
Acta Horticulturae Sinica ›› 2023, Vol. 50 ›› Issue (3): 657-668.doi: 10.16420/j.issn.0513-353x.2021-1057
• Research Notes • Previous Articles Next Articles
SUN Zeshuo1,2, JIANG Dongyue2, LIU Xinhong2, SHEN Xin2, LI Yingang2, QU Yufei3, LI Yonghua1,*( )
)
Received:2022-09-19
Revised:2022-12-01
Online:2023-03-25
Published:2023-04-03
Contact:
*(E-mail:liyhhany@henau.edu)
CLC Number:
SUN Zeshuo, JIANG Dongyue, LIU Xinhong, SHEN Xin, LI Yingang, QU Yufei, LI Yonghua. Cluster Analysis and Construction of DNA Fingerprinting of 42 Oriental Cultivars of Flowering Cherry Based on SSR Markers[J]. Acta Horticulturae Sinica, 2023, 50(3): 657-668.
Add to citation manager EndNote|Ris|BibTeX
URL: https://www.ahs.ac.cn/EN/10.16420/j.issn.0513-353x.2021-1057
| 遗传背景 Genetic background | 编号 Number | 品种 Cultivar | 花色 Flower color | 花型 Flower type |
|---|---|---|---|---|
| 江户彼岸樱系 Prunus spachiana | YH1 | 染井吉野樱 Prunus × yedoensis ‘Somei- yoshino’ | 淡红白色 Light white red | 单瓣 Single |
| series | YH3 | 美国樱 Prunus × yedoensis‘America’ | 淡粉红色 Rose pink | 单瓣 Single |
| YH5 | 江户彼岸樱 Prunus × subhirtella | 淡红色 Light red | 单瓣 Single | |
| YH7 | 八重红彼岸樱 Prunus × subhirtella‘Yaebeni- higan’ | 淡红色 Light red | 半重瓣 Semidouble | |
| YH10 | 越彼岸樱 Prunus × subhirtella‘Koshiensis’ | 深粉红色 Rediance | 单瓣 Single | |
| YH13 | 八重红枝垂樱 Prunus spachiana‘Plena Rosea’ | 淡红色 Light red | 半重瓣 Semidouble | |
| YH18 | 小松乙女樱 Prunus spachiana‘Komatsu- otome’ | 淡粉色 Light pink | 单瓣 Single | |
| YH35 | 御帝吉野樱 Prunus × yedoensis ‘Mikado- yoshino’ | 白色 White | 单瓣 Single | |
| YH37 | 思川樱 Prunus × subhirtella‘Omoigawa’ | 淡紫红色 Light purple red | 单瓣 Single | |
| 山樱系 Prunus | YH2 | 白妙樱 Prunus serrulata‘Sirotae’ | 白色 White | 半重瓣 Semidouble |
| serrulata series | YH4 | 苔清水樱 Prunus serrulata‘Kokeshimidsu’ | 淡粉红色 Rose pink | 单瓣 Single |
| YH6 | 蝴蝶樱 Prunus serrulata‘Kocho’ | 淡粉色 Light pink | 单瓣 Single | |
| YH8 | 郁金樱 Prunus serrulata‘Grandiflora’ | 淡黄绿色 Light yellowish green | 半重瓣 Semidouble | |
| YH9 | 红华樱 Prunus serrulata‘Kouka’ | 淡红色 Light red | 重瓣 Double | |
| YH14 | 麒麟樱 Prunus serrulata‘Kirin’ | 淡紫红色 Light purple red | 重瓣 Double | |
| YH15 | 松前早咲樱 Prunus serrulata‘Matsumae- hayazaki’ | 淡紫红色 Light purple red | 半重瓣 Semidouble | |
| YH19 | 银河山樱 Prunus serrulata‘Erecta’ | 淡红色 Light red | 半重瓣 Semidouble | |
| YH21 | 御衣黄樱 Prunus serrulata‘Gioiko’ | 黄绿色 Yellowish green | 半重瓣 Semidouble | |
| YH23 | 花笠樱 Prunus serrulata‘Hanagasa’ | 淡红色 Light red | 重瓣 Double | |
| YH24 | 福禄寿樱 Prunus serrulate‘Contorta’ | 淡紫红色 Light purple red | 半重瓣 Semidouble | |
| YH25 | 红时雨樱 Prunus serrulata‘Beni-shigure’ | 淡紫红色 Light purple red | 重瓣 Double | |
| YH26 | 仙台屋樱 Prunus jamasakura‘Sendaiya’ | 淡紫红色 Light purple red | 单瓣 Single | |
| YH29 | 霞樱 Prunus verecunda | 白色 White | 单瓣 Single | |
| YH31 | 八重红大岛樱 Prunus serrulata ‘Yaebeni- ohshima’ | 淡红色 Light red | 重瓣 Double | |
| YH32 | 御车返樱 Prunus serrulata‘Mikurumakaishi’ | 淡紫红色 Light purple red | 单瓣 Single | |
| YH33 | 一叶樱 Prunus serrulata‘Hisakura’ | 淡红色 Light red | 重瓣 Double | |
| YH38 | 太白樱 Prunus serrulata‘Taihaku’ | 白色 White | 单瓣 Single | |
| YH41 | 山樱花 Prunus serrulata | 白色 White | 单瓣 Single | |
| 豆樱系 Prunus | YH27 | 豆樱 Prunus incisa | 白色 White | 单瓣 Single |
| incisa series | YH28 | 绿樱 Prunus incisa‘Yamadei’ | 白色 White | 单瓣 Single |
| YH30 | 御殿场樱 Prunus × incisa‘Gotenba-zakura’ | 淡紫红色 Light purple red | 单瓣 Single | |
| 钟花樱系 Prunus | YH11 | 钟花樱1 Prunus campanulata | 玫红色 Rose red | 单瓣 Single |
| campanulata series | YH12 | 紫荆樱 Prunus cerasoides | 紫红色 Purplish red | 单瓣 Single |
| YH16 | 中国红樱 Prunus campanulata‘Zhongguohong’ | 深红色 Deep red | 单瓣 Single | |
| YH17 | 红粉佳人樱 Prunus sp. | 玫红色 Rose red | 单瓣 Single | |
| YH20 | 椿寒樱 Prunus pseudocerasus‘Introrsa’ | 粉红色 Pink | 单瓣 Single | |
| YH22 | 寒樱 Prunus × kanzakura‘Praecox’ | 淡红色 Light red | 单瓣 Single | |
| YH34 | 大渔樱 Prunus × kanzakura‘Tairyo-zakura’ | 淡紫红色 Light purple red | 单瓣 Single | |
| YH36 | 大寒樱 Prunus × kanzakura‘Oh-kanzakuru’ | 淡红色 Light red | 单瓣 Single | |
| YH39 | 雅樱 Prunus‘Miyabi’ | 紫红色 Purplish red | 单瓣 Single | |
| YH40 | 钟花樱2 Prunus campanulata | 玫红色 Rose red | 单瓣 Single | |
| 尾叶樱系 Prunus dielsiana series | YH42 | 尾叶樱 Prunus dielsiana | 粉白色 Pink white | 单瓣 Single |
Table 1 Forty-two oriental cultivars of flowering cherry materials
| 遗传背景 Genetic background | 编号 Number | 品种 Cultivar | 花色 Flower color | 花型 Flower type |
|---|---|---|---|---|
| 江户彼岸樱系 Prunus spachiana | YH1 | 染井吉野樱 Prunus × yedoensis ‘Somei- yoshino’ | 淡红白色 Light white red | 单瓣 Single |
| series | YH3 | 美国樱 Prunus × yedoensis‘America’ | 淡粉红色 Rose pink | 单瓣 Single |
| YH5 | 江户彼岸樱 Prunus × subhirtella | 淡红色 Light red | 单瓣 Single | |
| YH7 | 八重红彼岸樱 Prunus × subhirtella‘Yaebeni- higan’ | 淡红色 Light red | 半重瓣 Semidouble | |
| YH10 | 越彼岸樱 Prunus × subhirtella‘Koshiensis’ | 深粉红色 Rediance | 单瓣 Single | |
| YH13 | 八重红枝垂樱 Prunus spachiana‘Plena Rosea’ | 淡红色 Light red | 半重瓣 Semidouble | |
| YH18 | 小松乙女樱 Prunus spachiana‘Komatsu- otome’ | 淡粉色 Light pink | 单瓣 Single | |
| YH35 | 御帝吉野樱 Prunus × yedoensis ‘Mikado- yoshino’ | 白色 White | 单瓣 Single | |
| YH37 | 思川樱 Prunus × subhirtella‘Omoigawa’ | 淡紫红色 Light purple red | 单瓣 Single | |
| 山樱系 Prunus | YH2 | 白妙樱 Prunus serrulata‘Sirotae’ | 白色 White | 半重瓣 Semidouble |
| serrulata series | YH4 | 苔清水樱 Prunus serrulata‘Kokeshimidsu’ | 淡粉红色 Rose pink | 单瓣 Single |
| YH6 | 蝴蝶樱 Prunus serrulata‘Kocho’ | 淡粉色 Light pink | 单瓣 Single | |
| YH8 | 郁金樱 Prunus serrulata‘Grandiflora’ | 淡黄绿色 Light yellowish green | 半重瓣 Semidouble | |
| YH9 | 红华樱 Prunus serrulata‘Kouka’ | 淡红色 Light red | 重瓣 Double | |
| YH14 | 麒麟樱 Prunus serrulata‘Kirin’ | 淡紫红色 Light purple red | 重瓣 Double | |
| YH15 | 松前早咲樱 Prunus serrulata‘Matsumae- hayazaki’ | 淡紫红色 Light purple red | 半重瓣 Semidouble | |
| YH19 | 银河山樱 Prunus serrulata‘Erecta’ | 淡红色 Light red | 半重瓣 Semidouble | |
| YH21 | 御衣黄樱 Prunus serrulata‘Gioiko’ | 黄绿色 Yellowish green | 半重瓣 Semidouble | |
| YH23 | 花笠樱 Prunus serrulata‘Hanagasa’ | 淡红色 Light red | 重瓣 Double | |
| YH24 | 福禄寿樱 Prunus serrulate‘Contorta’ | 淡紫红色 Light purple red | 半重瓣 Semidouble | |
| YH25 | 红时雨樱 Prunus serrulata‘Beni-shigure’ | 淡紫红色 Light purple red | 重瓣 Double | |
| YH26 | 仙台屋樱 Prunus jamasakura‘Sendaiya’ | 淡紫红色 Light purple red | 单瓣 Single | |
| YH29 | 霞樱 Prunus verecunda | 白色 White | 单瓣 Single | |
| YH31 | 八重红大岛樱 Prunus serrulata ‘Yaebeni- ohshima’ | 淡红色 Light red | 重瓣 Double | |
| YH32 | 御车返樱 Prunus serrulata‘Mikurumakaishi’ | 淡紫红色 Light purple red | 单瓣 Single | |
| YH33 | 一叶樱 Prunus serrulata‘Hisakura’ | 淡红色 Light red | 重瓣 Double | |
| YH38 | 太白樱 Prunus serrulata‘Taihaku’ | 白色 White | 单瓣 Single | |
| YH41 | 山樱花 Prunus serrulata | 白色 White | 单瓣 Single | |
| 豆樱系 Prunus | YH27 | 豆樱 Prunus incisa | 白色 White | 单瓣 Single |
| incisa series | YH28 | 绿樱 Prunus incisa‘Yamadei’ | 白色 White | 单瓣 Single |
| YH30 | 御殿场樱 Prunus × incisa‘Gotenba-zakura’ | 淡紫红色 Light purple red | 单瓣 Single | |
| 钟花樱系 Prunus | YH11 | 钟花樱1 Prunus campanulata | 玫红色 Rose red | 单瓣 Single |
| campanulata series | YH12 | 紫荆樱 Prunus cerasoides | 紫红色 Purplish red | 单瓣 Single |
| YH16 | 中国红樱 Prunus campanulata‘Zhongguohong’ | 深红色 Deep red | 单瓣 Single | |
| YH17 | 红粉佳人樱 Prunus sp. | 玫红色 Rose red | 单瓣 Single | |
| YH20 | 椿寒樱 Prunus pseudocerasus‘Introrsa’ | 粉红色 Pink | 单瓣 Single | |
| YH22 | 寒樱 Prunus × kanzakura‘Praecox’ | 淡红色 Light red | 单瓣 Single | |
| YH34 | 大渔樱 Prunus × kanzakura‘Tairyo-zakura’ | 淡紫红色 Light purple red | 单瓣 Single | |
| YH36 | 大寒樱 Prunus × kanzakura‘Oh-kanzakuru’ | 淡红色 Light red | 单瓣 Single | |
| YH39 | 雅樱 Prunus‘Miyabi’ | 紫红色 Purplish red | 单瓣 Single | |
| YH40 | 钟花樱2 Prunus campanulata | 玫红色 Rose red | 单瓣 Single | |
| 尾叶樱系 Prunus dielsiana series | YH42 | 尾叶樱 Prunus dielsiana | 粉白色 Pink white | 单瓣 Single |
| 引物名称 Primer name | 引物序列(5′-3′) Primer sequence | 退火温度/℃ Tm | 来源 Source |
|---|---|---|---|
| SSR5 | F:GATGTAGAGGGAGGTTCTGTATTTG | 56 | 基因组测序开发 |
| R:CGGATCTAGTGAGGGATGGGTATG | Genome sequencing development | ||
| SSR11 | F:TAGTCTCAGCAGAACCACCAAATAC | 56 | |
| R:ATGATACTGATTACGAGAGGCTTAG | |||
| BPPCT008 | F:ATGGTGTGTATGGACATGATGA | 55 | Dirlewanger et al., |
| R:CCTCAACCTAAGACACCTTCACT | |||
| BPPCT040 | F:ATGAGGACGTGTCTGAATGG | 55 | |
| R:AGCCAAACCCCTCTTATACG | |||
| CPSCT021 | F:GCCACTTCGGCTAAAAGAGA | 55 | Mnejja et al., |
| R:TCCATATCTCCTCCTGCTTGA | |||
| M9a | F:GCCGAAACCCTAGGTGAGCG | 58 | Yamamoto et al., |
| R:CAGGATGCTTGCGGTGCTTG | |||
| MA016b | F:TGGCTGGTGGAGACGGAGGA | 56 | |
| R:ATGATACCCAGCCTCCCGGG | |||
| MA007a | F:GTGCATCGTTAGGAACTGCC | 56 | |
| R:GCCCCTGAGATACAACTGCA | |||
| EMPaS11A | F:ACCACTTTGAGGAACTTGGG | 55 | Vaughan & Russell, |
| R:CTGCCTGGAAGAGCAATAAC | |||
| PceGA25 | F:GCAATTCGAGCTGTATTTCAGATG | 58 | Cantini et al., |
| R:CAGTTGGCGGCTATCATGTCTTAC | |||
| PS12A02 | F:GCCACCAATGGTTCTTCC | 56 | Downey & Lezzoni, |
| R:AGCACCAGATGCACCTGA | |||
| pchcms1 | F:GTTACACCTCTGTCACA | 56 | Sosinski et al., |
| R:CTTGGCTGGCATTCCTA | |||
| UDP97-403 | F:CTGGCTTACAACTCGCAAGC | 56 | Cipriani et al., |
| R:CGTCGACCAACTGAGACTCA | |||
| UDP96-005 | F:GTAACGCTCGCTACCACAAA | 56 | |
| R:CCTGCATATCACCACCCAG |
Table 2 Fourteen pairs of SSR primers in this study
| 引物名称 Primer name | 引物序列(5′-3′) Primer sequence | 退火温度/℃ Tm | 来源 Source |
|---|---|---|---|
| SSR5 | F:GATGTAGAGGGAGGTTCTGTATTTG | 56 | 基因组测序开发 |
| R:CGGATCTAGTGAGGGATGGGTATG | Genome sequencing development | ||
| SSR11 | F:TAGTCTCAGCAGAACCACCAAATAC | 56 | |
| R:ATGATACTGATTACGAGAGGCTTAG | |||
| BPPCT008 | F:ATGGTGTGTATGGACATGATGA | 55 | Dirlewanger et al., |
| R:CCTCAACCTAAGACACCTTCACT | |||
| BPPCT040 | F:ATGAGGACGTGTCTGAATGG | 55 | |
| R:AGCCAAACCCCTCTTATACG | |||
| CPSCT021 | F:GCCACTTCGGCTAAAAGAGA | 55 | Mnejja et al., |
| R:TCCATATCTCCTCCTGCTTGA | |||
| M9a | F:GCCGAAACCCTAGGTGAGCG | 58 | Yamamoto et al., |
| R:CAGGATGCTTGCGGTGCTTG | |||
| MA016b | F:TGGCTGGTGGAGACGGAGGA | 56 | |
| R:ATGATACCCAGCCTCCCGGG | |||
| MA007a | F:GTGCATCGTTAGGAACTGCC | 56 | |
| R:GCCCCTGAGATACAACTGCA | |||
| EMPaS11A | F:ACCACTTTGAGGAACTTGGG | 55 | Vaughan & Russell, |
| R:CTGCCTGGAAGAGCAATAAC | |||
| PceGA25 | F:GCAATTCGAGCTGTATTTCAGATG | 58 | Cantini et al., |
| R:CAGTTGGCGGCTATCATGTCTTAC | |||
| PS12A02 | F:GCCACCAATGGTTCTTCC | 56 | Downey & Lezzoni, |
| R:AGCACCAGATGCACCTGA | |||
| pchcms1 | F:GTTACACCTCTGTCACA | 56 | Sosinski et al., |
| R:CTTGGCTGGCATTCCTA | |||
| UDP97-403 | F:CTGGCTTACAACTCGCAAGC | 56 | Cipriani et al., |
| R:CGTCGACCAACTGAGACTCA | |||
| UDP96-005 | F:GTAACGCTCGCTACCACAAA | 56 | |
| R:CCTGCATATCACCACCCAG |
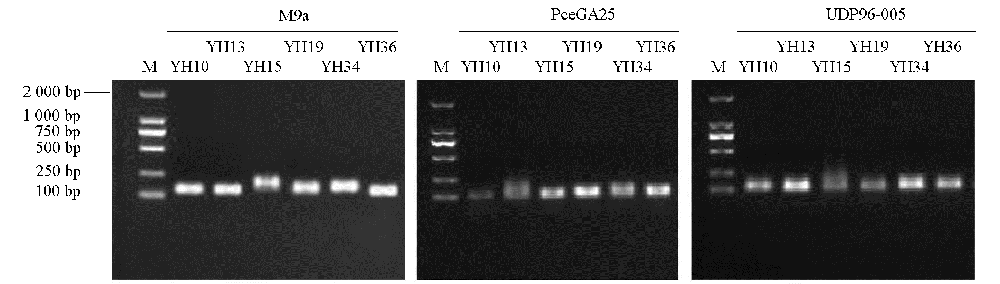
Fig. 1 2% agarose gel electrophoresis of part of the primer YH10:Prunus × subhirtella‘Koshiensis’;YH13:P. spachiana‘Plena Rosea’;YH15:P. serrulata‘Matsumae- hayazaki’;YH19:P. serrulata‘Erecta’;YH34:P. × kanzakura‘Tairyo-zakura’;YH36:P. × kanzakura‘Oh-kanzakuru’. The same below.
| 引物名称 Primer name | 产物大小/bp Production size | 等位基因 Na | 有效等位基因 Ne | 观察杂合度 Ho | 期望杂合度 He | Shannon’s指数 I | 多态信息含量 PIC |
|---|---|---|---|---|---|---|---|
| SSR5 | 209 ~ 331 | 9 | 3.13 | 0.03 | 0.69 | 1.53 | 0.65 |
| SSR11 | 57 ~ 266 | 12 | 5.56 | 0.29 | 0.83 | 2.02 | 0.80 |
| BPPCT008 | 51 ~ 120 | 12 | 5.70 | 0.64 | 0.83 | 1.99 | 0.80 |
| BPPCT040 | 115 ~ 154 | 15 | 9.07 | 0.07 | 0.90 | 2.41 | 0.88 |
| CPSCT021 | 127 ~ 164 | 20 | 8.38 | 0.60 | 0.89 | 2.47 | 0.87 |
| M9a | 141 ~ 167 | 10 | 4.95 | 0.23 | 0.81 | 1.85 | 0.77 |
| EMPAS11A | 63 ~ 129 | 13 | 5.75 | 0.20 | 0.84 | 2.06 | 0.81 |
| PceGA25 | 60 ~ 175 | 7 | 2.25 | 0.50 | 0.56 | 1.16 | 0.52 |
| MA016b | 101 ~ 114 | 8 | 3.97 | 0.04 | 0.76 | 1.71 | 0.72 |
| PS12A02 | 145 ~ 187 | 23 | 12.90 | 0.35 | 0.94 | 2.84 | 0.92 |
| MA007a | 48 ~ 123 | 8 | 2.68 | 0.08 | 0.63 | 1.23 | 0.56 |
| pchcms1 | 142 ~ 175 | 14 | 6.16 | 0.81 | 0.85 | 2.18 | 0.82 |
| UDP97-403 | 102 ~ 144 | 20 | 5.22 | 0.50 | 0.82 | 2.26 | 0.80 |
| UDP96-005 | 100 ~ 193 | 16 | 9.69 | 0.02 | 0.91 | 2.49 | 0.89 |
| 平均Mean | 13.36 | 6.10 | 0.31 | 0.80 | 2.01 | 0.77 |
Table 3 Genetic information of 14 pairs of SSR primers in 42 oriental cultivars of flowering cherry
| 引物名称 Primer name | 产物大小/bp Production size | 等位基因 Na | 有效等位基因 Ne | 观察杂合度 Ho | 期望杂合度 He | Shannon’s指数 I | 多态信息含量 PIC |
|---|---|---|---|---|---|---|---|
| SSR5 | 209 ~ 331 | 9 | 3.13 | 0.03 | 0.69 | 1.53 | 0.65 |
| SSR11 | 57 ~ 266 | 12 | 5.56 | 0.29 | 0.83 | 2.02 | 0.80 |
| BPPCT008 | 51 ~ 120 | 12 | 5.70 | 0.64 | 0.83 | 1.99 | 0.80 |
| BPPCT040 | 115 ~ 154 | 15 | 9.07 | 0.07 | 0.90 | 2.41 | 0.88 |
| CPSCT021 | 127 ~ 164 | 20 | 8.38 | 0.60 | 0.89 | 2.47 | 0.87 |
| M9a | 141 ~ 167 | 10 | 4.95 | 0.23 | 0.81 | 1.85 | 0.77 |
| EMPAS11A | 63 ~ 129 | 13 | 5.75 | 0.20 | 0.84 | 2.06 | 0.81 |
| PceGA25 | 60 ~ 175 | 7 | 2.25 | 0.50 | 0.56 | 1.16 | 0.52 |
| MA016b | 101 ~ 114 | 8 | 3.97 | 0.04 | 0.76 | 1.71 | 0.72 |
| PS12A02 | 145 ~ 187 | 23 | 12.90 | 0.35 | 0.94 | 2.84 | 0.92 |
| MA007a | 48 ~ 123 | 8 | 2.68 | 0.08 | 0.63 | 1.23 | 0.56 |
| pchcms1 | 142 ~ 175 | 14 | 6.16 | 0.81 | 0.85 | 2.18 | 0.82 |
| UDP97-403 | 102 ~ 144 | 20 | 5.22 | 0.50 | 0.82 | 2.26 | 0.80 |
| UDP96-005 | 100 ~ 193 | 16 | 9.69 | 0.02 | 0.91 | 2.49 | 0.89 |
| 平均Mean | 13.36 | 6.10 | 0.31 | 0.80 | 2.01 | 0.77 |
| 引物名称 Primer name | 基因型 Genotype | 可区分品种数量 Cultivar No. which can be distinguished | 区分率/% Identified rate |
|---|---|---|---|
| SSR5 | 9 | 5 | 11.90 |
| SSR11 | 13 | 7 | 16.67 |
| BPPCT008 | 19 | 10 | 23.81 |
| BPPCT040 | 16 | 8 | 19.05 |
| CPSCT021 | 25 | 16 | 38.10 |
| M9a | 12 | 4 | 9.52 |
| EMPAS11A | 16 | 11 | 26.19 |
| PceGA25 | 9 | 5 | 11.90 |
| MA016b | 9 | 4 | 9.52 |
| PS12A02 | 25 | 21 | 50.00 |
| MA007a | 8 | 5 | 11.90 |
| pchcms1 | 19 | 10 | 23.81 |
| UDP97-403 | 22 | 15 | 35.71 |
| UDP96-005 | 17 | 9 | 21.43 |
| 总计(total) | 219 | — | — |
| 均值(Mean) | 15.64 | 9.29 | 22.11 |
Table 4 Amplification genotype of 14 pairs of SSR primers
| 引物名称 Primer name | 基因型 Genotype | 可区分品种数量 Cultivar No. which can be distinguished | 区分率/% Identified rate |
|---|---|---|---|
| SSR5 | 9 | 5 | 11.90 |
| SSR11 | 13 | 7 | 16.67 |
| BPPCT008 | 19 | 10 | 23.81 |
| BPPCT040 | 16 | 8 | 19.05 |
| CPSCT021 | 25 | 16 | 38.10 |
| M9a | 12 | 4 | 9.52 |
| EMPAS11A | 16 | 11 | 26.19 |
| PceGA25 | 9 | 5 | 11.90 |
| MA016b | 9 | 4 | 9.52 |
| PS12A02 | 25 | 21 | 50.00 |
| MA007a | 8 | 5 | 11.90 |
| pchcms1 | 19 | 10 | 23.81 |
| UDP97-403 | 22 | 15 | 35.71 |
| UDP96-005 | 17 | 9 | 21.43 |
| 总计(total) | 219 | — | — |
| 均值(Mean) | 15.64 | 9.29 | 22.11 |
| 品种编号 Number | DNA数字指纹图谱 DNA fingerprinting | 品种编号 Number | DNA数字指纹图谱 |
|---|---|---|---|
| DNA fingerprinting | |||
| YH1 | A53/53-B129/142-C104/116 | YH22 | A52/108-B140/146-C104/109 |
| YH2 | A51/108-B131/142-C104/104 | YH23 | A52/120-B140/146-C104/126 |
| YH3 | A51/51-B130/142-C104/116 | YH24 | A101/109-B146/146-C104/127 |
| YH4 | A52/52-B130/130-C104/104 | YH25 | A108/108-B134/164-C131/144 |
| YH5 | A52/108-B128/141-C104/116 | YH26 | A52/108-B142/142-C104/129 |
| YH6 | A52/108-B129/135-C104/104 | YH27 | A52/101-B134/134-C104/104 |
| YH7 | A51/96-B130/145-C110/110 | YH28 | A51/51-B134/134-C104/116 |
| YH8 | A51/108-B136/141-C102/102 | YH29 | A52/109-B133/146-C104/116 |
| YH9 | A52/108-B130/151-C127/127 | YH30 | A51/51-B134/134-C104/115 |
| YH10 | A52/52-B128/147-C110/110 | YH31 | A51/108-B134/134-C104/115 |
| YH11 | A52/102-B127/127-C106/106 | YH32 | A51/108-B134/134-C103/103 |
| YH12 | A52/104-B130/139-C125/135 | YH33 | A108/108-B134/140-C104/104 |
| YH13 | A96/116-B132/148-C107/107 | YH34 | A51/108-B130/146-C104/104 |
| YH14 | A108/108-B134/146-C127/144 | YH35 | A51/120-B134/148-C110/127 |
| YH15 | A52/100-B146/146-C104/104 | YH36 | A51/109-B130/146-C104/104 |
| YH16 | A52/102-B130/130-C127/127 | YH37 | A51/108-B134/164-C104/104 |
| YH17 | A100/100-B130/130-C112/129 | YH38 | A51/108-B140/140-C104/127 |
| YH18 | A52/120-B133/146-C104/108 | YH39 | A52/109-B132/132-C127/127 |
| YH19 | A52/108-B134/146-C104/127 | YH40 | A102/102-B130/130-C103/130 |
| YH20 | A102/102-B130/130-C113/113 | YH41 | A51/51-B140/146-C112/112 |
| YH21 | A108/108-B140/146-C104/127 | YH42 | A102/102-B132/132-C112/112 |
Table 5 The DNA fingerprint codes of 42 oriental cultivars of flowering cherry
| 品种编号 Number | DNA数字指纹图谱 DNA fingerprinting | 品种编号 Number | DNA数字指纹图谱 |
|---|---|---|---|
| DNA fingerprinting | |||
| YH1 | A53/53-B129/142-C104/116 | YH22 | A52/108-B140/146-C104/109 |
| YH2 | A51/108-B131/142-C104/104 | YH23 | A52/120-B140/146-C104/126 |
| YH3 | A51/51-B130/142-C104/116 | YH24 | A101/109-B146/146-C104/127 |
| YH4 | A52/52-B130/130-C104/104 | YH25 | A108/108-B134/164-C131/144 |
| YH5 | A52/108-B128/141-C104/116 | YH26 | A52/108-B142/142-C104/129 |
| YH6 | A52/108-B129/135-C104/104 | YH27 | A52/101-B134/134-C104/104 |
| YH7 | A51/96-B130/145-C110/110 | YH28 | A51/51-B134/134-C104/116 |
| YH8 | A51/108-B136/141-C102/102 | YH29 | A52/109-B133/146-C104/116 |
| YH9 | A52/108-B130/151-C127/127 | YH30 | A51/51-B134/134-C104/115 |
| YH10 | A52/52-B128/147-C110/110 | YH31 | A51/108-B134/134-C104/115 |
| YH11 | A52/102-B127/127-C106/106 | YH32 | A51/108-B134/134-C103/103 |
| YH12 | A52/104-B130/139-C125/135 | YH33 | A108/108-B134/140-C104/104 |
| YH13 | A96/116-B132/148-C107/107 | YH34 | A51/108-B130/146-C104/104 |
| YH14 | A108/108-B134/146-C127/144 | YH35 | A51/120-B134/148-C110/127 |
| YH15 | A52/100-B146/146-C104/104 | YH36 | A51/109-B130/146-C104/104 |
| YH16 | A52/102-B130/130-C127/127 | YH37 | A51/108-B134/164-C104/104 |
| YH17 | A100/100-B130/130-C112/129 | YH38 | A51/108-B140/140-C104/127 |
| YH18 | A52/120-B133/146-C104/108 | YH39 | A52/109-B132/132-C127/127 |
| YH19 | A52/108-B134/146-C104/127 | YH40 | A102/102-B130/130-C103/130 |
| YH20 | A102/102-B130/130-C113/113 | YH41 | A51/51-B140/146-C112/112 |
| YH21 | A108/108-B140/146-C104/127 | YH42 | A102/102-B132/132-C112/112 |
| [1] |
Cantini C, Iezzoni A F, Lamboy W F. 2001. DNA fingerprinting of tetraploid cherry germplasm using simple sequence repeats. Journal of the American Society for Horticultural Science, 126 (2):205-209.
doi: 10.21273/JASHS.126.2.205 URL |
| [2] |
Cipriani G, Lot G, Huang W G, Huang W G, Marrazzo M T, Peterlunger E, Testolin R. 1999. AC/GT and AG/CT microsatellite repeats in peach [Prunus persica(L.)Batch]:isolation,characterization and cross-species amplification in Prunus. Theoretical and Applied Genetics, 99:65-72.
doi: 10.1007/s001220051209 URL |
| [3] | Cong Rui, Zhang Kai-wen, Yi Xian-gui, Li Meng, Wang Xian-rong. 2020. Construction of DNA fingerprinting of 201 Prunus tatsienensis L. resources based on SSR markers. Molecular Plant Breeding, 18 (20):6776-6784. (in Chinese) |
| 从睿, 张开文, 伊贤贵, 李蒙, 王贤荣. 2020. 基于SSR标记的201份康定樱桃种质资源DNA指纹图谱构建. 分子植物育种, 18 (20):6776-6784. | |
| [4] |
Dirlewanger E, Cosson P, Tavaud M. 2002. Development of microsatellite markers in peach[Prunus persica(L.)Batsch] and their use in genetic diversity analysis in peach and sweet cherry(Prunus avium L.). Theoretical and Applied Genetics, 105 (1):127-138.
pmid: 12582570 |
| [5] |
Downey S L, Lezzoni A F. 2000. Polymorphic DNA markers in black cherry(Prunus serotina)are idenitified using sequences from sweet cherry,peach,and sour cherry. Journal of the American Society for Horticultural Science, 125 (1):76-80.
doi: 10.21273/JASHS.125.1.76 URL |
| [6] |
Fu Tao, Wang Zhi-long, Lin Li, Lin Le-jing. 2015. Study on systematic identification methods of primrose germplasm resources. Acta Horticulturae Sinica, 42 (12):2455-2468. (in Chinese)
doi: 10.16420/j.issn.0513-353x.2015-0468 |
|
付涛, 王志龙, 林立, 林乐静. 2015. 樱属植物种质资源系统鉴定方法的研究. 园艺学报, 42 (12):2455-2468.
doi: 10.16420/j.issn.0513-353x.2015-0468 URL |
|
| [7] |
Gao Tian-xiang, Cai Yu-liang, Feng Ying, Zhao Xiao-jun. 2016. Genetic diversity and genetic structure of 14 natural populations of cherry in China based on SSR markers. Acta Horticulturae Sinica, 43 (6):1148-1156. (in Chinese)
doi: 10.16420/j.issn.0513-353x.2015-0815 |
|
高天翔, 蔡宇良, 冯瑛, 赵晓军. 2016. 中国樱桃14个自然居群遗传多样性和遗传结构的SSR评价. 园艺学报, 43 (6):1148-1156.
doi: 10.16420/j.issn.0513-353x.2015-0815 URL |
|
| [8] |
Gao Yuan, Wang Kun, Wang Da-jiang, Gong Xin, Liu Li-jun, Liu Feng-zhi. 2016. Molecular identification of apple cultivars using TP-M13-SSR markers. Acta Horticulturae Sinica, 43 (1):25-37. (in Chinese)
doi: 10.16420/j.issn.0513-353x.2015-0324 |
|
高源, 王昆, 王大江, 龚欣, 刘立军, 刘凤之. 2016. 利用TP-M13-SSR标记构建苹果栽培品种的分子身份证. 园艺学报, 43 (1):25-37.
doi: 10.16420/j.issn.0513-353x.2015-0324 URL |
|
| [9] |
Guan C F, Zhang P X, Hu C Q, Chachar S, Riaz A, Wang R Z, Yang Y. 2019. Genetic diversity,germplasm identification and population structure of Diospyros kaki Thunb. from different geographic regions in China using SSR markers. Scientia Horticulturae, 251:233-240.
doi: 10.1016/j.scienta.2019.02.062 URL |
| [10] |
Guo L L, Guo D L, Zhao W, Hou X G. 2018. Newly developed SSR markers reveal genetic diversity and geographical clustering in Paeonia suffruticosa based on flower colour. The Journal of Horticultural Science and Biotechnology, 93 (4):416-424.
doi: 10.1080/14620316.2017.1373039 URL |
| [11] | Huang Xing-fa, Yin Yue, Zhao Jian-hua, Yao Jia-yi, Qin Xiao-ya, Wang Shu-fen, Cao You-long, Zhan Xiang-qiang. 2021. Development and genetic diversity analysis of genomic SSR markers in Lycium ruthenicum Murr. Journal of Northwest A & F University(Natural Science), 49 (1):126-135. (in Chinese) |
| 黄兴发, 尹跃, 赵建华, 姚佳依, 秦小雅, 汪淑芬, 曹有龙, 战祥强. 2021. 黑果枸杞基因组SSR标记开发及遗传多样性分析. 西北农林科技大学学报(自然科学版), 49 (1):126-135. | |
| [12] |
Jiang Shuang, Zhang Xueying, An Haishan, Xu Fangjie, Zhang Jiaying. 2021. Development and polymorphism of SSR markers in loquat (Eriobotrya japonica)genome. Acta Horticulturae Sinica, 48 (5):1013-1022. (in Chinese)
doi: 10.16420/j.issn.0513-353x.2020-0497 |
|
蒋爽, 张学英, 安海山, 徐芳杰, 章加应. 2021. 枇杷全基因组SSR标记开发及其多态性研究. 园艺学报, 48 (5):1013-1022.
doi: 10.16420/j.issn.0513-353x.2020-0497 URL |
|
| [13] | Kato S, Matsumoto A, Yoshimura K, Katsuki T, Iwamoto K, Kawahara T, Mukai Y, Tsuda Y, Ishio S, Nakamura K, Moriwaki K, Shiroishi T, Gojobori T, Yoshimaru H. 2014. Origins of Japanese flowering cherry(Prunus subgenus Cerasus)cultivars revealed using nuclear SSR markers. Tree Genetics & Genomes, 10 (3):477-487. |
| [14] |
Kato S, Matsumoto A, Yoshimura K, Katsuki T, Iwamoto K, Tsuda Y, Ishio S, Nakamura K, Moriwaki K, Shiroishi T, Gojobori T, Yoshimaru H. 2012. Clone identification in Japanese flowering cherry(Prunus subgenus Cerasus) cultivars using nuclear SSR markers. Breeding Science, 62 (3):248-255.
doi: 10.1270/jsbbs.62.248 URL |
| [15] |
Khadivi-Khub A, Zamani Z, Fattahi R, Wvnsch A. 2014. Genetic variation in wild Prunus L. subgenus Cerasus germplasm from Iran characterized by nuclear and chloroplast SSR markers. Trees, 28 (2):471-485.
doi: 10.1007/s00468-013-0964-z URL |
| [16] | Liu Shuo, Liu Ning, Zhang Qiuping, Zhang Yuping, Zhang Yujun, Xu Ming, Ma Xiaoxue, Liu Weisheng. 2019. Genetic diversity analysis of apricot germplasm resources in North and Northeast China. Acta Horticulturae Sinica, 46 (6):1045-1056. (in Chinese) |
|
刘硕, 刘宁, 章秋平, 张玉萍, 张玉君, 徐铭, 马小雪, 刘威生. 2019. 中国华北和东北地区杏种质资源遗传多样性分析. 园艺学报, 46 (6):1045-1056.
doi: 10.16420/j.issn.0513-353x.2018-0477 URL |
|
| [17] | Li Hai-bo, Ding Hong-mei, Chen You-wu, Xu Liang, Li Nan, Hu Chuan-jiu. 2017. SSR fingerprint identification of 12 Camellia oleifera varieties. Journal of the China Cereals and Oils Society, 32 (10):171-178. (in Chinese) |
| 李海波, 丁红梅, 陈友吾, 徐梁, 李楠, 胡传久. 2017. 12个油茶品种的SSR特征指纹鉴别. 中国粮油学报, 32 (10):171-178. | |
| [18] |
Ma S N, Han C Y, Zhou J, Hu R C, Jiang X, Wu F F, Tian K, Nie G, Zhang X Q. 2020. Fingerprint identification of white clover cultivars based on SSR molecular markers. Molecular Biology Reports, 47 (11):8513-8521.
doi: 10.1007/s11033-020-05893-7 pmid: 33040266 |
| [19] |
Mnejja M, Garcia-Mas J, Howad W, Badenes M L, Arus P. 2004. Simple-sequence repeat(SSR)markers of Japanese plum (Prunus salicina Lindl.) are highly polymorphic and transferable to peach and almond. Molecular Ecology Notes, 4 (2):163-166.
doi: 10.1111/men.2004.4.issue-2 URL |
| [20] |
Shahi-Gharahlar A, Zamani Z, Fatahi R, Bouzari N. 2011. Estimation of genetic diversity in some Iranian wild Prunus subgenus Cerasus accessions using inter-simple sequence repeat(ISSR)markers. Biochemical Systematics and Ecology, 39 (4-6):826-833.
doi: 10.1016/j.bse.2011.07.018 URL |
| [21] |
Nie Xinghua, Li Yiran, Tian Shoule, Wang Xuefeng, Su Shuchai, Cao Qingqin, Xing Yu, Qin Ling. 2022. Construction of DNA fingerprint map and analysis of genetic diversity for Chinese Chestnut cultivars(lines). Acta Horticulturae Sinica, 49 (11):2313-2324. (in Chinese)
doi: 10.16420/j.issn.0513-353x.2021-0863 URL |
|
聂兴华, 李伊然, 田寿乐, 王雪峰, 苏淑钗, 曹庆芹, 邢宇, 秦岭. 2022. 中国板栗品种(系)DNA 指纹图谱构建及其遗传多样性分析. 园艺学报, 49 (11):2313-2324.
doi: 10.16420/j.issn.0513-353x.2021-0863 |
|
| [22] |
Sosinski B, Gannavarapu M, Hager L D, Beck L E, King G J, Ryder C D, Rajapakse S, Baird W V, Ballard R E, Abbott A G. 2000. Characterization of microsatellite markers in peach [Prunus persica (L.) Batsch]. Theoretical and Applied Genetics, 101 (3):421-428.
doi: 10.1007/s001220051499 URL |
| [23] | Su Qun, Yang Ya-han, Tian Min, Bu Chao-yang, Mao Li-yan, Zhang Jin-zhong, Pan Jie-chun, Lu Jia-shi. 2020. Genetic diversity analysis and DNA fingerprinting construction of Nymphaea germplasm resources. Journal of Tropical Crops, 41 (2):258-266. (in Chinese) |
| 苏群, 杨亚涵, 田敏, 卜朝阳, 毛立彦, 张进忠, 潘介春, 卢家仕. 2020. 睡莲种质资源遗传多样性分析及DNA指纹图谱构建. 热带作物学报, 41 (2):258-266. | |
| [24] | Vaughan S P, Russell K. 2004. Characterization of novel microsatellites and development of multiplex PCR for large-scale population studies in wild cherry,Prunus avium. Molecular Ecology Notes, 3 (4):429-431. |
| [25] | Wang Jue, Wang Yan, Zhang Jing, Chen Tao, Wang Lei, Chen Qing, Tang Haoru, Wang Xiaorong. 2020. InDel marker development of Chinese cherry and evaluation of its universality in Rosaceae trees. Acta Horticulturae Sinica, 47 (1):98-110. (in Chinese) |
|
王珏, 王燕, 张静, 陈涛, 王磊, 陈清, 汤浩茹, 王小蓉. 2020. 中国樱桃InDel标记开发及其在蔷薇科果树中通用性评价. 园艺学报, 47 (1):98-110.
doi: 10.16420/j.issn.0513-353x.2019-0062 URL |
|
| [26] | Wang Lin, Ao Dun, Bao Wen-quan, Zhang Shu-ning, Chen Jun-xing, Li Feng-ming, Meng Fan-qing, Yang Yu-ying, Bai Yu-e. 2021. Identification and fingerprint construction of peach cultivars based on SSR molecular markers. Journal of Central South University of Forestry and Technology, 41 (6):131-138. (in Chinese) |
| 王淋, 敖敦, 包文泉, 张淑宁, 陈俊兴, 李凤鸣, 孟繁庆, 杨钰莹, 白玉娥. 2021. 基于SSR分子标记的桃品种鉴别及指纹图谱构建. 中南林业科技大学学报, 41 (6):131-138. | |
| [27] | Wu Bao-huan, Huang Wen-xin, Shi Wen-ting, Yang Hai-jun, Cui Da-fang. 2018. Quantitative classification of Prunus L. subgenus Cerasus(Mill.)A. Gray in China. Journal of Sun Yatsen University(Natural Science), 57 (1):36-43. (in Chinese) |
| 吴保欢, 黄文鑫, 石文婷, 羊海军, 崔大方. 2018. 中国李属樱亚属Prunus L. subgenus Cerasus(Mill.)A. Gray的数量分类. 中山大学学报(自然科学版), 57 (1):36-43. | |
| [28] | Wu Fan, Liu Xin-hong, Jiang Dong-yue, Li Yin-gang, Yang Shao-zong, Liu Hua-hong, Ni Sui. 2017. Study on improved genomic DNA extraction method of typical subgenus Cerasus. Chinese Journal of Wild Plant Resources, 36 (2):24-27. (in Chinese) |
| 吴帆, 柳新红, 蒋冬月, 李因刚, 杨少宗, 刘华红, 倪穗. 2017. 典型樱亚属植物基因组DNA改良提取方法研究. 中国野生植物资源, 36 (2):24-27. | |
| [29] |
Yamamoto T, Mochida K, Imai T, Shi Y Z, Ogiwara I, Hayashi T. 2002. Microsatellite markers in peach [Prunus persica(L.)Batsch] derived from an enriched genomic and cDNA libraries. Molecular Ecology Notes, 2:298-301.
doi: 10.1046/j.1471-8286.2002.00242.x URL |
| [30] | Yan Jia-wen, Li Jian-hui, Bai Wen-fu, Yu Lin, Nie Dong-ling, Xiong Ying, Li Bai-hai, Wu Si-zheng. 2022. Genotyping and genetic diversity analysis of 47 cherry germplasm resources. Molecular Plant Breeding, 20 (3):1026-1036. |
| 严佳文, 李建挥, 柏文富, 禹霖, 聂东伶, 熊颖, 李柏海, 吴思政. 2022. 47份樱花种质资源的基因分型和遗传多样性分析. 分子植物育种, 20 (3):1026-1036. | |
| [31] | Zhao Qing-jie, Li Hai-bo, Qu Yan, Wang Qing-hua, Guo Jia, Xu Liang. 2016. Molecular identification of 20 cherry cultivars based on SCAR marker. Journal of Nanjing Forestry University(Natural Science Edition), 40 (5):34-40. (in Chinese) |
| 赵庆杰, 李海波, 屈燕, 王青华, 郭佳, 徐梁. 2016. 基于SCAR标记的20个樱花品种的分子鉴别. 南京林业大学学报(自然科学版), 40 (5):34-40. | |
| [32] | Zhou Cheng-cheng, Yang De-ming, Li Shi-kun, Wan Jia-yi, Rong Jun-dong, Zheng Yu-shan. 2020. ISSR analysis of genetic relationships of 18 cherry species. Forest & Environment, 40 (1):46-53. (in Chinese) |
| 周成城, 杨德明, 李士坤, 万佳艺, 荣俊冬, 郑郁善. 2020. 18份樱属材料亲缘关系的ISSR分析. 森林与环境报, 40 (1):46-53. | |
| [33] | Zhou Yu-bo, Zhu Peng, Gong Wei, Wang Jing-yan, Yan Si-yu, Wu Kai-zhi. 2018. Construction of SSR fingerprinting and genetic diversity analysis of walnut cultivars in Sichuan Province. Acta Botanica Boreali-Occidentalia Sinica, 38 (7):1254-1261. (in Chinese) |
| 周于波, 朱鹏, 龚伟, 王景燕, 闫思宇, 吴开志. 2018. 四川核桃良种SSR指纹图谱构建及遗传多样性分析. 西北植物学报, 38 (7):1254-1261. | |
| [34] | Zong Yu, Wang Yue, Zhu You-yin, Shao Xu, Li Yong-qiang, Guo Wei-dong. 2016. Development and identification of SSR molecular markers based on Chinese cherry transcriptome. Acta Horticulturae Sinica, 43 (8):1566-1576. (in Chinese) |
|
宗宇, 王月, 朱友银, 邵姁, 李永强, 郭卫东. 2016. 基于中国樱桃转录组的SSR分子标记开发与鉴定. 园艺学报, 43 (8):1566-1576.
doi: 10.16420/j.issn.0513-353x.2016-0005 |
| [1] | SUN Lei, YAN Ailing, ZHANG Guojun, WANG Huiling, WANG Xiaoyue, REN Jiancheng, XU Haiying. A New Table Grape Cultivar‘Ruidu Mozhi’ [J]. Acta Horticulturae Sinica, 2023, 50(3): 685-686. |
| [2] | ZHENG Xiaoming, LIU Xiaoxi, LUO Jianning, WU Haibin, LI Junxing, ZHAO Gangjun, LUO Shaobo, GONG Hao. A Bitter Gourd Cultivar‘Qiaolü Pearl Bitter Gourd’ [J]. Acta Horticulturae Sinica, 2023, 50(3): 687-688. |
| [3] | ZHANG Huijun, YUAN Yushu, ZHANG Yan, WU Qibo, DAI Zuyun. A New Melon Cultivar‘Huaitian 1’ [J]. Acta Horticulturae Sinica, 2023, 50(3): 689-690. |
| [4] | XU Shenping, YUAN Xiuyun, LIANG Fang, ZHANG Yan, JIANG Suhua, NIU Suyan, CUI Bo. A New Phalaenopsis Cultivar‘Zhengshi Zhaoxia’ [J]. Acta Horticulturae Sinica, 2023, 50(3): 691-692. |
| [5] | TAO Dayan, GUAN Shikai, SONG Qian, ZHOU Jinye, LUO Shuming, YAN Haixia. A New Primulina Cultivar‘Return With Honor’ [J]. Acta Horticulturae Sinica, 2023, 50(3): 693-694. |
| [6] | WANG Rui, HONG Wenjuan, LUO Hua, ZHAO Lina, CHEN Ying, WANG Jun. Construction of SSR Fingerprints of Pomegranate Cultivars and Male Parent Identification of Hybrids [J]. Acta Horticulturae Sinica, 2023, 50(2): 265-278. |
| [7] | MENG Qiufeng, WANG Bingliang, WANG Yuhong, HUANG Yunping, WANG Jie. A New Tuber Mustard Cultivar‘Yongzha 805’ [J]. Acta Horticulturae Sinica, 2023, 50(2): 451-452. |
| [8] | YU Hailong, ZHANG Meiyan, LI Qiaozhen, ZHANG Lujun, SHANG Xiaodong, TAN Qi, ZHOU Feng, LI Yu. A New Panus giganteus Cultivar‘Shenxun 1’ [J]. Acta Horticulturae Sinica, 2023, 50(2): 453-454. |
| [9] | WANG Xiqing, JIA Yunhe, YAN Wen, FU Yongkai, YOU Haibo, LI Dongyan, ZHAO Jingchao. A New Watermelon Cultivar‘Longsheng Jiali’with High Resistance to Fusarium Wilt [J]. Acta Horticulturae Sinica, 2023, 50(2): 455-456. |
| [10] | ZHU Wei, CAO Jinjin, CHEN Xi, ZHANG Wei, SUN Rongze, ZHU Shaocai, ZHAO Jiageng, CUI Yaqi, WANG Yuxuan, YU Xiaonan. New Herbaceous Peony Cultivars‘Fluttering Pink’‘Fairy’s Cheek’ ‘Blushing Smile’and‘Tiny Lotus’ [J]. Acta Horticulturae Sinica, 2023, 50(2): 457-458. |
| [11] | LIU Yiping, NI Menghui, WU Fangfang, LIU Hongli, HE Dan, KONG Dezheng. Association Analysis of Organ Traits with SSR Markers in Lotus(Nelumbo nucifera) [J]. Acta Horticulturae Sinica, 2023, 50(1): 103-115. |
| [12] | XING Zhudong, LÜ Futang, GUO Shangjing, ZHANG Yanyi. A New Peach Cultivar‘Liaoda Hongjin’ [J]. Acta Horticulturae Sinica, 2023, 50(1): 225-226. |
| [13] | LI Zhiliang, LI Zhenxing, LI Tao, SUN Baojuan, LI Ying, XU Xiaowan, WANG Hengming, HENG Zhou, GONG Chao. A New Eggplant Cultivar‘Xinfeng 3’ [J]. Acta Horticulturae Sinica, 2023, 50(1): 227-228. |
| [14] | CAI Peng, FANG Chao, LI Yuejian, LIU Duchen, LIU Xiaojun, LIANG Genyun. A New Eggplant Cultivar‘Tianjiao’ [J]. Acta Horticulturae Sinica, 2023, 50(1): 229-230. |
| [15] | ZHANG Hongmei, CUI Jiawei, YU Jizhu, DING Xiaotao, HE Lizhong, JIN Haijun. A New Bitter Gourd Cultivar‘Qingcui 1’ [J]. Acta Horticulturae Sinica, 2023, 50(1): 231-232. |
| Viewed | ||||||
|
Full text |
|
|||||
|
Abstract |
|
|||||
Copyright © 2012 Acta Horticulturae Sinica 京ICP备10030308号-2 国际联网备案号 11010802023439
Tel: 010-82109523 E-Mail: yuanyixuebao@126.com
Support by: Beijing Magtech Co.Ltd