
Acta Horticulturae Sinica ›› 2023, Vol. 50 ›› Issue (4): 815-830.doi: 10.16420/j.issn.0513-353x.2021-1238
• Research Papers • Previous Articles Next Articles
WANG Tonghuan1, WU Yuxin1, WU Yiyuan1, LI Xinxin1, LIU Mengyang1, YANG Lianlian1,2, LI Jiapeng1,2, ZHANG Zhongshan1,2, CAO Fang1, ZHONG Xueting1,2, WANG Zhanqi1,2,*( )
)
Received:2022-08-13
Revised:2022-12-11
Online:2023-04-25
Published:2023-04-27
Contact:
*(E-mail:zhqwang@zjhu.edu.cn)
CLC Number:
WANG Tonghuan, WU Yuxin, WU Yiyuan, LI Xinxin, LIU Mengyang, YANG Lianlian, LI Jiapeng, ZHANG Zhongshan, CAO Fang, ZHONG Xueting, WANG Zhanqi. Genome-wide Identification and Expression Analysis of the GRAS Gene Family in Response to Cold Stress in Chrysanthemum nankingense[J]. Acta Horticulturae Sinica, 2023, 50(4): 815-830.
Add to citation manager EndNote|Ris|BibTeX
URL: https://www.ahs.ac.cn/EN/10.16420/j.issn.0513-353x.2021-1238
| 基因名 Gene name | 基因ID Gene ID | 方向 Strand | 开放阅读框/bp ORF | 外显子数 Number of exons | 蛋白质/aa Protein | 等电点 pI | 分子量/kD MW | ||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| CnGRAS1 | CHR00004937-RA | 反向 Reverse | 1 401 | 6 | 466 | 4.6 | 52.8 | ||||||
| CnGRAS2 | CHR00004939-RA | 反向 Reverse | 948 | 6 | 315 | 4.5 | 35.8 | ||||||
| CnGRAS3 | CHR00004941-RA | 反向 Reverse | 1 305 | 4 | 434 | 4.6 | 48.9 | ||||||
| CnGRAS4 | CHR00004943-RA | 反向 Reverse | 1 005 | 5 | 334 | 5.0 | 37.9 | ||||||
| CnGRAS5 | CHR00010605-RA | 反向 Reverse | 1 464 | 3 | 487 | 5.7 | 54.8 | ||||||
| CnGRAS6 | CHR00010606-RA | 反向 Reverse | 1 551 | 6 | 516 | 5.5 | 57.9 | ||||||
| CnGRAS7 | CHR00030831-RA | 反向 Reverse | 1 317 | 6 | 438 | 5.3 | 49.3 | ||||||
| CnGRAS8 | CHR00009270-RA | 反向 Reverse | 882 | 3 | 293 | 5.2 | 33.7 | ||||||
| CnGRAS9 | CHR00003961-RA | 反向 Reverse | 3 216 | 3 | 1 071 | 5.5 | 121.1 | ||||||
| CnGRAS10 | CHR00055273-RA | 反向 Reverse | 762 | 4 | 253 | 5.7 | 28.5 | ||||||
| CnGRAS11 | CHR00032218-RA | 反向 Reverse | 1 407 | 2 | 468 | 6.1 | 52.6 | ||||||
| CnGRAS12 | CHR00031862-RA | 正向 Forward | 624 | 3 | 207 | 8.9 | 22.7 | ||||||
| CnGRAS13 | CHR00060013-RA | 正向 Forward | 2 187 | 7 | 728 | 5.1 | 81.5 | ||||||
| CnGRAS14 | CHR00049603-RA | 反向 Reverse | 1 965 | 3 | 654 | 5.5 | 71.8 | ||||||
| CnGRAS15 | CHR00044507-RA | 正向 Forward | 1 587 | 2 | 528 | 5.9 | 59.0 | ||||||
| CnGRAS16 | CHR00088841-RA | 正向 Forward | 684 | 2 | 227 | 6.4 | 25.3 | ||||||
| CnGRAS17 | CHR00057450-RA | 反向 Reverse | 1 809 | 4 | 602 | 6.4 | 66.5 | ||||||
| CnGRAS18 | CHR00032070-RA | 正向 Forward | 1 605 | 4 | 534 | 5.8 | 61.0 | ||||||
| CnGRAS19 | CHR00001839-RA | 正向 Forward | 1 656 | 1 | 551 | 6.2 | 62.0 | ||||||
| CnGRAS20 | CHR00003428-RA | 正向 Forward | 1 560 | 3 | 519 | 4.9 | 58.8 | ||||||
| CnGRAS21 | CHR00080196-RA | 反向 Reverse | 1 863 | 4 | 620 | 5.8 | 69.5 | ||||||
| CnGRAS22 | CHR00056344-RA | 反向 Reverse | 1 509 | 5 | 502 | 5.2 | 56.5 | ||||||
| CnGRAS23 | CHR00043261-RA | 反向 Reverse | 1 617 | 6 | 538 | 5.4 | 61.0 | ||||||
| CnGRAS24 | CHR00007360-RA | 正向 Forward | 1 647 | 6 | 548 | 5.7 | 61.3 | ||||||
| CnGRAS25 | CHR00084234-RA | 反向 Reverse | 1 971 | 5 | 656 | 6.3 | 75.5 | ||||||
| CnGRAS26 | CHR00026458-RA | 正向 Forward | 1 587 | 7 | 528 | 5.1 | 59.7 | ||||||
| CnGRAS27 | CHR00012285-RA | 正向 Forward | 1 392 | 5 | 463 | 5.8 | 53.1 | ||||||
| CnGRAS28 | CHR00012287-RA | 正向 Forward | 1 389 | 2 | 462 | 6.2 | 53.3 | ||||||
| CnGRAS29 | CHR00001229-RA | 反向 Reverse | 1 659 | 5 | 552 | 6.4 | 62.5 | ||||||
| CnGRAS30 | CHR00090421-RA | 正向 Forward | 1 326 | 5 | 441 | 5.8 | 50.0 | ||||||
| CnGRAS31 | CHR00090422-RA | 正向 Forward | 1 770 | 12 | 589 | 8.2 | 67.0 | ||||||
| CnGRAS32 | CHR00006827-RA | 反向 Reverse | 1 989 | 4 | 662 | 6.5 | 73.5 | ||||||
| CnGRAS33 | CHR00080708-RA | 反向 Reverse | 1 845 | 7 | 614 | 5.7 | 68.2 | ||||||
| CnGRAS34 | CHR00009185-RA | 正向 Forward | 1 761 | 2 | 586 | 7.2 | 66.5 | ||||||
| CnGRAS35 | CHR00014432-RA | 反向 Reverse | 1 284 | 4 | 427 | 7.3 | 48.9 | ||||||
| CnGRAS36 | CHR00027166-RA | 正向 Forward | 1 995 | 5 | 664 | 5.8 | 73.2 | ||||||
| CnGRAS37 | CHR00030490-RA | 反向 Reverse | 1 221 | 3 | 406 | 6.1 | 46.3 | ||||||
| CnGRAS38 | CHR00089224-RA | 反向 Reverse | 1 389 | 8 | 462 | 5.8 | 51.9 | ||||||
| CnGRAS39 | CHR00032024-RA | 反向 Reverse | 840 | 2 | 279 | 7.2 | 31.8 | ||||||
| CnGRAS40 | CHR00059102-RA | 反向 Reverse | 1 176 | 5 | 391 | 5.1 | 43.2 | ||||||
| CnGRAS41 | CHR00020653-RA | 反向 Reverse | 660 | 7 | 219 | 9.1 | 25.6 | ||||||
| CnGRAS42 | CHR00023444-RA | 正向 Forward | 1 461 | 4 | 486 | 5.6 | 54.6 | ||||||
| CnGRAS43 | CHR00047978-RA | 正向 Forward | 2 112 | 5 | 703 | 7.6 | 79.9 | ||||||
| CnGRAS44 | CHR00037746-RA | 反向(reverse) | 1 350 | 8 | 449 | 5.7 | 50.2 | ||||||
| CnGRAS45 | CHR00050945-RA | 正向 Forward | 1 464 | 3 | 487 | 5.3 | 54.8 | ||||||
| CnGRAS46 | CHR00043205-RA | 反向(reverse) | 1 656 | 6 | 551 | 5.3 | 62.2 | ||||||
| CnGRAS47 | CHR00020406-RA | 正向 Forward | 1 047 | 3 | 348 | 7.7 | 38.9 | ||||||
| CnGRAS48 | CHR00034025-RA | 反向 Reverse | 1 608 | 3 | 535 | 5.2 | 59.6 | ||||||
| CnGRAS49 | CHR00074287-RA | 正向 Forward | 1 500 | 2 | 499 | 5.4 | 55.5 | ||||||
| CnGRAS50 | CHR00022194-RA | 正向 Forward | 1 269 | 3 | 422 | 5.7 | 48.2 | ||||||
| CnGRAS51 | CHR00013495-RA | 正向 Forward | 771 | 4 | 256 | 5.9 | 28.4 | ||||||
| CnGRAS52 | CHR00043827-RA | 反向 Reverse | 1 221 | 4 | 406 | 5.9 | 46.8 | ||||||
| CnGRAS53 | CHR00043828-RA | 反向 Reverse | 2 172 | 5 | 723 | 5.4 | 82.3 | ||||||
| CnGRAS54 | CHR00082830-RA | 反向 Reverse | 1 779 | 4 | 592 | 5.7 | 101.7 | ||||||
| CnGRAS55 | CHR00031262-RA | 正向 Forward | 1 413 | 3 | 470 | 8.0 | 53.8 | ||||||
| CnGRAS56 | CHR00066643-RA | 正向 Forward | 2 175 | 7 | 724 | 5.3 | 82.6 | ||||||
| CnGRAS57 | CHR00036461-RA | 正向 Forward | 1 665 | 5 | 554 | 6.3 | 61.9 | ||||||
| CnGRAS58 | CHR00037682-RA | 反向 Reverse | 1 548 | 3 | 515 | 6.0 | 56.4 | ||||||
| CnGRAS59 | CHR00081984-RA | 正向 Forward | 963 | 2 | 320 | 8.6 | 37.1 | ||||||
| CnGRAS60 | CHR00076473-RA | 反向 Reverse | 1 209 | 2 | 402 | 5.4 | 44.2 | ||||||
| CnGRAS61 | CHR00077126-RA | 正向 Forward | 1 326 | 6 | 441 | 5.2 | 49.7 | ||||||
| CnGRAS62 | CHR00021855-RA | 正向 Forward | 1 356 | 4 | 451 | 5.9 | 50.9 | ||||||
| CnGRAS63 | CHR00039693-RA | 反向 Reverse | 1 584 | 4 | 527 | 5.6 | 58.6 | ||||||
| CnGRAS64 | CHR00022872-RA | 正向 Forward | 1 023 | 4 | 340 | 5.0 | 37.8 | ||||||
| CnGRAS65 | CHR00086408-RA | 反向 Reverse | 1 323 | 3 | 440 | 5.1 | 49.7 | ||||||
| CnGRAS66 | CHR00081508-RA | 正向 Forward | 1 632 | 4 | 543 | 6.3 | 61.2 | ||||||
| CnGRAS67 | CHR00087940-RA | 反向 Reverse | 1 878 | 5 | 625 | 5.4 | 69.6 | ||||||
| CnGRAS68 | CHR00058033-RA | 反向 Reverse | 1 488 | 3 | 495 | 7.5 | 54.8 | ||||||
| CnGRAS69 | CHR00087040-RA | 反向 Reverse | 1 032 | 4 | 343 | 6.0 | 39.3 | ||||||
| CnGRAS70 | CHR00087041-RA | 反向 Reverse | 1 914 | 10 | 637 | 5.4 | 71.9 | ||||||
| CnGRAS71 | CHR00087042-RA | 反向 Reverse | 1 446 | 3 | 481 | 5.9 | 54.8 | ||||||
| CnGRAS72 | CHR00087043-RA | 反向 Reverse | 2 208 | 4 | 735 | 4.9 | 83.8 | ||||||
| CnGRAS73 | CHR00087044-RA | 反向 Reverse | 969 | 8 | 322 | 8.2 | 36.3 | ||||||
| CnGRAS74 | CHR00087600-RA | 反向 Reverse | 1 371 | 4 | 456 | 6.2 | 51.3 | ||||||
| CnGRAS75 | CHR00091555-RA | 反向 Reverse | 1 257 | 4 | 418 | 5.3 | 46.5 | ||||||
| CnGRAS76 | CHR00065020-RA | 反向 Reverse | 1 581 | 5 | 526 | 4.8 | 57.4 | ||||||
| CnGRAS77 | CHR00065743-RA | 正向 Forward | 1 515 | 6 | 504 | 6.1 | 57.8 | ||||||
| CnGRAS78 | CHR00069367-RA | 正向 Forward | 1 488 | 6 | 495 | 5.3 | 54.2 | ||||||
| CnGRAS79 | CHR00093834-RA | 反向 Reverse | 705 | 3 | 234 | 4.6 | 26.1 | ||||||
| CnGRAS80 | CHR00074223-RA | 正向 Forward | 735 | 5 | 244 | 9.1 | 28.6 | ||||||
Table 1 Structural,physical and chemical features of GRAS gene family in Chrysanthemum nankingense
| 基因名 Gene name | 基因ID Gene ID | 方向 Strand | 开放阅读框/bp ORF | 外显子数 Number of exons | 蛋白质/aa Protein | 等电点 pI | 分子量/kD MW | ||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| CnGRAS1 | CHR00004937-RA | 反向 Reverse | 1 401 | 6 | 466 | 4.6 | 52.8 | ||||||
| CnGRAS2 | CHR00004939-RA | 反向 Reverse | 948 | 6 | 315 | 4.5 | 35.8 | ||||||
| CnGRAS3 | CHR00004941-RA | 反向 Reverse | 1 305 | 4 | 434 | 4.6 | 48.9 | ||||||
| CnGRAS4 | CHR00004943-RA | 反向 Reverse | 1 005 | 5 | 334 | 5.0 | 37.9 | ||||||
| CnGRAS5 | CHR00010605-RA | 反向 Reverse | 1 464 | 3 | 487 | 5.7 | 54.8 | ||||||
| CnGRAS6 | CHR00010606-RA | 反向 Reverse | 1 551 | 6 | 516 | 5.5 | 57.9 | ||||||
| CnGRAS7 | CHR00030831-RA | 反向 Reverse | 1 317 | 6 | 438 | 5.3 | 49.3 | ||||||
| CnGRAS8 | CHR00009270-RA | 反向 Reverse | 882 | 3 | 293 | 5.2 | 33.7 | ||||||
| CnGRAS9 | CHR00003961-RA | 反向 Reverse | 3 216 | 3 | 1 071 | 5.5 | 121.1 | ||||||
| CnGRAS10 | CHR00055273-RA | 反向 Reverse | 762 | 4 | 253 | 5.7 | 28.5 | ||||||
| CnGRAS11 | CHR00032218-RA | 反向 Reverse | 1 407 | 2 | 468 | 6.1 | 52.6 | ||||||
| CnGRAS12 | CHR00031862-RA | 正向 Forward | 624 | 3 | 207 | 8.9 | 22.7 | ||||||
| CnGRAS13 | CHR00060013-RA | 正向 Forward | 2 187 | 7 | 728 | 5.1 | 81.5 | ||||||
| CnGRAS14 | CHR00049603-RA | 反向 Reverse | 1 965 | 3 | 654 | 5.5 | 71.8 | ||||||
| CnGRAS15 | CHR00044507-RA | 正向 Forward | 1 587 | 2 | 528 | 5.9 | 59.0 | ||||||
| CnGRAS16 | CHR00088841-RA | 正向 Forward | 684 | 2 | 227 | 6.4 | 25.3 | ||||||
| CnGRAS17 | CHR00057450-RA | 反向 Reverse | 1 809 | 4 | 602 | 6.4 | 66.5 | ||||||
| CnGRAS18 | CHR00032070-RA | 正向 Forward | 1 605 | 4 | 534 | 5.8 | 61.0 | ||||||
| CnGRAS19 | CHR00001839-RA | 正向 Forward | 1 656 | 1 | 551 | 6.2 | 62.0 | ||||||
| CnGRAS20 | CHR00003428-RA | 正向 Forward | 1 560 | 3 | 519 | 4.9 | 58.8 | ||||||
| CnGRAS21 | CHR00080196-RA | 反向 Reverse | 1 863 | 4 | 620 | 5.8 | 69.5 | ||||||
| CnGRAS22 | CHR00056344-RA | 反向 Reverse | 1 509 | 5 | 502 | 5.2 | 56.5 | ||||||
| CnGRAS23 | CHR00043261-RA | 反向 Reverse | 1 617 | 6 | 538 | 5.4 | 61.0 | ||||||
| CnGRAS24 | CHR00007360-RA | 正向 Forward | 1 647 | 6 | 548 | 5.7 | 61.3 | ||||||
| CnGRAS25 | CHR00084234-RA | 反向 Reverse | 1 971 | 5 | 656 | 6.3 | 75.5 | ||||||
| CnGRAS26 | CHR00026458-RA | 正向 Forward | 1 587 | 7 | 528 | 5.1 | 59.7 | ||||||
| CnGRAS27 | CHR00012285-RA | 正向 Forward | 1 392 | 5 | 463 | 5.8 | 53.1 | ||||||
| CnGRAS28 | CHR00012287-RA | 正向 Forward | 1 389 | 2 | 462 | 6.2 | 53.3 | ||||||
| CnGRAS29 | CHR00001229-RA | 反向 Reverse | 1 659 | 5 | 552 | 6.4 | 62.5 | ||||||
| CnGRAS30 | CHR00090421-RA | 正向 Forward | 1 326 | 5 | 441 | 5.8 | 50.0 | ||||||
| CnGRAS31 | CHR00090422-RA | 正向 Forward | 1 770 | 12 | 589 | 8.2 | 67.0 | ||||||
| CnGRAS32 | CHR00006827-RA | 反向 Reverse | 1 989 | 4 | 662 | 6.5 | 73.5 | ||||||
| CnGRAS33 | CHR00080708-RA | 反向 Reverse | 1 845 | 7 | 614 | 5.7 | 68.2 | ||||||
| CnGRAS34 | CHR00009185-RA | 正向 Forward | 1 761 | 2 | 586 | 7.2 | 66.5 | ||||||
| CnGRAS35 | CHR00014432-RA | 反向 Reverse | 1 284 | 4 | 427 | 7.3 | 48.9 | ||||||
| CnGRAS36 | CHR00027166-RA | 正向 Forward | 1 995 | 5 | 664 | 5.8 | 73.2 | ||||||
| CnGRAS37 | CHR00030490-RA | 反向 Reverse | 1 221 | 3 | 406 | 6.1 | 46.3 | ||||||
| CnGRAS38 | CHR00089224-RA | 反向 Reverse | 1 389 | 8 | 462 | 5.8 | 51.9 | ||||||
| CnGRAS39 | CHR00032024-RA | 反向 Reverse | 840 | 2 | 279 | 7.2 | 31.8 | ||||||
| CnGRAS40 | CHR00059102-RA | 反向 Reverse | 1 176 | 5 | 391 | 5.1 | 43.2 | ||||||
| CnGRAS41 | CHR00020653-RA | 反向 Reverse | 660 | 7 | 219 | 9.1 | 25.6 | ||||||
| CnGRAS42 | CHR00023444-RA | 正向 Forward | 1 461 | 4 | 486 | 5.6 | 54.6 | ||||||
| CnGRAS43 | CHR00047978-RA | 正向 Forward | 2 112 | 5 | 703 | 7.6 | 79.9 | ||||||
| CnGRAS44 | CHR00037746-RA | 反向(reverse) | 1 350 | 8 | 449 | 5.7 | 50.2 | ||||||
| CnGRAS45 | CHR00050945-RA | 正向 Forward | 1 464 | 3 | 487 | 5.3 | 54.8 | ||||||
| CnGRAS46 | CHR00043205-RA | 反向(reverse) | 1 656 | 6 | 551 | 5.3 | 62.2 | ||||||
| CnGRAS47 | CHR00020406-RA | 正向 Forward | 1 047 | 3 | 348 | 7.7 | 38.9 | ||||||
| CnGRAS48 | CHR00034025-RA | 反向 Reverse | 1 608 | 3 | 535 | 5.2 | 59.6 | ||||||
| CnGRAS49 | CHR00074287-RA | 正向 Forward | 1 500 | 2 | 499 | 5.4 | 55.5 | ||||||
| CnGRAS50 | CHR00022194-RA | 正向 Forward | 1 269 | 3 | 422 | 5.7 | 48.2 | ||||||
| CnGRAS51 | CHR00013495-RA | 正向 Forward | 771 | 4 | 256 | 5.9 | 28.4 | ||||||
| CnGRAS52 | CHR00043827-RA | 反向 Reverse | 1 221 | 4 | 406 | 5.9 | 46.8 | ||||||
| CnGRAS53 | CHR00043828-RA | 反向 Reverse | 2 172 | 5 | 723 | 5.4 | 82.3 | ||||||
| CnGRAS54 | CHR00082830-RA | 反向 Reverse | 1 779 | 4 | 592 | 5.7 | 101.7 | ||||||
| CnGRAS55 | CHR00031262-RA | 正向 Forward | 1 413 | 3 | 470 | 8.0 | 53.8 | ||||||
| CnGRAS56 | CHR00066643-RA | 正向 Forward | 2 175 | 7 | 724 | 5.3 | 82.6 | ||||||
| CnGRAS57 | CHR00036461-RA | 正向 Forward | 1 665 | 5 | 554 | 6.3 | 61.9 | ||||||
| CnGRAS58 | CHR00037682-RA | 反向 Reverse | 1 548 | 3 | 515 | 6.0 | 56.4 | ||||||
| CnGRAS59 | CHR00081984-RA | 正向 Forward | 963 | 2 | 320 | 8.6 | 37.1 | ||||||
| CnGRAS60 | CHR00076473-RA | 反向 Reverse | 1 209 | 2 | 402 | 5.4 | 44.2 | ||||||
| CnGRAS61 | CHR00077126-RA | 正向 Forward | 1 326 | 6 | 441 | 5.2 | 49.7 | ||||||
| CnGRAS62 | CHR00021855-RA | 正向 Forward | 1 356 | 4 | 451 | 5.9 | 50.9 | ||||||
| CnGRAS63 | CHR00039693-RA | 反向 Reverse | 1 584 | 4 | 527 | 5.6 | 58.6 | ||||||
| CnGRAS64 | CHR00022872-RA | 正向 Forward | 1 023 | 4 | 340 | 5.0 | 37.8 | ||||||
| CnGRAS65 | CHR00086408-RA | 反向 Reverse | 1 323 | 3 | 440 | 5.1 | 49.7 | ||||||
| CnGRAS66 | CHR00081508-RA | 正向 Forward | 1 632 | 4 | 543 | 6.3 | 61.2 | ||||||
| CnGRAS67 | CHR00087940-RA | 反向 Reverse | 1 878 | 5 | 625 | 5.4 | 69.6 | ||||||
| CnGRAS68 | CHR00058033-RA | 反向 Reverse | 1 488 | 3 | 495 | 7.5 | 54.8 | ||||||
| CnGRAS69 | CHR00087040-RA | 反向 Reverse | 1 032 | 4 | 343 | 6.0 | 39.3 | ||||||
| CnGRAS70 | CHR00087041-RA | 反向 Reverse | 1 914 | 10 | 637 | 5.4 | 71.9 | ||||||
| CnGRAS71 | CHR00087042-RA | 反向 Reverse | 1 446 | 3 | 481 | 5.9 | 54.8 | ||||||
| CnGRAS72 | CHR00087043-RA | 反向 Reverse | 2 208 | 4 | 735 | 4.9 | 83.8 | ||||||
| CnGRAS73 | CHR00087044-RA | 反向 Reverse | 969 | 8 | 322 | 8.2 | 36.3 | ||||||
| CnGRAS74 | CHR00087600-RA | 反向 Reverse | 1 371 | 4 | 456 | 6.2 | 51.3 | ||||||
| CnGRAS75 | CHR00091555-RA | 反向 Reverse | 1 257 | 4 | 418 | 5.3 | 46.5 | ||||||
| CnGRAS76 | CHR00065020-RA | 反向 Reverse | 1 581 | 5 | 526 | 4.8 | 57.4 | ||||||
| CnGRAS77 | CHR00065743-RA | 正向 Forward | 1 515 | 6 | 504 | 6.1 | 57.8 | ||||||
| CnGRAS78 | CHR00069367-RA | 正向 Forward | 1 488 | 6 | 495 | 5.3 | 54.2 | ||||||
| CnGRAS79 | CHR00093834-RA | 反向 Reverse | 705 | 3 | 234 | 4.6 | 26.1 | ||||||
| CnGRAS80 | CHR00074223-RA | 正向 Forward | 735 | 5 | 244 | 9.1 | 28.6 | ||||||
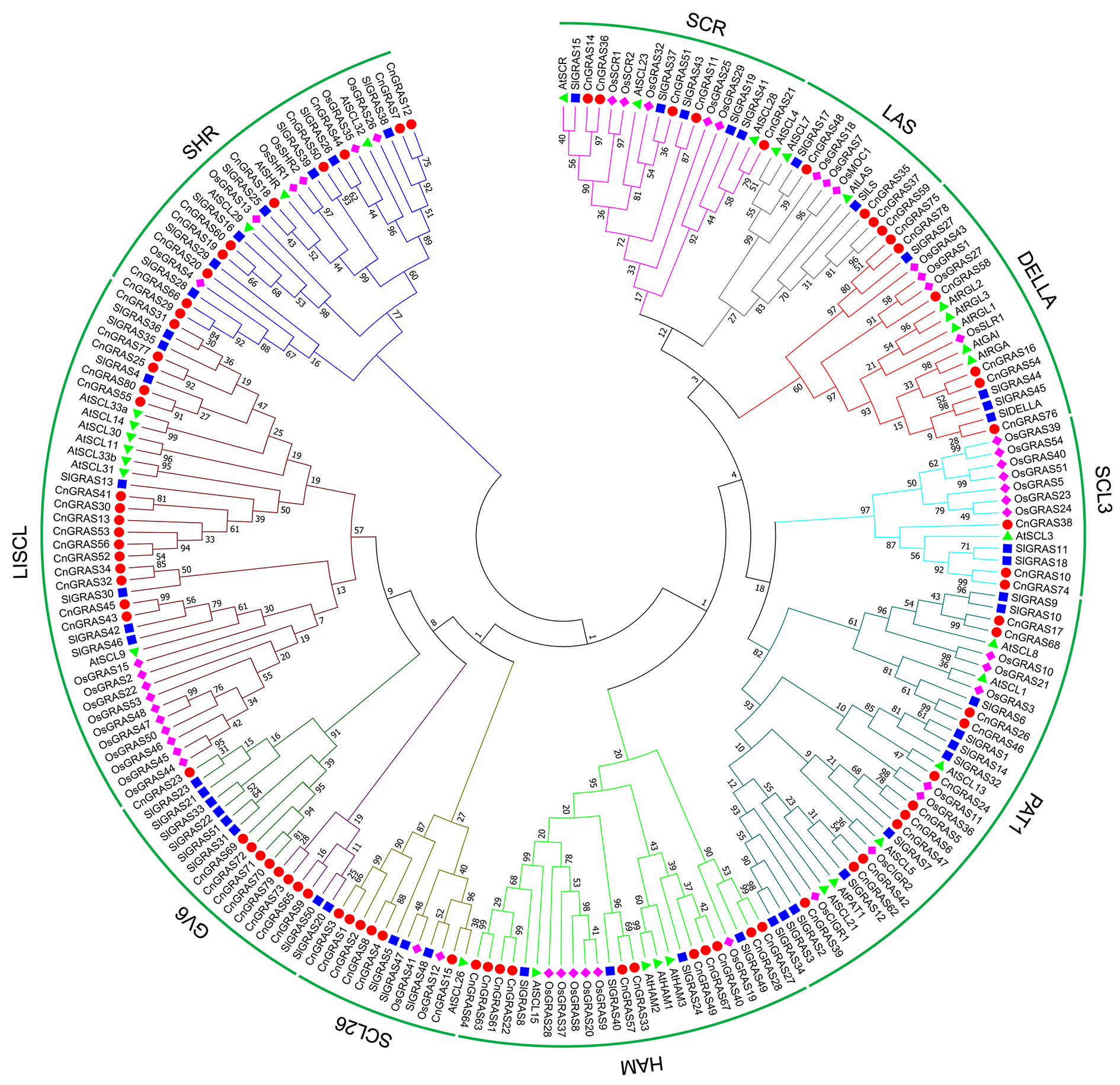
Fig. 1 Phylogenetic analysis of GRAS gene family in Chrysanthemum nankingense(Cn),Arabidopsis thaliana(At),Solanum lycopersicum(Sl)and Oryza sativa(Os)
| 基因对 Gene pair | 非同义替换率(Ka) Non-synonymous substitution rate | 同义替换率(Ks) Synonymous substitution rate | Ka/Ks | 复制时间/Myr Duplication date | 纯化选择 Purifying selection | ||
|---|---|---|---|---|---|---|---|
| CnGRAS2 | CnGRAS3 | 0.038 | 0.087 | 0.438 | 2.9 | Yes | |
| CnGRAS5 | CnGRAS6 | 0.151 | 0.555 | 0.272 | 18.5 | Yes | |
| CnGRAS27 | CnGRAS28 | 0.024 | 0.080 | 0.305 | 2.7 | Yes | |
| CnGRAS30 | CnGRAS31 | 0.470 | 2.249 | 0.209 | 75.0 | Yes | |
| CnGRAS52 | CnGRAS53 | 0.142 | 0.415 | 0.344 | 13.8 | Yes | |
| CnGRAS69 | CnGRAS70 | 0.275 | 0.821 | 0.335 | 27.4 | Yes | |
| CnGRAS70 | CnGRAS71 | 0.116 | 0.150 | 0.769 | 5.0 | Yes | |
| CnGRAS71 | CnGRAS72 | 0.145 | 0.271 | 0.534 | 9.0 | Yes | |
| CnGRAS72 | CnGRAS73 | 0.630 | 2.365 | 0.266 | 78.8 | Yes | |
Table 2 Divergence between duplicated GRAS gene pairs in Chrysanthemum nankingense
| 基因对 Gene pair | 非同义替换率(Ka) Non-synonymous substitution rate | 同义替换率(Ks) Synonymous substitution rate | Ka/Ks | 复制时间/Myr Duplication date | 纯化选择 Purifying selection | ||
|---|---|---|---|---|---|---|---|
| CnGRAS2 | CnGRAS3 | 0.038 | 0.087 | 0.438 | 2.9 | Yes | |
| CnGRAS5 | CnGRAS6 | 0.151 | 0.555 | 0.272 | 18.5 | Yes | |
| CnGRAS27 | CnGRAS28 | 0.024 | 0.080 | 0.305 | 2.7 | Yes | |
| CnGRAS30 | CnGRAS31 | 0.470 | 2.249 | 0.209 | 75.0 | Yes | |
| CnGRAS52 | CnGRAS53 | 0.142 | 0.415 | 0.344 | 13.8 | Yes | |
| CnGRAS69 | CnGRAS70 | 0.275 | 0.821 | 0.335 | 27.4 | Yes | |
| CnGRAS70 | CnGRAS71 | 0.116 | 0.150 | 0.769 | 5.0 | Yes | |
| CnGRAS71 | CnGRAS72 | 0.145 | 0.271 | 0.534 | 9.0 | Yes | |
| CnGRAS72 | CnGRAS73 | 0.630 | 2.365 | 0.266 | 78.8 | Yes | |
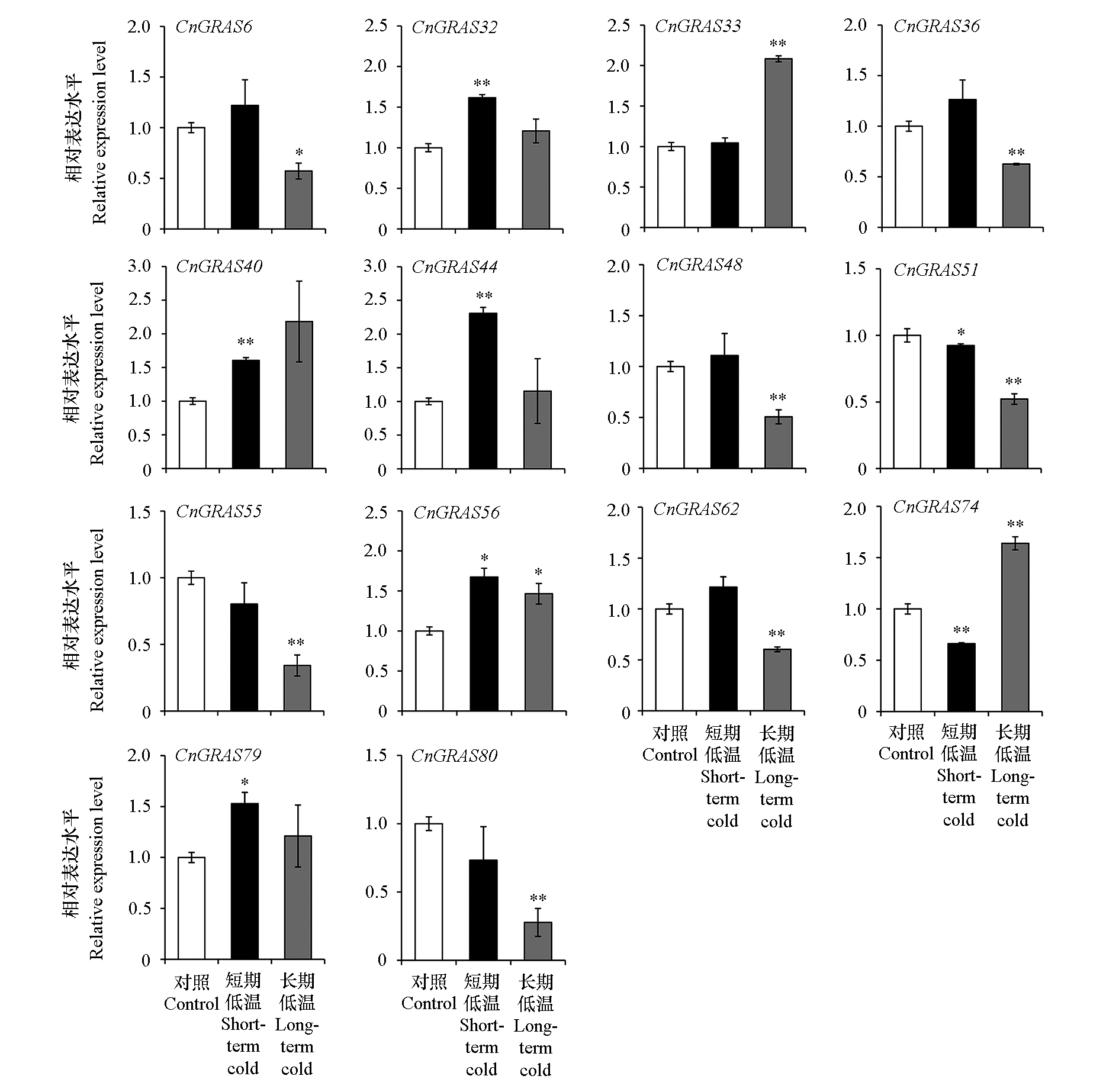
Fig. 7 Expression of GRAS gene family in Chrysanthemum nankingense under short-term cold or long-term cold(STC or LTC)stress Student’s t-test,*P < 0.05,**P < 0.01,ns,not significant.
| [1] |
Bailey T L, Johnson J, Grant C E, Noble W S. 2015. The MEME suite. Nucleic Acids Research, 43 (W1):W39-W49.
doi: 10.1093/nar/gkv416 URL |
| [2] | Blum M, Chang H Y, Chuguransky S, Grego T, Kandasaamy S, Mitchell A, Nuka G, Paysan-Lafosse T, Qureshi M, Raj S, Richardson L, Salazar G A, Williams L, Bork P, Bridge A, Gough J, Haft D H, Letunic I, Marchler-Bauer A, Mi H, Natale D A, Necci M, Orengo C A, Pandurangan A P, Rivoire C, Sigrist C J A, Sillitoe I, Thanki N, Thomas P D, Tosatto S C E, Wu C H, Bateman A, Finn R D. 2021. The InterPro protein families and domains database:20 years on. Nucleic Acids Research, 49 (D1):D344-D354. |
| [3] |
Chen C, Chen H, Zhang Y, Thomas H R, Frank M H, He Y, Xia R. 2020. TBtools:an integrative toolkit developed for interactive analyses of big biological data. Molecular Plant, 13 (8):1194-1202.
doi: 10.1016/j.molp.2020.06.009 URL |
| [4] |
Chen H, Li H, Lu X, Chen L, Liu J, Wu H. 2019. Identification and expression analysis of GRAS transcription factors to elucidate candidate genes related to stolons,fruit ripening and abiotic stresses in woodland strawberry(Fragaria vesca). International Journal of Molecular Sciences, 20 (18):4593.
doi: 10.3390/ijms20184593 URL |
| [5] |
Grimplet J, Agudelo-Romero P, Teixeira R T, Martinez-Zapater J M, Fortes A M. 2016. Structural and functional analysis of the GRAS gene family in grapevine indicates a role of GRAS proteins in the control of development and stress responses. Frontiers in Plant Science, 7:353.
doi: 10.3389/fpls.2016.00353 pmid: 27065316 |
| [6] |
Hu B, Jin J, Guo A Y, Zhang H, Luo J, Gao G. 2015. GSDS 2.0:an upgraded gene feature visualization server. Bioinformatics, 31 (8):1296-1297.
doi: 10.1093/bioinformatics/btu817 URL |
| [7] |
Huang W, Hu B, Liu J, Zhou Y, Liu S. 2020. Identification and characterization of tonoplast sugar transporter(TST)gene family in cucumber. Horticultural Plant Journal, 6 (3),145-157.
doi: 10.1016/j.hpj.2020.03.005 URL |
| [8] |
Huerta-Cepas J, Forslund K, Coelho L P, Szklarczyk D, Jensen L J, von Mering C, Bork P. 2017. Fast genome-wide functional annotation through orthology assignment by eggNOG-Mapper. Molecular Biology and Evolution, 34 (8):2115-2122.
doi: 10.1093/molbev/msx148 pmid: 28460117 |
| [9] |
Jin J F, Wang Z Q, He Q Y, Wang J Y, Li P F, Xu J M, Zheng S J, Fan W, Yang J L. 2020. Genome-wide identification and expression analysis of the NAC transcription factor family in tomato(Solanum lycopersicum)during aluminum stress. BMC Genomics, 21 (1):288.
doi: 10.1186/s12864-020-6689-7 |
| [10] |
Koch M A, Haubold B, Mitchell-Olds T. 2000. Comparative evolutionary analysis of chalcone synthase and alcohol dehydrogenase loci in Arabidopsis,Arabis,and related genera(Brassicaceae). Molecular Biology and Evolution, 17 (10):1483-1498.
doi: 10.1093/oxfordjournals.molbev.a026248 pmid: 11018155 |
| [11] |
Kovaka S, Zimin A V, Pertea G M, Razaghi R, Salzberg S L, Pertea M. 2019. Transcriptome assembly from long-read RNA-seq alignments with StringTie2. Genome Biology, 20 (1):278.
doi: 10.1186/s13059-019-1910-1 pmid: 31842956 |
| [12] |
Kumar S, Stecher G, Tamura K. 2016. MEGA7:Molecular evolutionary genetics analysis version 7.0 for bigger datasets. Molecular Biology and Evolution, 33 (7):1870-1874.
doi: 10.1093/molbev/msw054 URL |
| [13] |
Landi S, Capasso G, Esposito S. 2021. Different G6PDH isoforms show specific roles in acclimation to cold stress at various growth stages of barley(Hordeum vulgare)and Arabidopsis thaliana. Plant Physiology and Biochemistry, 169:190-202.
doi: 10.1016/j.plaphy.2021.11.017 URL |
| [14] |
Lee M H, Kim B, Song S K, Heo J O, Yu N I, Lee S A, Kim M, Kim D G, Sohn S O, Lim C E, Chang K S, Lee M M, Lim J. 2008. Large-scale analysis of the GRAS gene family in Arabidopsis thaliana. Plant Molecular Biology, 67 (6):659-670.
doi: 10.1007/s11103-008-9345-1 URL |
| [15] | Lescot M. 2002. PlantCARE,a database of plant cis-acting regulatory elements and a portal to tools for in silico analysis of promoter sequences. Nucleic Acids Research, 30 (1):325-327. |
| [16] |
Li M, Sun B, Xie F, Gong R, Luo Y, Zhang F, Yan Z, Tang H. 2019. Identification of the GRAS gene family in the Brassica juncea genome provides insight into its role in stem swelling in stem mustard. PeerJ, 7:e6682.
doi: 10.7717/peerj.6682 URL |
| [17] |
Li Y, Liang J, Zeng X, Guo H, Luo Y, Kear P, Zhang S, Zhu G. 2021. Genome-wide analysis of MYB gene family in potato provides insights into tissue-specific regulation of anthocyanin biosynthesis. Horticultural Plant Journal, 7 (2):129-141.
doi: 10.1016/j.hpj.2020.12.001 URL |
| [18] |
Liu B, Sun Y, Xue J, Jia X, Li R. 2018. Genome-wide characterization and expression analysis of GRAS gene family in pepper(Capsicum annuum L.). PeerJ, 6:e4796.
doi: 10.7717/peerj.4796 URL |
| [19] |
Liu M, Huang L, Ma Z, Sun W, Wu Q, Tang Z, Bu T, Li C, Chen H. 2019. Genome-wide identification,expression analysis and functional study of the GRAS gene family in Tartary buckwheat(Fagopyrum tataricum). BMC Plant Biology, 19 (1):342.
doi: 10.1186/s12870-019-1951-3 |
| [20] |
Liu X, Widmer A. 2014. Genome-wide comparative analysis of the GRAS gene family in Populus,Arabidopsis and rice. Plant Molecular Biology Reporter, 32 (6):1129-1145.
doi: 10.1007/s11105-014-0721-5 URL |
| [21] |
Liu Y, Shi Y, Zhu N, Zhong S, Bouzayen M, Li Z. 2020. SlGRAS 4 mediates a novel regulatory pathway promoting chilling tolerance in tomato. Plant Biotechnology Journal, 18 (7):1620-1633.
doi: 10.1111/pbi.v18.7 URL |
| [22] |
Lu X, Liu W, Xiang C, Li X, Wang Q, Wang T, Liu Z, Zhang J, Gao L, Zhang W. 2020. Genome-wide characterization of GRAS family and their potential roles in cold tolerance of cucumber(Cucumis sativus L.). International Journal of Molecular Sciences, 21 (11):3857.
doi: 10.3390/ijms21113857 URL |
| [23] | Mistry J, Chuguransky S, Williams L, Qureshi M, Salazar G A, Sonnhammer E L L, Tosatto S C E, Paladin L, Raj S, Richardson L J, Finn R D, Bateman A. 2021. Pfam:The protein families database in 2021. Nucleic Acids Research, 49 (D1):D412-D419. |
| [24] |
Mistry J, Finn R D, Eddy S R, Bateman A, Punta M. 2013. Challenges in homology search:HMMER3 and convergent evolution of coiled-coil regions. Nucleic Acids Research, 41 (12):e121.
doi: 10.1093/nar/gkt263 URL |
| [25] |
Niu Y, Zhao T, Xu X, Li J. 2017. Genome-wide identification and characterization of GRAS transcription factors in tomato(Solanum lycopersicum). PeerJ, 5(11):e3955.
doi: 10.7717/peerj.3955 URL |
| [26] |
Pei Q, Yu T, Wu T, Yang Q, Gong K, Zhou R, Cui C, Yu Y, Zhao W, Kang X, Cao R, Song X. 2021. Comprehensive identification and analyses of the Hsf gene family in the whole-genome of three Apiaceae species. Horticultural Plant Journal, 7 (5),457-468.
doi: 10.1016/j.hpj.2020.08.005 URL |
| [27] |
Ren L, Sun J, Chen S, Gao J, Dong B, Liu Y, Xia X, Wang Y, Liao Y, Teng N, Fang W, Guan Z, Chen F, Jiang J. 2014. A transcriptomic analysis of Chrysanthemum nankingense provides insights into the basis of low temperature tolerance. BMC Genomics, 15 (1):844.
doi: 10.1186/1471-2164-15-844 |
| [28] |
Rozas J, Ferrer-Mata A, Sánchez-DelBarrio J C, Guirao-Rico S, Librado P, Ramos-Onsins S E, Sánchez-Gracia A. 2017. DnaSP 6:DNA sequence polymorphism analysis of large data sets. Molecular Biology and Evolution, 34 (12):3299-3302.
doi: 10.1093/molbev/msx248 URL |
| [29] |
Shan Z, Luo X, Wu M, Wei L, Fan Z, Zhu Y. 2020. Genome-wide identification and expression of GRAS gene family members in cassava. BMC Plant Biology, 20 (1):46.
doi: 10.1186/s12870-020-2242-8 |
| [30] |
Shang Q M, Li L, Dong C J. 2012. Multiple tandem duplication of the phenylalanine ammonia-lyase genes in Cucumis sativus L.. Planta, 236 (4):1093-1105.
doi: 10.1007/s00425-012-1659-1 URL |
| [31] |
Shannon P, Markiel A, Ozier O, Baliga N S, Wang J T, Ramage D, Amin N, Schwikowski B, Ideker T. 2003. Cytoscape:a software environment for integrated models of biomolecular interaction networks. Genome Research, 13 (11):2498-2504.
doi: 10.1101/gr.1239303 pmid: 14597658 |
| [32] |
Sidhu N S, Pruthi G, Singh S, Bishnoi R, Singla D. 2020. Genome-wide identification and analysis of GRAS transcription factors in the bottle gourd genome. Scientific Reports, 10 (1):14338.
doi: 10.1038/s41598-020-71240-2 pmid: 32868844 |
| [33] |
Song C, Liu Y, Song A, Dong G, Zhao H, Sun W, Ramakrishnan S, Wang Y, Wang S, Li T, Niu Y, Jiang J, Dong B, Xia Y, Chen S, Hu Z, Chen F, Chen S. 2018. The Chrysanthemum nankingense genome provides insights into the evolution and diversification of chrysanthemum flowers and medicinal traits. Molecular Plant, 11 (12):1482-1491.
doi: S1674-2052(18)30308-3 pmid: 30342096 |
| [34] |
Szklarczyk D, Gable A L, Lyon D, Junge A, Wyder S, Huerta-Cepas J, Simonovic M, Doncheva N T, Morris J H, Bork P, Jensen L J, Mering C V. 2019. STRING v11:protein-protein association networks with increased coverage,supporting functional discovery in genome-wide experimental datasets. Nucleic Acids Research, 47 (D1):D607-D613.
doi: 10.1093/nar/gky1131 |
| [35] |
Tang Mingjia, Xu Jin, Lin Rui, Song Jianing, Yu Jingquan, Zhou Yanhong. 2022. Advances in physiological and molecular mechanism of tomato responses to light and temperature stress. Acta Horticulturae Sinica, 49 (10):2174-2188. (in Chinese)
doi: 10.16420/j.issn.0513-353x.2022-0600 URL |
|
唐明佳, 徐进, 林锐, 宋珈凝, 喻景权, 周艳虹. 2022. 番茄响应光温逆境的生理分子机制研究进展. 园艺学报, 49 (10):2174-2188.
doi: 10.16420/j.issn.0513-353x.2022-0600 URL |
|
| [36] |
To V T, Shi Q, Zhang Y, Shi J, Shen C, Zhang D, Cai W. 2020. Genome-wide analysis of the GRAS gene family in barley(Hordeum vulgare L.). Genes, 11 (5):553.
doi: 10.3390/genes11050553 URL |
| [37] |
Upadhyay R K, Mattoo A K. 2018. Genome-wide identification of tomato(Solanum lycopersicum L.)lipoxygenases coupled with expression profiles during plant development and in response to methyl-jasmonate and wounding. Journal of Plant Physiology, 231:318-328.
doi: S0176-1617(18)30562-5 pmid: 30368230 |
| [38] |
Wang Y, Lü J, Chen D, Zhang J, Qi K, Cheng R, Zhang H, Zhang S. 2018a. Genome-wide identification,evolution,and expression analysis of the KT/HAK/KUP family in pear. Genome, 61 (10):755-765.
doi: 10.1139/gen-2017-0254 URL |
| [39] |
Wang Y, Tang H, Debarry J D, Tan X, Li J, Wang X, Lee T H, Jin H, Marler B, Guo H, Kissinger J C, Paterson A H. 2012. MCScanX:a toolkit for detection and evolutionary analysis of gene synteny and collinearity. Nucleic Acids Research, 40 (7):e49.
doi: 10.1093/nar/gkr1293 URL |
| [40] |
Wang Y X, Liu Z W, Wu Z J, Li H, Wang W L, Cui X, Zhuang J. 2018b. Genome-wide identification and expression analysis of GRAS family transcription factors in tea plant(Camellia sinensis). Scientific Reports, 8 (1):3949.
doi: 10.1038/s41598-018-22275-z |
| [41] |
Wang Z, Li S, Jian S, Ye F, Wang T, Gong L, Li X. 2021a. Low temperature tolerance is impaired by polystyrene nanoplastics accumulated in cells of barley(Hordeum vulgare L.)plants. Journal of Hazardous Materials, 426:127826.
doi: 10.1016/j.jhazmat.2021.127826 URL |
| [42] |
Wang Z, Wong D C J, Wang Y, Xu G, Ren C, Liu Y, Kuang Y, Fan P, Li S, Xin H, Liang Z. 2021b. GRAS-domain transcription factor PAT 1 regulates jasmonic acid biosynthesis in grape cold stress response. Plant Physiology, 186 (3):1660-1678.
doi: 10.1093/plphys/kiab142 URL |
| [43] |
Won S Y, Jung J A, Kim J S. 2021. Genome-wide analysis of the MADS-Box gene family in chrysanthemum. Computational Biology and Chemistry, 90:107424.
doi: 10.1016/j.compbiolchem.2020.107424 URL |
| [44] | Yang M, Derbyshire M K, Yamashita R A, Marchler-Bauer A. 2020. NCBI’s conserved domain database and tools for protein domain analysis. Current Protocols in Bioinformatics, 69 (1):e90. |
| [45] |
Yuan Y, Fang L, Karungo S K, Zhang L, Gao Y, Li S, Xin H. 2016. Overexpression of VaPAT1,a GRAS transcription factor from Vitis amurensis,confers abiotic stress tolerance in Arabidopsis. Plant Cell Reports, 35 (3):655-666.
doi: 10.1007/s00299-015-1910-x URL |
| [46] |
Zhang H, Liu X, Wang X, Sun M, Song R, Mao P, Jia S. 2021. Genome-wide identification of GRAS gene family and their responses to abiotic stress in Medicago sativa. International Journal of Molecular Sciences, 22 (14):7729.
doi: 10.3390/ijms22147729 URL |
| [47] | Zhang Z L, Ogawa M, Fleet C M, Zentella R, Hu J, Heo J O, Lim J, Kamiya Y, Yamaguchi S, Sun T P. 2011. Scarecrow-like 3 promotes gibberellin signaling by antagonizing master growth repressor DELLA in Arabidopsis. Proceedings of the National Academy of Sciences of the United States of American, 108 (5):2160-2165. |
| [1] | ZHENG Jiarui, YANG Xiaoyan, YE Jiabao, LIAO Yongling, XU Feng. Advances in the Functional Studies of MYC2 Transcription Factor in Plants [J]. Acta Horticulturae Sinica, 2023, 50(4): 896-908. |
| [2] | LIU Yunuo, CAO Ya, WANG Shuai, DU Meixia, ZHENG Lin, CHEN Shanchun, ZOU Xiuping. Expression Analysis of CsMYB41 and CsMYB63 Genes in Response to Citrus Canker [J]. Acta Horticulturae Sinica, 2023, 50(3): 495-507. |
| [3] | YU Qinhan, LI Junduo, CUI Ying, WANG Jiahui, ZHENG Qiaoling, XU Weirong. Screening and Identification of Interacting Protein of VaMYB4a from Vitis amurensis [J]. Acta Horticulturae Sinica, 2023, 50(3): 508-522. |
| [4] | YE Zimao, SHEN Wanxia, LIU Mengyu, WANG Tong, ZHANG Xiaonan, YU Xin, LIU Xiaofeng, ZHAO Xiaochun. Effect of R2R3-MYB Transcription Factor CitMYB21 on Flavonoids Biosynthesis in Citrus [J]. Acta Horticulturae Sinica, 2023, 50(2): 250-264. |
| [5] | SONG Yanhong, CHEN Yaduo, ZHANG Xiaoyu, SONG Pan, LIU Lifeng, LI Gang, ZHAO Xia, ZHOU Houcheng. The Transcription Factor FvbHLH130 Activates Flowering in Fragaria vesca [J]. Acta Horticulturae Sinica, 2023, 50(2): 295-306. |
| [6] | HAN Rui, ZHONG Xionghui, CHEN Denghui, CUI Jian, YUE Xiangqing, XIE Jianming, KANG Jungen. Cloning and Functional Analysis of BobHLH34 Gene in Cabbage that Interacts with XopR from Xanthomonas [J]. Acta Horticulturae Sinica, 2023, 50(2): 319-330. |
| [7] | TIAN Mingkang, XU Zhixiang, LIU Xiuqun, SUI Shunzhao, LI Mingyang, LI Zhineng. Identification of the AP2 Subfamily Transcription Factors in Chimonanthus praecox and the Functional Study of CpAP2-L11 [J]. Acta Horticulturae Sinica, 2023, 50(2): 382-396. |
| [8] | YU Tingting, LI Huan, NING Yuansheng, SONG Jianfei, PENG Lulin, JIA Junqi, ZHANG Weiwei, YANG Hongqiang. Genome-wide Identification of GRAS Gene Family in Apple and Expression Analysis of Its Response to Auxin [J]. Acta Horticulturae Sinica, 2023, 50(2): 397-409. |
| [9] | LIN Haijiao, LIANG Yuchen, LI Ling, MA Jun, ZHANG Lu, LAN Zhenying, YUAN Zening. Exploration and Regulation Network Analysis of CBF Pathway Related Cold Tolerance Genes in Lavandula angustifolia [J]. Acta Horticulturae Sinica, 2023, 50(1): 131-144. |
| [10] | ZHAO Xueyan, WANG Qi, WANG Li, WANG Fangyuan, WANG Qing, LI Yan. Comparative Transcriptome Analysis of Differential Expression in Different Tissues of Corydalis yanhusuo [J]. Acta Horticulturae Sinica, 2023, 50(1): 177-187. |
| [11] | JIA Xin, ZENG Zhen, CHEN Yue, FENG Hui, LÜ Yingmin, ZHAO Shiwei. Cloning and Expression Analysis of RcDREB2A Gene in Rosa chinensis‘Old Blush’ [J]. Acta Horticulturae Sinica, 2022, 49(9): 1945-1956. |
| [12] | GAO Yanlong, WU Yuxia, ZHANG Zhongxing, WANG Shuangcheng, ZHANG Rui, ZHANG De, WANG Yanxiu. Bioinformatics Analysis of Apple ELO Gene Family and Its Expression Analysis Under Low Temperature Stress [J]. Acta Horticulturae Sinica, 2022, 49(8): 1621-1636. |
| [13] | QIU Ziwen, LIU Linmin, LIN Yongsheng, LIN Xiaojie, LI Yongyu, WU Shaohua, YANG Chao. Cloning and Functional Analysis of the MbEGS Gene from Melaleuca bracteata [J]. Acta Horticulturae Sinica, 2022, 49(8): 1747-1760. |
| [14] | XU Haifeng, WANG Zhongtang, CHEN Xin, LIU Zhiguo, WANG Lihu, LIU Ping, LIU Mengjun, ZHANG Qiong. The Analyses of Target Metabolomics in Flavonoid and Its Potential MYB Regulation Factors During Coloring Period of Winter Jujube [J]. Acta Horticulturae Sinica, 2022, 49(8): 1761-1771. |
| [15] | ZHENG Lin, WANG Shuai, LIU Yunuo, DU Meixia, PENG Aihong, HE Yongrui, CHEN Shanchun, ZOU Xiuping. Gene Cloning and Expression Analysis of NAC Gene in Citrus in Response to Huanglongbing [J]. Acta Horticulturae Sinica, 2022, 49(7): 1441-1457. |
| Viewed | ||||||
|
Full text |
|
|||||
|
Abstract |
|
|||||
Copyright © 2012 Acta Horticulturae Sinica 京ICP备10030308号-2 国际联网备案号 11010802023439
Tel: 010-82109523 E-Mail: yuanyixuebao@126.com
Support by: Beijing Magtech Co.Ltd