21] |
|
| [22] |
|
| [23] |
|
| [24] |
|
| [25] |
|
| [26] |
|
| [27] |
|
|
李晓玉, 江海波, 江海洋, 朱苏文. 2013. 玉米全基因组谷胱苷肽-S-转移酶基因家族的分析. 安徽农业大学学报, 40 (3):350-356.
|
|
| [28] |
|
|
李永生, 方永丰, 李玥, 张同祯, 慕平, 王芳, 彭云玲, 王威, 张金文, 王汉宁. 2016. 玉米逆境响应基因ZmGST23克隆和表达分析. 农业生物技术学报, 24 (5):667-677.
|
|
| [29] |
|
|
李濯雪, 陈信波. 2015. 植物组织特异性启动子及相关顺式作用元件研究进展. 生物学杂志, 32 (6):91-95.
|
|
| [30] |
|
| [31] |
|
| [32] |
|
|
马凯恒, 谢牧洪, 郑欣悦, 贾彩霞, 洪心悦, 孙宇栋, 杨桂燕. 2021. 脱落酸和赤霉素对核桃JrGSTU8基因响应炭疽病的表达调控. 中南林业科技大学学报, 41 (12):121-129.
|
|
| [33] |
doi: 10.3724/SP.J.1006.2023.14207 |
|
马骊, 白静, 赵玉红, 孙柏林, 侯献飞, 方彦, 王旺田, 蒲媛媛, 刘丽君, 徐佳, 陶肖蕾, 孙万仓, 武军艳. 2023. 冷胁迫下甘蓝型冬油菜表达蛋白及BnGSTs基因家族的鉴定与分析. 作物学报, 49 (1):153-169.
doi: 10.3724/SP.J.1006.2023.14207 |
|
| [34] |
doi: 10.1104/pp.123.4.1561 pmid: 10938372 |
| [35] |
|
| [36] |
doi: 10.1111/j.1365-313X.2008.03761.x pmid: 19067976 |
| [37] |
|
| [38] |
|
| [39] |
|
| [40] |
|
| [41] |
|
|
王丽萍, 戚元成, 赵彦修, 张慧. 2002. 盐地碱蓬GST基因的克隆、序列分析及其表达特征. 植物生理与分子生物学学报,(2):133-136.
|
|
| [42] |
|
| [43] |
|
| [44] |
|
| [45] |
|
| [46] |
doi: 10.16288/j.yczz.16-435 pmid: 28903901 |
|
许磊, 陈文, 司国阳, 黄艺园, 林毅, 蔡永萍, 高俊山. 2017. 陆地棉GST基因家族全基因组分析. 遗传, 39 (8):737-752.
pmid: 28903901 |
|
| [47] |
|
|
张创娟, 程斌, 杨乐, 冷艳, 李师翁. 2022. 基于全基因组和转录组的绿豆谷胱甘肽转移酶基因及其对镉胁迫的响应. 西北农业学报, 31 (6):703-717.
|
|
| [48] |
|
| [49] |
|
| [50] |
pmid: 16622324 |
|
赵凤云, 王晓云, 赵彦修, 张慧. 2006. 转入盐地碱蓬谷胱甘肽转移酶和过氧化氢酶基因增强水稻幼苗对低温胁迫的抗性. 植物生理与分子生物学学报, 32 (2):231-238.
pmid: 16622324 |
|
| [51] |
doi: 10.7525/j.issn.1673-5102.2022.05.010 |
|
钟鑫爱, 孟诗琪, 周婉婷, 姚琦, 张琼, 兴旺, 刘大丽. 2022. 甜菜谷胱甘肽S-转移酶基因家族鉴定及在镉胁迫下的响应分析. 植物研究, 42 (5):790-801.
doi: 10.7525/j.issn.1673-5102.2022.05.010 |
| [1] | LIU Jianhao, JING Yanfu, LIU Yuexin, XU Yaoguang, YU Yang, GE Xiuxiu, and XIE Hua, . Identification of Peach NAC Gene Family and Role of PpNAC050 in Promoting Fruit Fructose Accumulation [J]. Acta Horticulturae Sinica, 2024, 51(9): 1983-1996. |
| [2] | ZHAO Jiaying, ZENG Zhouting, CEN Xinying, SHI Jiaoqi, LI Xiaoxian, SHEN Xiaoxia, and YU Zhenming, . Genome-Wide Identification and Expression Analysis of CCO Gene Family in Dendrobium officinale During Flower Development [J]. Acta Horticulturae Sinica, 2024, 51(9): 2075-2088. |
| [3] | CHEN Jiayue, DUAN Yingming, ZHOU Yan, XIAO Yang, BIAN Yinbing, GONG Yuhua. Identification,Expression and Function Analysis of ALDH Gene Family in Lentinula edodes [J]. Acta Horticulturae Sinica, 2024, 51(5): 1033-1046. |
| [4] | LIU Jinhong, WANG Zheng, YU Hao, XIN Yirui, QI Guoning, LIU Shenkui, REN Huimin. Identification of SLAC Gene Family in Phyllostachys edulis and Characterization of PheSLAC1 [J]. Acta Horticulturae Sinica, 2024, 51(3): 545-559. |
| [5] | LUO Xinrui, ZHANG Xiaoxu, WANG Yuping, WANG Zhi, MA Yuanyuan, ZHOU Bingyue. Identification and Expression Analysis of Trihelix Gene Family in Common Bean [J]. Acta Horticulturae Sinica, 2024, 51(12): 2775-2790. |
| [6] | HAN Shiwen, LIU Tao, WANG Liping, LI Nanyang, WANG Suna, and WANG Xing, . Genome-Wide Identification and Stress-Responsive Expression Analysis of the Cucumber SRS Gene Family [J]. Acta Horticulturae Sinica, 2024, 51(10): 2281-2296. |
| [7] | LU Jing, WEI Suyun, YIN Tongming, CHEN Yingnan. Research Progress on the Cytokinin Type-A Response Regulator Gene Family [J]. Acta Horticulturae Sinica, 2023, 50(9): 1867-1888. |
| [8] | HUANG Zhihao, LIU Tingting, DONG Xujie, YAN Mingli, LIUZhixiang , ZENG Chaozhen. Identification and Expression Analysis of HMA Gene Family in Brassica juncea Under Cadmium Stress [J]. Acta Horticulturae Sinica, 2023, 50(6): 1230-1242. |
| [9] | HE Weizhi, LEI Weiqi, GUO Xiangxin, LI Ruilian, CHEN Guanqun. Identification of the MYB Gene Family and Functional Analysis of Key Genes Related to Blue Flower Coloration in Agapanthus praecox [J]. Acta Horticulturae Sinica, 2023, 50(6): 1255-1268. |
| [10] | YU Tingting, LI Huan, NING Yuansheng, SONG Jianfei, PENG Lulin, JIA Junqi, ZHANG Weiwei, YANG Hongqiang. Genome-wide Identification of GRAS Gene Family in Apple and Expression Analysis of Its Response to Auxin [J]. Acta Horticulturae Sinica, 2023, 50(2): 397-409. |
| [11] | SHEN Yuxiao, ZOU Jinyu, LUO Ping, SHANG Wenqian, LI Yonghua, HE Songlin, WANG Zheng, SHI Liyun. Genome-wide Identification and Abiotic Stress Response Analysis of PP2C Family Genes in Rosa chinensis‘Old Blush’ [J]. Acta Horticulturae Sinica, 2023, 50(10): 2139-2156. |
| [12] | ZHAI Hanhan, ZHAI Yujie, TIAN Yi, ZHANG Ye, YANG Li, WEN Zhiliang, CHEN Haijiang. Genome-wide Identification of Peach SAUR Gene Family and Characterization of PpSAUR5 Gene [J]. Acta Horticulturae Sinica, 2023, 50(1): 1-14. |
| [13] | GAO Yanlong, WU Yuxia, ZHANG Zhongxing, WANG Shuangcheng, ZHANG Rui, ZHANG De, WANG Yanxiu. Bioinformatics Analysis of Apple ELO Gene Family and Its Expression Analysis Under Low Temperature Stress [J]. Acta Horticulturae Sinica, 2022, 49(8): 1621-1636. |
| [14] | LIU Jinming, GUO Caihua, YUAN Xing, KANG Chao, QUAN Shaowen, NIU Jianxin. Genome-wide Identification of Dof Family Genes and Expression Analysis Sepal Persistent and Abscission in Pear [J]. Acta Horticulturae Sinica, 2022, 49(8): 1637-1649. |
| [15] | TAO Xin, ZHU Rongxiang, GONG Xin, WU Lei, ZHANG Shaoling, ZHAO Jianrong, ZHANG Huping. Fructokinase Gene PpyFRK5 Plays an Important Role in Sucrose Accumulation of Pear Fruit [J]. Acta Horticulturae Sinica, 2022, 49(7): 1429-1440. |
| Viewed | ||||||
|
Full text |
|
|||||
|
Abstract |
|
|||||
Copyright © 2012 Acta Horticulturae Sinica 京ICP备10030308号-2 国际联网备案号 11010802023439
Tel: 010-82109523 E-Mail: yuanyixuebao@126.com
Support by: Beijing Magtech Co.Ltd