
Acta Horticulturae Sinica ›› 2023, Vol. 50 ›› Issue (11): 2350-2364.doi: 10.16420/j.issn.0513-353x.2023-0355
• Genetic & Breeding·Germplasm Resources·Molecular Biology • Previous Articles Next Articles
XIE Qian1, ZHANG Shiyan1, JIANG Lai1, DING Mingyue1, LIU Lingling1, WU Rujian2, CHEN Qingxi1,*( )
)
Received:2023-05-22
Revised:2023-07-02
Online:2023-11-25
Published:2023-11-28
Contact:
CHEN Qingxi
XIE Qian, ZHANG Shiyan, JIANG Lai, DING Mingyue, LIU Lingling, WU Rujian, CHEN Qingxi. Analysis of Canarium album Transcriptome SSR Information and Molecular Marker Development and Application[J]. Acta Horticulturae Sinica, 2023, 50(11): 2350-2364.
Add to citation manager EndNote|Ris|BibTeX
URL: https://www.ahs.ac.cn/EN/10.16420/j.issn.0513-353x.2023-0355
| 种质来源 Germplasm source | 编号 Number | 代号 Code | 种质名称 Germplasm name |
|---|---|---|---|
| 福建福州闽侯 Minhou,Fuzhou,Fujian | 1 | MA1 | 福榄1号 Fulan 1 |
| 2 | MA2 | 惠圆 Huiyuan | |
| 3 | MA3 | 青皮长营 Qingpi Changying | |
| 4 | MA4 | 大长营 Dachangying | |
| 5 | MA5 | 黄皮长营 Huangpi Changying | |
| 6 | MA6 | 长营 Changying | |
| 7 | MA7 | 檀头 Tantou | |
| 8 | MA8 | 甜榄1号 Tianlan 1 | |
| 9 | MA9 | 自来圆1号 Zilaiyuan 1 | |
| 10 | MA10 | 自来圆2号 Zilaiyuan 2 | |
| 福建福州闽清 Minqing,Fuzhou,Fujian | 11 | MA11 | 福榄2号 Fulan 2 |
| 12 | MA12 | 灵峰Lingfeng | |
| 13 | MA13 | 闽清1号 Minqing 1 | |
| 14 | MA14 | 清榄1号 Qinglan 1 | |
| 福建福州晋安 Jin’an,Fuzhou,Fujian | 15 | MA15 | 实生1 Shisheng 1 |
| 16 | MA16 | 实生2 Shisheng 2 | |
| 17 | MA17 | 实生3 Shisheng 3 | |
| 18 | MA18 | 实生4 Shisheng 4 | |
| 福建莆田 Putian,Fujian | 19 | MB1 | 庄边1号 Zhuangbian 1 |
| 福建泉州南安 Nan’an,Quanzhou,Fujian | 20 | MC1 | 南安1号 Nan’an 1 |
| 21 | MC2 | 南安2号 Nan’an 2 | |
| 福建漳州云霄 Yunxiao,Zhangzhou,Fujian | 22 | ME1 | 穗橄榄 Suiganlan |
| 福建漳州漳浦 Zhangpu,Zhangzhou,Fujian | 23 | ME2 | 漳浦11号Zhangpu 11 |
| 24 | ME3 | 漳浦7号 Zhangpu 7 | |
| 福建龙岩永定 Yongding,Longyan,Fujian | 25 | MF1 | 永定1号Yongding 1 |
| 26 | MF2 | 永定2号Yongding 2 | |
| 福建龙岩长汀 Changting,Longyan,Fujian | 27 | MF3 | 长汀1号Changting 1 |
| 福建南平延平 Yanping,Nanping,Fujian | 28 | MH1 | 埔头1号 Putou 1 |
| 29 | MH2 | 剑州榄 Jianzhoulan | |
| 福建宁德福安 Fu’an,Ningde,Fujian | 30 | MJ1 | 葡萄橄榄 Grape Chinese Olive |
| 31 | MJ2 | 四季榄1号 Sijilan 1 | |
| 32 | MJ3 | 四季榄2号 Sijilan 2 | |
| 33 | MJ4 | 子弹橄榄 Bullet Chinese Olive | |
| 34 | MJ5 | 子阳1号 Ziyang 1 | |
| 广东茂名电白 Dianbai,Maoming,Guangdong | 35 | YK1 | 青皮榄 Green-Rind Chinese Olive |
| 广东茂名高州 Gaozhou,Maoming,Guangdong | 36 | YK2 | 高州1号 Gaozhou 1 |
| 37 | YK3 | 高州2号 Gaozhou 2 | |
| 38 | YK4 | 香榄 Xianglan | |
| 广东潮州饶平 Raoping,Chaozhou,Guangdong | 39 | YU1 | 棱尖 Lengjian |
| 广东揭阳揭西 Jiexi,Jieyang,Guangdong | 40 | YV1 | 大白榄 Dabailan |
| 41 | YV2 | 东山长穗 Dongshan Changsui | |
| 42 | YV3 | 三棱榄 Sanlenglan | |
| 广西钦州浦北 Pubei,Qinzhou,Guangxi | 43 | GN1 | 猪腰榄 Zhuyaolan |
| 四川泸州合江 Hejiang,Luzhou,Sichuan | 44 | CE1 | 大梭子Dasuozi |
| 45 | CE2 | 丁香榄 Dingxianglan | |
| 46 | CE3 | 青果Chinese White Olive | |
| 47 | CE4 | 三白圆 Sanbaiyuan | |
| 浙江温州平阳 Pingyang,Wenzhou,Zhejiang | 48 | ZC1 | 平阳1号 Pingyang 1 |
| 49 | ZC2 | 平阳2号 Pingyang 2 | |
| 50 | ZC3 | 平阳3号 Pingyang 3 | |
| 51 | ZC4 | 平阳4号 Pingyang 4 | |
| 52 | ZC5 | 平阳5号 Pingyang 5 | |
| 53 | ZC6 | 平阳6号 Pingyang 6 | |
| 54 | ZC7 | 平阳7号 Pingyang 7 | |
| 55 | ZC8 | 平阳8号 Pingyang 8 | |
| 浙江温州瑞安 Rui’an,Wenzhou,Zhejiang | 56 | ZC9 | 瑞安1号 Rui’an 1 |
| 57 | ZC10 | 瑞安2号Rui’an 2 | |
| 58 | ZC11 | 瑞安3号Rui’an 3 | |
| 59 | ZC12 | 瑞安4号Rui’an 4 |
Table 1 The information of Chinese olive
| 种质来源 Germplasm source | 编号 Number | 代号 Code | 种质名称 Germplasm name |
|---|---|---|---|
| 福建福州闽侯 Minhou,Fuzhou,Fujian | 1 | MA1 | 福榄1号 Fulan 1 |
| 2 | MA2 | 惠圆 Huiyuan | |
| 3 | MA3 | 青皮长营 Qingpi Changying | |
| 4 | MA4 | 大长营 Dachangying | |
| 5 | MA5 | 黄皮长营 Huangpi Changying | |
| 6 | MA6 | 长营 Changying | |
| 7 | MA7 | 檀头 Tantou | |
| 8 | MA8 | 甜榄1号 Tianlan 1 | |
| 9 | MA9 | 自来圆1号 Zilaiyuan 1 | |
| 10 | MA10 | 自来圆2号 Zilaiyuan 2 | |
| 福建福州闽清 Minqing,Fuzhou,Fujian | 11 | MA11 | 福榄2号 Fulan 2 |
| 12 | MA12 | 灵峰Lingfeng | |
| 13 | MA13 | 闽清1号 Minqing 1 | |
| 14 | MA14 | 清榄1号 Qinglan 1 | |
| 福建福州晋安 Jin’an,Fuzhou,Fujian | 15 | MA15 | 实生1 Shisheng 1 |
| 16 | MA16 | 实生2 Shisheng 2 | |
| 17 | MA17 | 实生3 Shisheng 3 | |
| 18 | MA18 | 实生4 Shisheng 4 | |
| 福建莆田 Putian,Fujian | 19 | MB1 | 庄边1号 Zhuangbian 1 |
| 福建泉州南安 Nan’an,Quanzhou,Fujian | 20 | MC1 | 南安1号 Nan’an 1 |
| 21 | MC2 | 南安2号 Nan’an 2 | |
| 福建漳州云霄 Yunxiao,Zhangzhou,Fujian | 22 | ME1 | 穗橄榄 Suiganlan |
| 福建漳州漳浦 Zhangpu,Zhangzhou,Fujian | 23 | ME2 | 漳浦11号Zhangpu 11 |
| 24 | ME3 | 漳浦7号 Zhangpu 7 | |
| 福建龙岩永定 Yongding,Longyan,Fujian | 25 | MF1 | 永定1号Yongding 1 |
| 26 | MF2 | 永定2号Yongding 2 | |
| 福建龙岩长汀 Changting,Longyan,Fujian | 27 | MF3 | 长汀1号Changting 1 |
| 福建南平延平 Yanping,Nanping,Fujian | 28 | MH1 | 埔头1号 Putou 1 |
| 29 | MH2 | 剑州榄 Jianzhoulan | |
| 福建宁德福安 Fu’an,Ningde,Fujian | 30 | MJ1 | 葡萄橄榄 Grape Chinese Olive |
| 31 | MJ2 | 四季榄1号 Sijilan 1 | |
| 32 | MJ3 | 四季榄2号 Sijilan 2 | |
| 33 | MJ4 | 子弹橄榄 Bullet Chinese Olive | |
| 34 | MJ5 | 子阳1号 Ziyang 1 | |
| 广东茂名电白 Dianbai,Maoming,Guangdong | 35 | YK1 | 青皮榄 Green-Rind Chinese Olive |
| 广东茂名高州 Gaozhou,Maoming,Guangdong | 36 | YK2 | 高州1号 Gaozhou 1 |
| 37 | YK3 | 高州2号 Gaozhou 2 | |
| 38 | YK4 | 香榄 Xianglan | |
| 广东潮州饶平 Raoping,Chaozhou,Guangdong | 39 | YU1 | 棱尖 Lengjian |
| 广东揭阳揭西 Jiexi,Jieyang,Guangdong | 40 | YV1 | 大白榄 Dabailan |
| 41 | YV2 | 东山长穗 Dongshan Changsui | |
| 42 | YV3 | 三棱榄 Sanlenglan | |
| 广西钦州浦北 Pubei,Qinzhou,Guangxi | 43 | GN1 | 猪腰榄 Zhuyaolan |
| 四川泸州合江 Hejiang,Luzhou,Sichuan | 44 | CE1 | 大梭子Dasuozi |
| 45 | CE2 | 丁香榄 Dingxianglan | |
| 46 | CE3 | 青果Chinese White Olive | |
| 47 | CE4 | 三白圆 Sanbaiyuan | |
| 浙江温州平阳 Pingyang,Wenzhou,Zhejiang | 48 | ZC1 | 平阳1号 Pingyang 1 |
| 49 | ZC2 | 平阳2号 Pingyang 2 | |
| 50 | ZC3 | 平阳3号 Pingyang 3 | |
| 51 | ZC4 | 平阳4号 Pingyang 4 | |
| 52 | ZC5 | 平阳5号 Pingyang 5 | |
| 53 | ZC6 | 平阳6号 Pingyang 6 | |
| 54 | ZC7 | 平阳7号 Pingyang 7 | |
| 55 | ZC8 | 平阳8号 Pingyang 8 | |
| 浙江温州瑞安 Rui’an,Wenzhou,Zhejiang | 56 | ZC9 | 瑞安1号 Rui’an 1 |
| 57 | ZC10 | 瑞安2号Rui’an 2 | |
| 58 | ZC11 | 瑞安3号Rui’an 3 | |
| 59 | ZC12 | 瑞安4号Rui’an 4 |
| 重复类型 Repeat types | 序列数量 Number of sequences | 位点数量 Number of loci | 占全部SSR/% Proportion of all SSRs | EST中出现频率/% Frequency in EST | 平均距离/ kb Average distance | 平均长度/ bp Average length |
|---|---|---|---|---|---|---|
| 单核苷酸Single nucleotide | 25 262 | 27 390 | 31.82 | 8.53 | 12.59 | 10.34 |
| 二核苷酸Dinucleotide | 9 370 | 9 742 | 11.32 | 3.16 | 35.39 | 13.97 |
| 三核苷酸Trinucleotide | 7 773 | 8 114 | 9.43 | 2.62 | 42.48 | 16.88 |
| 四核苷酸Tetranucleotide | 954 | 961 | 1.12 | 0.32 | 358.71 | 21.91 |
| 五核苷酸Pentanucleotide | 269 | 269 | 0.31 | 0.09 | 1 281.50 | 26.43 |
| 六核苷酸Hexanucleotide | 234 | 234 | 0.27 | 0.08 | 1 473.17 | 37.10 |
| 复合型核苷酸Compound nucleotides | 36 135 | 39 374 | 45.74 | 12.19 | 8.76 | 24.45 |
Table 2 Basic distribution characteristics of SSR sequence of Chinese olive transcriptome
| 重复类型 Repeat types | 序列数量 Number of sequences | 位点数量 Number of loci | 占全部SSR/% Proportion of all SSRs | EST中出现频率/% Frequency in EST | 平均距离/ kb Average distance | 平均长度/ bp Average length |
|---|---|---|---|---|---|---|
| 单核苷酸Single nucleotide | 25 262 | 27 390 | 31.82 | 8.53 | 12.59 | 10.34 |
| 二核苷酸Dinucleotide | 9 370 | 9 742 | 11.32 | 3.16 | 35.39 | 13.97 |
| 三核苷酸Trinucleotide | 7 773 | 8 114 | 9.43 | 2.62 | 42.48 | 16.88 |
| 四核苷酸Tetranucleotide | 954 | 961 | 1.12 | 0.32 | 358.71 | 21.91 |
| 五核苷酸Pentanucleotide | 269 | 269 | 0.31 | 0.09 | 1 281.50 | 26.43 |
| 六核苷酸Hexanucleotide | 234 | 234 | 0.27 | 0.08 | 1 473.17 | 37.10 |
| 复合型核苷酸Compound nucleotides | 36 135 | 39 374 | 45.74 | 12.19 | 8.76 | 24.45 |

Fig. 1 Number (A) and proportion (B) of repeated primitive types in Chinese olive transcriptome SSR p1-p6 represent single nucleotide to hexanucleotide.

Fig. 3 Amplification of partial primers in six Chinese olive cultivars(lines) The order of samples a-f were Zilaiyuan 2,Sijilan 1,Bullet Chinese Olive,Fulan 1,Sanlenglan,and Qingpi Changying. Red numbers represent valid primers,blue numbers represent invalid primers.
| 引物 代号 Primer code | 正向引物序列 Forward primer sequence | 反向引物序列 Reverse primer sequence | 产物大 小/bp Product size | 退火温度 (正/反)/℃ Annealing temperature (F/R) | 重复基元 Repeat primitive | 等位位 点数 Number of alleles |
|---|---|---|---|---|---|---|
| 1 | CTCCAAGGGGCAAGACAGAG | TCACTGTCGCCGTCGTTAAA | 174 ~ 178 | 60.04/59.97 | (A)10;(GA)6 | 2 |
| 8 | GCCAACAGGACGATTCTCCA | AGCTAGGAGGTGAGACAGGG | 119 ~ 125 | 60.04/60.03 | (A)10;(TT)6(T)13* | 2 |
| 15 | AGCTTCCCCACACATGTCAG | ACTGGTATGCCCTTCACGTG | 221 ~ 226 | 59.96/60.04 | (TA)6 | 2 |
| 38 | TCATGCTCCTCCTCTCTGCT | CAGTGCACTTGTTCAGCCAC | 291 ~ 294 | 60.03/59.97 | (AAA)5(A)17*(AA)8* | 3 |
| 49 | GGAGACTCAAGGTCACTGCC | TTTGCTCTCCAGTGACGGTC | 290 ~ 305 | 60.04/59.97 | (GAGA)5(GA)10* | 2 |
| 53 | AATCTCCCAAGCAGATGGCC | ATCAGGCCACATCACTTGGG | 147 ~ 153 | 60.11/60.03 | (A)10;(TT)8(TTT)5*(T)17* | 2 |
| 59 | CTCCCACCCACAACCACTAC | GCCGCAAATCTCGCAGAAAT | 156 ~ 159 | 59.96/59.90 | (TGGAGA)13 | 2 |
| 63 | ACACCACCAAGCTTGTCACA | ACATTCCAGTCAGCAGCTCC | 205 ~ 215 | 60.04/60.04 | (A)10;(TT)6(T)12* | 3 |
| 64 | GGCAGATACGGGGCTCTTAC | CCATTCTCGTCCGCTGATGA | 293 ~ 326 | 59.97/59.90 | (CCT)6;(T)12(TT)6* | 6 |
| 69 | TCCGGGTATTGGAAGGCAAC | GGCTCTCTGCAAGGCGATAT | 180 ~ 183 | 60.04/59.97 | (AGG)5;(AA)6(A)12*ttcatctag aagg(A)10gtattagaataacaaaaagctgagctagccaatcctagatgaagatgtgtacagaatagtgtgtga(T)12(TT)6* | 2 |
| 72 | GGTCCAAAGAATCCGGGTGT | TGCTTTGATGGCAACGTGTG | 209 ~ 216 | 59.96/59.97 | (T)10 | 3 |
| 77 | GCCCCTTCCTGTTCTTCCTC | GGAACCGGAGTTTGGGCTAA | 177 ~ 194 | 60.04/59.96 | (CT)7 | 2 |
Table 3 The information of the developed 58 polymorphic EST
| 引物 代号 Primer code | 正向引物序列 Forward primer sequence | 反向引物序列 Reverse primer sequence | 产物大 小/bp Product size | 退火温度 (正/反)/℃ Annealing temperature (F/R) | 重复基元 Repeat primitive | 等位位 点数 Number of alleles |
|---|---|---|---|---|---|---|
| 1 | CTCCAAGGGGCAAGACAGAG | TCACTGTCGCCGTCGTTAAA | 174 ~ 178 | 60.04/59.97 | (A)10;(GA)6 | 2 |
| 8 | GCCAACAGGACGATTCTCCA | AGCTAGGAGGTGAGACAGGG | 119 ~ 125 | 60.04/60.03 | (A)10;(TT)6(T)13* | 2 |
| 15 | AGCTTCCCCACACATGTCAG | ACTGGTATGCCCTTCACGTG | 221 ~ 226 | 59.96/60.04 | (TA)6 | 2 |
| 38 | TCATGCTCCTCCTCTCTGCT | CAGTGCACTTGTTCAGCCAC | 291 ~ 294 | 60.03/59.97 | (AAA)5(A)17*(AA)8* | 3 |
| 49 | GGAGACTCAAGGTCACTGCC | TTTGCTCTCCAGTGACGGTC | 290 ~ 305 | 60.04/59.97 | (GAGA)5(GA)10* | 2 |
| 53 | AATCTCCCAAGCAGATGGCC | ATCAGGCCACATCACTTGGG | 147 ~ 153 | 60.11/60.03 | (A)10;(TT)8(TTT)5*(T)17* | 2 |
| 59 | CTCCCACCCACAACCACTAC | GCCGCAAATCTCGCAGAAAT | 156 ~ 159 | 59.96/59.90 | (TGGAGA)13 | 2 |
| 63 | ACACCACCAAGCTTGTCACA | ACATTCCAGTCAGCAGCTCC | 205 ~ 215 | 60.04/60.04 | (A)10;(TT)6(T)12* | 3 |
| 64 | GGCAGATACGGGGCTCTTAC | CCATTCTCGTCCGCTGATGA | 293 ~ 326 | 59.97/59.90 | (CCT)6;(T)12(TT)6* | 6 |
| 69 | TCCGGGTATTGGAAGGCAAC | GGCTCTCTGCAAGGCGATAT | 180 ~ 183 | 60.04/59.97 | (AGG)5;(AA)6(A)12*ttcatctag aagg(A)10gtattagaataacaaaaagctgagctagccaatcctagatgaagatgtgtacagaatagtgtgtga(T)12(TT)6* | 2 |
| 72 | GGTCCAAAGAATCCGGGTGT | TGCTTTGATGGCAACGTGTG | 209 ~ 216 | 59.96/59.97 | (T)10 | 3 |
| 77 | GCCCCTTCCTGTTCTTCCTC | GGAACCGGAGTTTGGGCTAA | 177 ~ 194 | 60.04/59.96 | (CT)7 | 2 |
| SSR位点代号 SSR locus code | Na 等位基因数 Allele number | Ne 有效等位基因数 Effective allele number | Ho 观测杂合度 Observed heterozygosity | He 期望杂合度 Expected heterozygosity | I 香农多样性指数 Shannon’s diversity index | PIC 多态性信息含量 Polymorphism information content |
|---|---|---|---|---|---|---|
| 1 | 3 | 2.068 | 0.390 | 0.521 | 0.767 | 0.399 |
| 8 | 4 | 2.487 | 1.000 | 0.603 | 1.040 | 0.515 |
| 15 | 2 | 2.000 | 1.000 | 0.504 | 0.693 | 0.375 |
| 38 | 3 | 2.636 | 1.000 | 0.626 | 1.031 | 0.549 |
| 49 | 4 | 2.208 | 1.000 | 0.552 | 0.881 | 0.444 |
| 53 | 4 | 1.932 | 0.458 | 0.487 | 0.901 | 0.438 |
| 59 | 4 | 2.446 | 0.102 | 0.589 | 1.022 | 0.496 |
| 63 | 6 | 3.118 | 1.000 | 0.685 | 1.359 | 0.633 |
| 64 | 10 | 4.497 | 0.661 | 0.784 | 1.766 | 0.748 |
| 69 | 3 | 2.068 | 1.000 | 0.521 | 0.767 | 0.399 |
| 72 | 3 | 1.108 | 0.102 | 0.099 | 0.233 | 0.095 |
| 77 | 2 | 1.017 | 0.017 | 0.017 | 0.049 | 0.017 |
| 平均值Average | 4 | 2.299 | 0.644 | 0.499 | 0.876 | 0.426 |
Table 4 Genetic diversity parameters of 12 pairs of EST-SSR markers in Chinese olive samples
| SSR位点代号 SSR locus code | Na 等位基因数 Allele number | Ne 有效等位基因数 Effective allele number | Ho 观测杂合度 Observed heterozygosity | He 期望杂合度 Expected heterozygosity | I 香农多样性指数 Shannon’s diversity index | PIC 多态性信息含量 Polymorphism information content |
|---|---|---|---|---|---|---|
| 1 | 3 | 2.068 | 0.390 | 0.521 | 0.767 | 0.399 |
| 8 | 4 | 2.487 | 1.000 | 0.603 | 1.040 | 0.515 |
| 15 | 2 | 2.000 | 1.000 | 0.504 | 0.693 | 0.375 |
| 38 | 3 | 2.636 | 1.000 | 0.626 | 1.031 | 0.549 |
| 49 | 4 | 2.208 | 1.000 | 0.552 | 0.881 | 0.444 |
| 53 | 4 | 1.932 | 0.458 | 0.487 | 0.901 | 0.438 |
| 59 | 4 | 2.446 | 0.102 | 0.589 | 1.022 | 0.496 |
| 63 | 6 | 3.118 | 1.000 | 0.685 | 1.359 | 0.633 |
| 64 | 10 | 4.497 | 0.661 | 0.784 | 1.766 | 0.748 |
| 69 | 3 | 2.068 | 1.000 | 0.521 | 0.767 | 0.399 |
| 72 | 3 | 1.108 | 0.102 | 0.099 | 0.233 | 0.095 |
| 77 | 2 | 1.017 | 0.017 | 0.017 | 0.049 | 0.017 |
| 平均值Average | 4 | 2.299 | 0.644 | 0.499 | 0.876 | 0.426 |
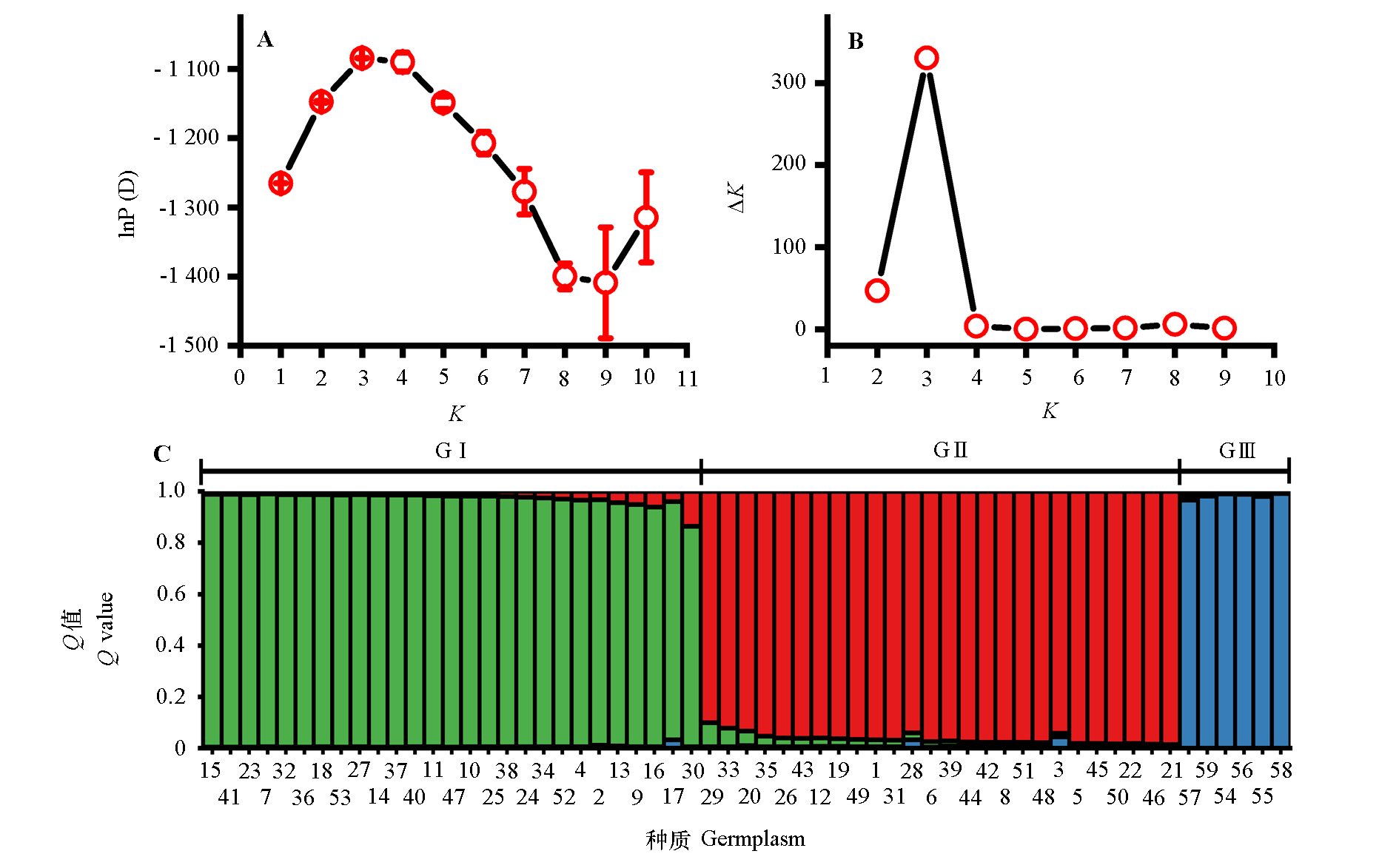
Fig. 4 Population structure of 59 Chinese olive germplasm A:The variation of logarithmic likelihood value lnP(D) with the number of subgroups K;B:The variation of ∆K value with the number of subgroups K;C:When K = 3,the population genetic structure of 59 Chinese olive germplasm materials obtained by structure calculation. The 59 germplasm were classified into three taxa,GⅠ,GⅡand GⅢ,based on the maximum Q value. The corresponding germplasm resources of Chinese olive sample number were shown in Table 1.
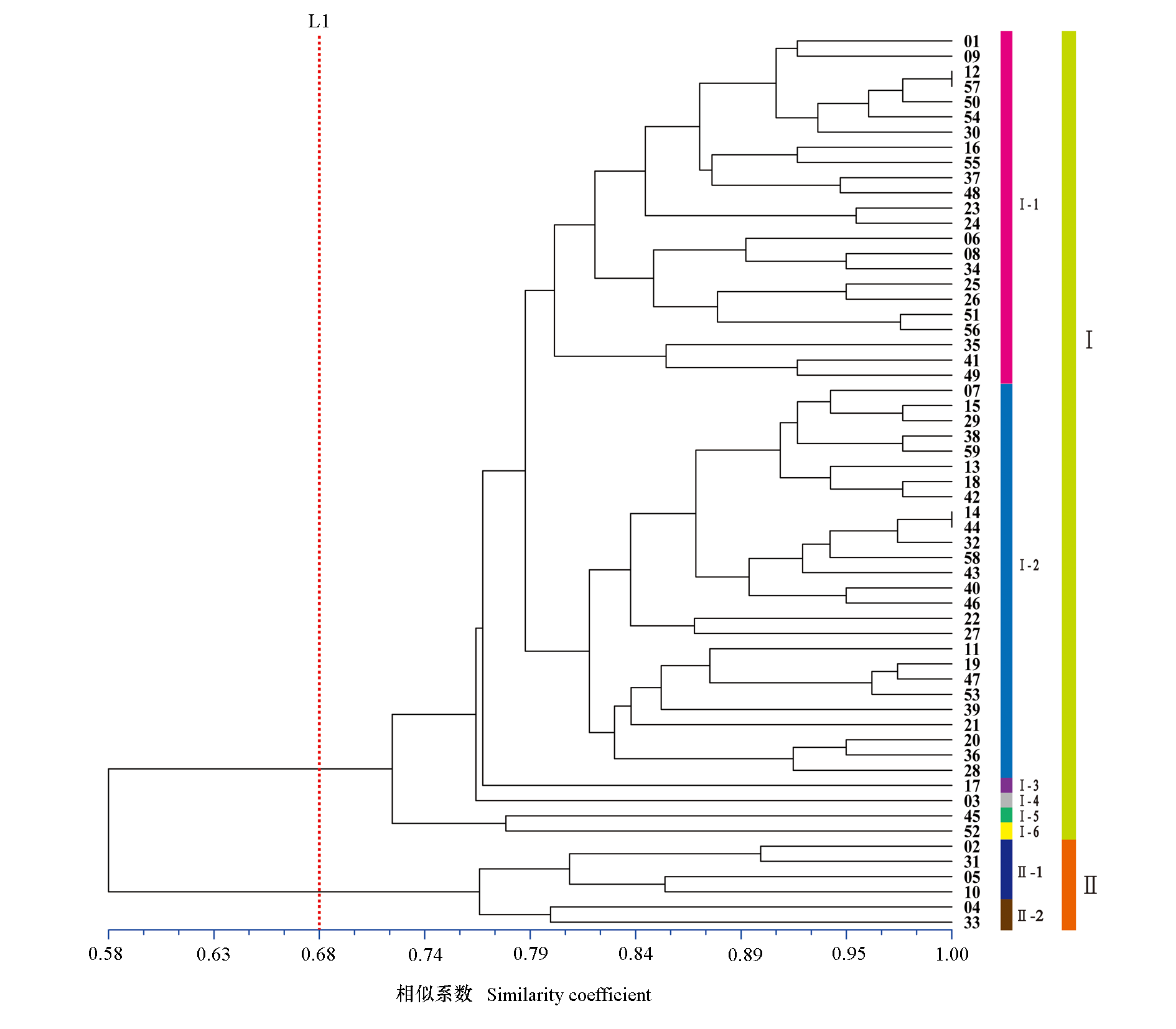
Fig. 6 Phylogenetic tree of 59 Chinese olive samples based on 12 EST-SSR markers developed The corresponding germplasm resources of Chinese olive sample number are shown in Table 1.
| [1] |
pmid: 6247908 |
| [2] |
|
|
陈杰忠. 2011. 橄榄. 果树栽培学各论:南方本(第四版). 北京: 中国农业出版社.
|
|
| [3] |
doi: 10.16420/j.issn.0513-353x.2021-0312 URL |
|
陈明堃, 陈璐, 孙维红, 马山虎, 兰思仁, 彭东辉, 刘仲健, 艾叶. 2022. 建兰种质资源遗传多样性分析及核心种质构建. 园艺学报, 49 (1):175-186.
doi: 10.16420/j.issn.0513-353x.2021-0312 URL |
|
| [4] |
|
|
池毓斌, 谢倩, 陈清西. 2016. 几个鲜食橄榄品种(系)及良种繁育方法简介. 中国南方果树, 45 (3):154-156,166.
|
|
| [5] |
Chinese Pharmacopoeia Commission. 2020. Canarii fructus. Pharmacopoeia of the people's republic of China:2020 ed. Beijing: The Medicine Science and Technology Press of China. (in Chinese)
|
|
国家药典委员会. 2020. 青果. 中华人民共和国药典: 2020年版. 北京: 中国医药科技出版社.
|
|
| [6] |
doi: 10.1186/s12870-015-0589-z URL |
| [7] |
|
|
杜庆章. 2014. 利用连锁与连锁不平衡联合作图解析毛白杨重要性状的等位遗传变异[博士论文]. 北京: 北京林业大学.
|
|
| [8] |
Editorial Committee of flora of China,Chinese Academy of Sciences. 1997. Burseraceae. Flora of China. Vol. 43. Beijing: Science Press. (in Chinese)
|
|
中国科学院中国植物志编辑委员会. 1997. 橄榄科. 中国植物志:第四十三卷. 北京: 科学出版社.
|
|
| [9] |
pmid: 13679975 |
| [10] |
doi: 10.1111/j.1365-294X.2005.02553.x pmid: 15969739 |
| [11] |
|
|
谷振军, 杨春霞, 丁伟, 李康琴, 黄宝祥. 2019. 南酸枣转录组SSR序列特征分析及其分子标记开发. 南方林业科学, 47 (3):12-15.
|
|
| [12] |
doi: 10.16420/j.issn.0513-353x.2019-0840 URL |
|
郭俊, 朱婕, 谢尚潜, 张叶, 叶蓓蕾, 郑丽燕, 凌鹏. 2020. 油梨转录组SSR分子标记开发与种质资源亲缘关系分析. 园艺学报, 47 (8):1552-1564.
doi: 10.16420/j.issn.0513-353x.2019-0840 URL |
|
| [13] |
doi: 10.16420/j.issn.0513-353x.2018-0753 URL |
|
胡文舜, 陈秀萍, 郑少泉. 2019. 龙眼EST-SSR标记开发及无患子科5个属种质遗传多样性分析. 园艺学报, 46 (7):1359-1372.
doi: 10.16420/j.issn.0513-353x.2018-0753 URL |
|
| [14] |
doi: 10.16420/j.issn.0513-353x.2020-0497 URL |
|
蒋爽, 张学英, 安海山, 徐芳杰, 章加应. 2021. 枇杷全基因组SSR标记开发及其多态性研究. 园艺学报, 48 (5):1013-1022.
doi: 10.16420/j.issn.0513-353x.2020-0497 URL |
|
| [15] |
|
|
赖瑞联, 沈朝贵, 冯新, 陈义挺, 韦晓霞, 吴如健. 2023. 橄榄果实转录组SSR和SNP/InDel位点特征. 热带作物学报, 44 (4):681-688.
|
|
| [16] |
|
|
李荷蓉. 2012. 苹果EST-SSR标记的开发与电子表达平台建立[硕士论文]. 南京: 南京农业大学.
|
|
| [17] |
doi: 10.1093/molbev/msh073 URL |
| [18] |
|
| [19] |
doi: 10.1007/s11033-020-05893-7 pmid: 33040266 |
| [20] |
|
|
毛娟, 梁国平, 卢世雄, 马宗桓, 王萍, 陈佰鸿. 2019. 葡萄EST-SSR标记的开发及其在遗传多样性分析中的应用. 中外葡萄与葡萄酒,(6):12-19.
|
|
| [21] |
doi: 10.1111/men.2006.6.issue-1 URL |
| [22] |
|
| [23] |
doi: 10.1007/s00438-021-01844-4 pmid: 35039933 |
| [24] |
doi: 10.1093/genetics/155.2.945 pmid: 10835412 |
| [25] |
doi: 10.1111/jse.v57.5 URL |
| [26] |
doi: 10.1101/gr.184001 pmid: 11483586 |
| [27] |
doi: 10.1139/x03-283 URL |
| [28] |
doi: 10.1270/jsbbs.66.204 pmid: 27162492 |
| [29] |
doi: 10.1111/pbr.v140.1 URL |
| [30] |
|
|
王小敏, 张春红, 胡丽超, 吴文龙, 李维林. 2022. 基于高通量测序的蓝莓果实转录组SSR信息分析. 北方园艺,(13):49-54.
|
|
| [31] |
doi: 10.1007/s11033-012-2404-3 pmid: 23275197 |
| [32] |
doi: 10.1186/1471-2164-12-451 |
| [33] |
|
|
卫尊征. 2010. 小叶杨遗传资源评价及重要性状的SSRs关联分析[博士论文]. 北京: 北京林业大学.
|
|
| [34] |
doi: 10.1111/jfds.1979.44.issue-1 URL |
|
谢倩, 李易易, 张诗艳, 束燕萍, 王威, 陈清西. 2023. 基于模糊数学感官评价、理化特性与电子舌的橄榄鲜食品质分析. 食品科学, 44 (3):69-78.
|
|
| [35] |
doi: 10.16420/j.issn.0513-353x.2021-0943 URL |
|
徐婉, 林雅君, 赵荘, 周庄. 2022. 兰属植物资源与育种研究进展. 园艺学报, 49 (12):2722-2742.
doi: 10.16420/j.issn.0513-353x.2021-0943 URL |
|
| [36] |
doi: 10.1016/j.scienta.2013.06.043 URL |
| [37] |
doi: 10.1186/1471-2164-11-94 |
| [38] |
doi: 10.16420/j.issn.0513-353x.2021-0203 URL |
|
赵青, 都真真, 李锡香, 宋江萍, 张晓辉, 阳文龙, 贾会霞, 王海平. 2021. 利用SSRseq分子标记的大蒜种质资源遗传多样性研究. 园艺学报, 48 (7):1397-1408.
doi: 10.16420/j.issn.0513-353x.2021-0203 URL |
|
| [39] |
|
|
郑世茂, 周希希, 贺乔乔, 张羽. 2022. 基于美味猕猴桃转录组的SSR特征分析. 分子植物育种, http://kns.cnki.net/kcms/detail/46.1068.S.20221129.1037.012.html:2023-05-19
|
|
| [40] |
doi: 10.1007/s11032-013-9923-z URL |
| [1] | NIE Xinghua, ZHANG Yu, LIU Song, YANG Jiabin, HAO Yaqiong, LIU Yang, QIN Ling, XING Yu. Study on Genetic Characteristics and Taxonomic Status of Wild Chinese Chestnut Based on Genome Re-sequencing [J]. Acta Horticulturae Sinica, 2023, 50(8): 1622-1636. |
| [2] | LU Yanqing, LIN Yanjin, LU Xinkun. Cell Wall Material Metabolism and the Response to High Temperature and Water Deficiency Stresses in Pericarp are Associated with Fruit Cracking of‘Duwei’Pummelo [J]. Acta Horticulturae Sinica, 2023, 50(8): 1747-1768. |
| [3] | MEI Yulin, XU Jinjian, JIANG Mengyue, YU Junhai, ZONG Yu, CHEN Wenrong, LIAO Fanglei, GUO Weidong. Identification of F1 Hybrids and Analysis of Fruit Shape Difference Between Fingered Citron and Citron [J]. Acta Horticulturae Sinica, 2023, 50(7): 1402-1418. |
| [4] | RUAN Ruoxin, LUO Huifeng, ZHANG Chen, HUANG Kangkang, XI Dujun, PEI Jiabo, XING Mengyun, LIU Hui. Transcriptome Analysis of Flower Buds of Sweet Cherry Cultivars with Different Chilling Requirements During Dormancy Stages in Hangzhou [J]. Acta Horticulturae Sinica, 2023, 50(6): 1187-1202. |
| [5] | XU Yue, LI Zixiong, CHEN Jie, SUN Liang. Transcriptional Bases of the Regulation of 2,4-D on Tomato Fruit Shape [J]. Acta Horticulturae Sinica, 2023, 50(4): 802-814. |
| [6] | WANG Zehan, YU Wentao, WANG Pengjie, LIU Caiguo, FAN Xiaojing, GU Mengya, CAI Chunping, WANG Pan, YE Naixing. WGCNA Analysis of Differentially Expressed Genes in Floral Organs of Tea Germplasms with Ovary-glabrous and Ovary-trichome [J]. Acta Horticulturae Sinica, 2023, 50(3): 620-634. |
| [7] | SUN Zeshuo, JIANG Dongyue, LIU Xinhong, SHEN Xin, LI Yingang, QU Yufei, LI Yonghua. Cluster Analysis and Construction of DNA Fingerprinting of 42 Oriental Cultivars of Flowering Cherry Based on SSR Markers [J]. Acta Horticulturae Sinica, 2023, 50(3): 657-668. |
| [8] | WANG Rui, HONG Wenjuan, LUO Hua, ZHAO Lina, CHEN Ying, WANG Jun. Construction of SSR Fingerprints of Pomegranate Cultivars and Male Parent Identification of Hybrids [J]. Acta Horticulturae Sinica, 2023, 50(2): 265-278. |
| [9] | JIANG Yu, TU Xunliang, HE Junrong. Analysis of Differential Expression Genes in Leaves of Leaf Color Mutant of Chinese Orchid [J]. Acta Horticulturae Sinica, 2023, 50(2): 371-381. |
| [10] | WANG Mengmeng, SUN Deling, CHEN Rui, YANG Yingxia, ZHANG Guan, LÜ Mingjie, WANG Qian, XIE Tianyu, NIU Guobao, SHAN Xiaozheng, TAN Jin, YAO Xingwei. Construction and Evaluation of Cauliflower Core Collection [J]. Acta Horticulturae Sinica, 2023, 50(2): 421-431. |
| [11] | DUAN Kaihang, WANG Xiaoling, MAO Yongmin, WANG Yao, REN Yongxiang, REN Liuliu, and SHEN Lianying. Analysis of Genetic Diversity of Wild Jujube Germplasm Resources Based on Quantitative Characters [J]. Acta Horticulturae Sinica, 2023, 50(12): 2568-2576. |
| [12] | ZHANG Yongqi, WANG Chao, XU Linlin, WU E’jiao, LI Tianhong, ZHAO Mizhen, YUAN Huazhao. Development of SSR Molecular Markers and Construction of Core Collection of Wild Diploid Strawberry [J]. Acta Horticulturae Sinica, 2023, 50(11): 2365-2375. |
| [13] | SU Qun, WANG Hongyan, LIU Jun, LI Chunniu, BU Zhaoyang, LIN Yuling, LU Jiashi, LAI Zhongxiong. Construction of Core Collection of Nymphaea Based on SSR Fluorescent Markers [J]. Acta Horticulturae Sinica, 2023, 50(10): 2128-2138. |
| [14] | LIU Yiping, NI Menghui, WU Fangfang, LIU Hongli, HE Dan, KONG Dezheng. Association Analysis of Organ Traits with SSR Markers in Lotus(Nelumbo nucifera) [J]. Acta Horticulturae Sinica, 2023, 50(1): 103-115. |
| [15] | ZHAO Xueyan, WANG Qi, WANG Li, WANG Fangyuan, WANG Qing, LI Yan. Comparative Transcriptome Analysis of Differential Expression in Different Tissues of Corydalis yanhusuo [J]. Acta Horticulturae Sinica, 2023, 50(1): 177-187. |
| Viewed | ||||||
|
Full text |
|
|||||
|
Abstract |
|
|||||
Copyright © 2012 Acta Horticulturae Sinica 京ICP备10030308号-2 国际联网备案号 11010802023439
Tel: 010-82109523 E-Mail: yuanyixuebao@126.com
Support by: Beijing Magtech Co.Ltd