
园艺学报 ›› 2022, Vol. 49 ›› Issue (7): 1441-1457.doi: 10.16420/j.issn.0513-353x.2021-0553
郑林, 王帅, 刘语诺, 杜美霞, 彭爱红, 何永睿, 陈善春( ), 邹修平(
), 邹修平( )
)
收稿日期:2022-02-08
修回日期:2022-04-25
出版日期:2022-07-25
发布日期:2022-07-29
通讯作者:
陈善春,邹修平
E-mail:chenshanchun@cric.cn;zouxiuping@cric.cn
基金资助:
ZHENG Lin, WANG Shuai, LIU Yunuo, DU Meixia, PENG Aihong, HE Yongrui, CHEN Shanchun( ), ZOU Xiuping(
), ZOU Xiuping( )
)
Received:2022-02-08
Revised:2022-04-25
Online:2022-07-25
Published:2022-07-29
Contact:
CHEN Shanchun,ZOU Xiuping
E-mail:chenshanchun@cric.cn;zouxiuping@cric.cn
摘要:
挖掘柑橘抗黄龙病(Huanglongbing,HLB)基因是抗病育种的基础和关键。以感染黄龙病菌亚洲种Candidatus Liberibacter asiaticus(CLas)早期(2个月)锦橙根和叶片中脉比较转录组数据为基础,筛选到9个响应柑橘黄龙病侵染诱导的NAC基因,从中选3个差异表达水平较高的基因克隆,分别命名为CsNAC21/22、CsNAC68和CsNAC78。生物信息分析表明3个基因均符合NAC基因家族的特征;烟草亚细胞定位结果表明,CsNAC68定位在细胞核,CsNAC21/22和CsNAC78定位在细胞核和细胞质。实时荧光定量PCR(qRT-PCR)分析表明,3个候选基因在易感HLB的锦橙、耐病的马蜂柑和高耐病的九里香的组织和病原菌诱导表达特征呈现明显差异。以健康植株为对照,CsNAC68和CsNAC78主要在锦橙的根中响应CLas感染,显著上调表达,CsNAC68在九里香叶肉和马蜂柑根中响应CLas感染,显著上调表达;CsNAC21/22主要在锦橙根和马蜂柑叶肉中显著下调表达。以‘锦橙’叶片为试材通过qRT-PCR分析候选基因响应SA、JA、ABA、ETH诱导的表达特征,结果表明,3个基因可能参与ABA的信号转导途径,CsNAC68可能参与SA和JA的信号转导途径,而CsNAC78可能参与ETH的信号转导途径。
中图分类号:
郑林, 王帅, 刘语诺, 杜美霞, 彭爱红, 何永睿, 陈善春, 邹修平. 柑橘响应黄龙病菌侵染的NAC基因的克隆及表达分析[J]. 园艺学报, 2022, 49(7): 1441-1457.
ZHENG Lin, WANG Shuai, LIU Yunuo, DU Meixia, PENG Aihong, HE Yongrui, CHEN Shanchun, ZOU Xiuping. Gene Cloning and Expression Analysis of NAC Gene in Citrus in Response to Huanglongbing[J]. Acta Horticulturae Sinica, 2022, 49(7): 1441-1457.
| 用途Amplification | 名称Name | 序列(5′-3′)Sequence |
|---|---|---|
| 黄龙病常规 PCR 检测 Huanglongbing routine PCR test | OI1 OI2C | F:GCGCGTATGCAATACGAGCGGCA |
| R:GCCTCGCGACTTCGCAACCCAT | ||
| 基因克隆Gene cloning | CsNAC21/22 | F:CGCGGATCCATGGGGCTAAGAGATATTGGAGCTAC |
| R:CCGGAATTCTCATAGGAAAACCATGCTGTTATCCAAT | ||
| CsNAC68 | F:TCCCCCGGGATGCCCAAAGAGGACCGAG | |
| R:GCGTCGACTCAATGAAGATACGCCATT | ||
| CsNAC78 | F:CGCGGATCCATGGACTATAATTGCCTGGGATACAG | |
| R:CCGGAATTCCTATATGAAGAAAAACAAGTTGTATAGAACAGAAG | ||
| 亚细胞定位Subcellular localization | CsNAC21/22 | F:GGGGTACCATGGGGCTAAGAGATATTGG |
| R:ACGCGTCGACTAGGAAAACCATGCTGTTAT | ||
| CsNAC68 | F:GGGGTACCATGCCCAAAGAGGACCGAGT | |
| R:ACGCGTCGACATGAAGATACGCCATTTCCC | ||
| CsNAC78 | F:GGGGTACCATGGACTATAATTGCCTGGG | |
| R:ACGCGTCGACTATGAAGAAAAACAAGTTGT |
表1 普通PCR引物
Table 1 The sequences of common PCR primer pairs
| 用途Amplification | 名称Name | 序列(5′-3′)Sequence |
|---|---|---|
| 黄龙病常规 PCR 检测 Huanglongbing routine PCR test | OI1 OI2C | F:GCGCGTATGCAATACGAGCGGCA |
| R:GCCTCGCGACTTCGCAACCCAT | ||
| 基因克隆Gene cloning | CsNAC21/22 | F:CGCGGATCCATGGGGCTAAGAGATATTGGAGCTAC |
| R:CCGGAATTCTCATAGGAAAACCATGCTGTTATCCAAT | ||
| CsNAC68 | F:TCCCCCGGGATGCCCAAAGAGGACCGAG | |
| R:GCGTCGACTCAATGAAGATACGCCATT | ||
| CsNAC78 | F:CGCGGATCCATGGACTATAATTGCCTGGGATACAG | |
| R:CCGGAATTCCTATATGAAGAAAAACAAGTTGTATAGAACAGAAG | ||
| 亚细胞定位Subcellular localization | CsNAC21/22 | F:GGGGTACCATGGGGCTAAGAGATATTGG |
| R:ACGCGTCGACTAGGAAAACCATGCTGTTAT | ||
| CsNAC68 | F:GGGGTACCATGCCCAAAGAGGACCGAGT | |
| R:ACGCGTCGACATGAAGATACGCCATTTCCC | ||
| CsNAC78 | F:GGGGTACCATGGACTATAATTGCCTGGG | |
| R:ACGCGTCGACTATGAAGAAAAACAAGTTGT |
| 目的基因 Target gene | 引物序列(5′-3′) Primer sequence |
|---|---|
| Cs1g09640.1 | F:AGCCATGGGACTTACCAACG;R:ACCAGTAAGGCCGTTTCCAG |
| Cs4g12500.1 | F:GATCATCAGCTTCCGGGGTT;R:TTCGATGCTGAGCGGTTTCT |
| Cs5g29650.1(CsNAC21/22) | F:GGTGGCAAAGCTCAATGCAA;R:GCGTCTTCCTCATCCCTACG |
| Cs6g14200.1 | F:AAGGGAAGTGCCTCAACGAG;R:TCGTCTCCGGAGGATGAACT |
| Cs8g14700.1 | F:GTCAAGGAGACACCGAGCAA;R:AGCCTGTGGCCTTCCAATAC |
| orange1.1t00587.1 | F:AGGAATTTGTCCTCCTTCGTT;R:AGCCGAAGGTTGAGGAGTTG |
| orange1.1t00588.1 | F:TACAAAGGTCGTGGCTCCAG;R:TGCAGCTCGGGTATAGTCCT |
| orange1.1t00589.1(CsNAC68) | F:CGAAGGAATTTGTCCTCTTCCG;R:AGACCGCGGTTTATCTGTGG |
| orange1.1t00590.1(CsNAC78) | F:ACTGGTGGCGATCGTCAAAT;R:AGAACAGAAGAGATTGCCTGATGA |
表2 qRT-PCR引物
Table 2 The sequences of qRT-PCR primer pairs
| 目的基因 Target gene | 引物序列(5′-3′) Primer sequence |
|---|---|
| Cs1g09640.1 | F:AGCCATGGGACTTACCAACG;R:ACCAGTAAGGCCGTTTCCAG |
| Cs4g12500.1 | F:GATCATCAGCTTCCGGGGTT;R:TTCGATGCTGAGCGGTTTCT |
| Cs5g29650.1(CsNAC21/22) | F:GGTGGCAAAGCTCAATGCAA;R:GCGTCTTCCTCATCCCTACG |
| Cs6g14200.1 | F:AAGGGAAGTGCCTCAACGAG;R:TCGTCTCCGGAGGATGAACT |
| Cs8g14700.1 | F:GTCAAGGAGACACCGAGCAA;R:AGCCTGTGGCCTTCCAATAC |
| orange1.1t00587.1 | F:AGGAATTTGTCCTCCTTCGTT;R:AGCCGAAGGTTGAGGAGTTG |
| orange1.1t00588.1 | F:TACAAAGGTCGTGGCTCCAG;R:TGCAGCTCGGGTATAGTCCT |
| orange1.1t00589.1(CsNAC68) | F:CGAAGGAATTTGTCCTCTTCCG;R:AGACCGCGGTTTATCTGTGG |
| orange1.1t00590.1(CsNAC78) | F:ACTGGTGGCGATCGTCAAAT;R:AGAACAGAAGAGATTGCCTGATGA |
| 基因ID Gene ID | 基因 Gene | CLas vs. 空白对照 CLas vs. Mock | |
|---|---|---|---|
| 叶片中脉Midrib | 根Root | ||
| Cs1g09640.1 | CsNTL9-1 | 1.252 | 4.283 |
| Cs4g12500.1 | CsNAC | — | — |
| Cs5g29650.1 | CsNAC21/22 | — | — |
| Cs6g14240.1 | CsNAC090-1 | 5.469 | — |
| Cs8g14700.1 | CsNAC090-2 | -1.172 | — |
| orange1.1t00587.1 | CsNAC050 | 2.315 | 5.735 |
| orange1.1t00588.1 | CsNTL9-2 | — | 20.474 |
| orange1.1t00589.1 | CsNAC68 | — | 4.163 |
| orange1.1t00590.1 | CsNAC78 | 1.633 | 11.316 |
表3 CsNAC家族在锦橙中应答CLas侵染的转录组比较
Table 3 Comparative analysis of CsNAC family transcriptome in response to CLas infection in Jincheng
| 基因ID Gene ID | 基因 Gene | CLas vs. 空白对照 CLas vs. Mock | |
|---|---|---|---|
| 叶片中脉Midrib | 根Root | ||
| Cs1g09640.1 | CsNTL9-1 | 1.252 | 4.283 |
| Cs4g12500.1 | CsNAC | — | — |
| Cs5g29650.1 | CsNAC21/22 | — | — |
| Cs6g14240.1 | CsNAC090-1 | 5.469 | — |
| Cs8g14700.1 | CsNAC090-2 | -1.172 | — |
| orange1.1t00587.1 | CsNAC050 | 2.315 | 5.735 |
| orange1.1t00588.1 | CsNTL9-2 | — | 20.474 |
| orange1.1t00589.1 | CsNAC68 | — | 4.163 |
| orange1.1t00590.1 | CsNAC78 | 1.633 | 11.316 |
| 基因 Gene | 根 Root | 叶脉 Midrib | 基因 Gene | 根 Root | 叶脉 Midrib | |
|---|---|---|---|---|---|---|
| CsNTL9-1 | 2.42 ± 0.31 | 1.16 ± 0.24 | CsNAC050 | 4.24 ± 0.37 | 2.63 ± 0.44 | |
| CsNAC | 0.93 ± 0.09 | 2.01 ± 0.32 | CsNTL9-2 | 1.49 ± 0.15 | 1.68 ± 0.18 | |
| CsNAC21/22 | 0.49 ± 0.13 | 0.68 ± 0.10 | CsNAC68 | 8.62 ± 0.39 | 2.32 ± 0.18 | |
| CsNAC090-1 | 0.75 ± 0.12 | 1.38 ± 0.22 | CsNAC78 | 6.62 ± 0.29 | 1.86 ± 0.04 | |
| CsNAC090-2 | 0.97 ± 0.04 | 0.63 ± 0.10 |
表4 柑橘黄龙病菌诱导锦橙9个NAC基因的相对表达量
Table 4 Relative expression levels of 9 NAC genes in Jincheng induced by HLB
| 基因 Gene | 根 Root | 叶脉 Midrib | 基因 Gene | 根 Root | 叶脉 Midrib | |
|---|---|---|---|---|---|---|
| CsNTL9-1 | 2.42 ± 0.31 | 1.16 ± 0.24 | CsNAC050 | 4.24 ± 0.37 | 2.63 ± 0.44 | |
| CsNAC | 0.93 ± 0.09 | 2.01 ± 0.32 | CsNTL9-2 | 1.49 ± 0.15 | 1.68 ± 0.18 | |
| CsNAC21/22 | 0.49 ± 0.13 | 0.68 ± 0.10 | CsNAC68 | 8.62 ± 0.39 | 2.32 ± 0.18 | |
| CsNAC090-1 | 0.75 ± 0.12 | 1.38 ± 0.22 | CsNAC78 | 6.62 ± 0.29 | 1.86 ± 0.04 | |
| CsNAC090-2 | 0.97 ± 0.04 | 0.63 ± 0.10 |
| 名称 Name | CAP号 CAP ID | ORF/bp | 氨基酸数 Number of amino acid | 分子量 Molecular weight | 等电点 pI | 亲水性平均值 Grand average of hydropathicity | 脂肪指数 Aliphatic index | 不稳定系数 Instability index | 亚细胞定位 Subcellular localization |
|---|---|---|---|---|---|---|---|---|---|
| CsNAC21/22 | Cs5g29650.1 | 921 | 306 | 35 107.14 | 5.43 | -0.712 | 66.84 | 43.83 | 细胞核 Nuclear |
| CsNAC68 | orange1.1t00589.1 | 1 512 | 503 | 56 809.43 | 6.59 | -0.730 | 58.99 | 41.30 | 细胞核 Nuclear |
| CsNAC78 | orange1.1t00590.1 | 459 | 152 | 17 691.99 | 8.39 | -0.543 | 71.84 | 32.27 | 细胞质 Cytoplasm |
表5 柑橘3个NAC基因基因编码的氨基酸序列分析
Table 5 Amino acid sequence analysis of three NAC genes in citrus
| 名称 Name | CAP号 CAP ID | ORF/bp | 氨基酸数 Number of amino acid | 分子量 Molecular weight | 等电点 pI | 亲水性平均值 Grand average of hydropathicity | 脂肪指数 Aliphatic index | 不稳定系数 Instability index | 亚细胞定位 Subcellular localization |
|---|---|---|---|---|---|---|---|---|---|
| CsNAC21/22 | Cs5g29650.1 | 921 | 306 | 35 107.14 | 5.43 | -0.712 | 66.84 | 43.83 | 细胞核 Nuclear |
| CsNAC68 | orange1.1t00589.1 | 1 512 | 503 | 56 809.43 | 6.59 | -0.730 | 58.99 | 41.30 | 细胞核 Nuclear |
| CsNAC78 | orange1.1t00590.1 | 459 | 152 | 17 691.99 | 8.39 | -0.543 | 71.84 | 32.27 | 细胞质 Cytoplasm |

图2 柑橘CsNAC与拟南芥AtNAC、水稻OsNAC氨基酸的多序列比对
Fig. 2 Multi-sequence alignment of citrus CsNAC amino acid sequences with Arabidopsis thaliana AtNAC and Oryza sativa OsNAC amino acid sequences
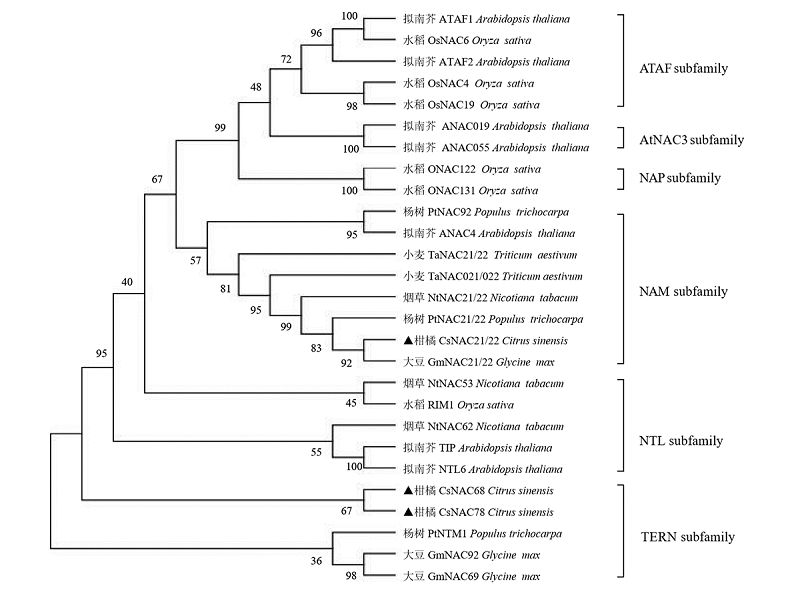
图3 CsNAC系统进化树分析采用蛋白的全长序列进行邻接法构建,并在每个节点指示1 000次复制的bootstrap值,分支上的数字为自展值(%)。
Fig. 3 CsNAC phylogenetic tree analysisConstructed by Neighbor-Joining method with the full-length sequence of the protein,and the Bootstrap value of 1 000 copies is indicated on each node,and the number on the branch is the self-spreading value(%).
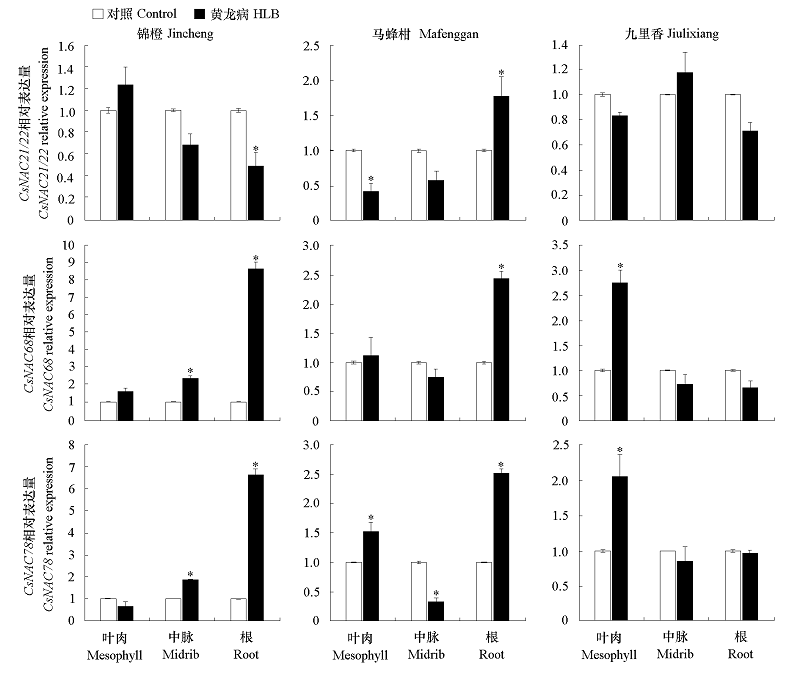
图5 不同抗性柑橘锦橙、马蜂柑和九里香接种Clas后3个NAC基因在不同组织中的相对表达量“*”表示与同一品种健康植株对照的差异显著性(P < 0.05)。
Fig. 5 Analysis of the relative expression levels of three NAC genes in different tissues of Jincheng,Mafenggan and Jiulixiang“*”Represents the significance of the difference with healthy plants of the same cultivar(P < 0.05).
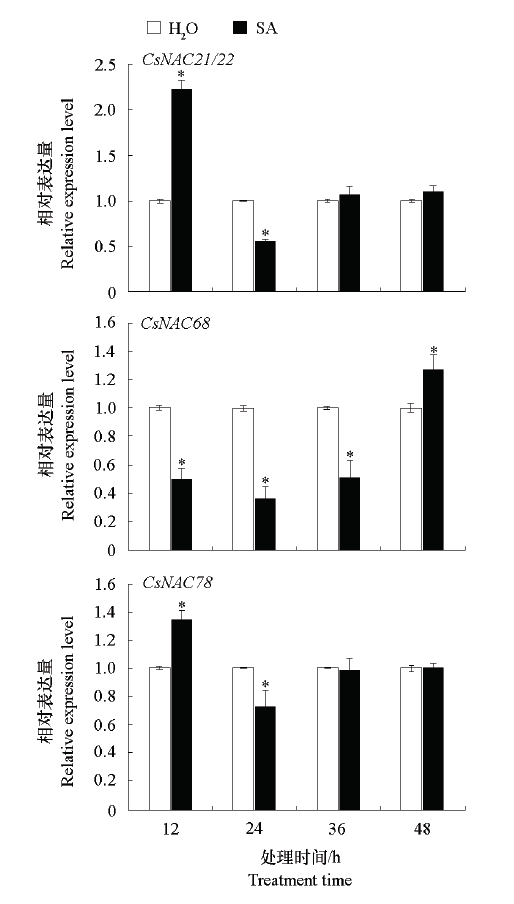
图6 SA诱导下CsNAC的表达量“*”表示与同一时期水处理对照的差异显著性(P < 0.05)。
Fig. 6 CsNAC expression induced by SA“*”indicates the significant difference between water treatment control in the same period(P < 0.05).
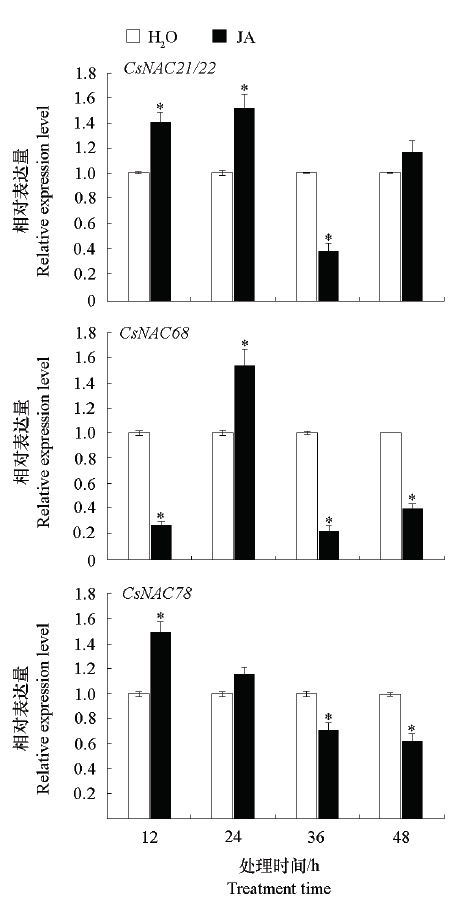
图7 JA诱导下CsNAC的表达量“*”表示与同一时期水处理对照的差异显著性(P < 0.05)。
Fig. 7 CsNAC expression induced by JA“*”indicates the significant difference between water treatment control in the same period(P < 0.05).
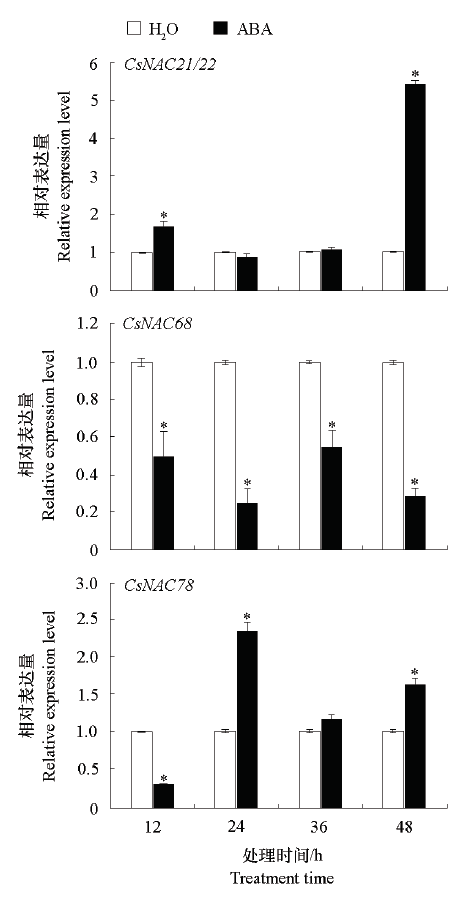
图8 ABA诱导下CsNAC的表达量“*”表示与同一时期水处理对照的差异显著性(P < 0.05)。
Fig. 8 CsNAC expression induced by ABA“*”indicates the significant difference between water treatment control in the same period(P < 0.05).

图9 ETH诱导下CsNAC的表达量“*”表示与同一时期水处理对照的差异显著性(P < 0.05)。
Fig. 9 CsNAC expression induced by ETH“*”indicates the significant difference between water treatment control in the same period(P < 0.05).
| [1] |
Aida M, Tshida T, Fukaki H, Fujisawa H, Tasaka M. 1997. Genes involved in organ separation in Arabidopsis:an analysis of the cup-shaped cotyledon mutant. Plant Cell, 9:841-857.
pmid: 9212461 |
| [2] |
Artimo P, Jonnalagedda M, Arnold K, Baratin D, Csardi G, De Castro E, Duvaud S, Flegel V, Fortier A, Gasteiger E, Grosdidier A, Hernandez C, Ioannidis V, Kuznetsov d, Liechti r, Moretti S, Mostaguir K, Redaschi N, Rossier G, Xenarios I, Stockinger H. 2012. ExPASy:SIB bioinformatics resource portal. Nucleic Acids Research, 40 (W1):W597-603.
doi: 10.1093/nar/gks400 URL |
| [3] |
Baldwin E, Plotto A, Bai J, Manthey J, Zhao W, Raithore S, Trey M. 2018. Effect of abscission zone formation on orange(Citrus sinensis) fruit/juice quality for trees affected by Huanglongbing(HLB). Journal of Agricultural and Food Chemistry, 66 (11):2877-2890.
doi: 10.1021/acs.jafc.7b05635 URL |
| [4] | Bove J M. 2006. Huanglongbing:a destructive,newly-emerging,century-old disease of citrus. Journal of Plant Pathology, 88 (1):7-37. |
| [5] |
Bu Q Y, Jiang H L, Li C B, Zhai Q Z, Zhang J, Wu X Y, Sun J Q, Xie Q, Li C Y. 2008. Role of the Arabidopsis thaliana NAC transcription factors ANAC019 and ANAC055 in regulating jasmonic acid-signaled defense responses. Cell Research, 18 (7):756-767.
doi: 10.1038/cr.2008.53 URL |
| [6] |
Chen N, Wu S H, Fu J L, Cao B H, Lei J J, Chen C M, Jiang J. 2016. Overexpression of the eggplant(Solanum melongena)NAC family transcription factor SmNAC suppresses resistance to bacterial wilt. Scientific Reports, 6 (1):1-20.
doi: 10.1038/s41598-016-0001-8 URL |
| [7] | Chen Qian, You Shuang-mei, Xing Le-hua, Xu Fan, Luo Ming, Guo Qi-gao. 2021. Research progress of NAC transcription factors in fruit trees. Molecular Plant Breeding, 19 (19):6396-6405. (in Chinese) |
| 陈倩, 游双梅, 邢乐华, 徐凡, 罗明, 郭启高. 2021. 果树NAC转录因子的研究进展. 分子植物育种, 19 (19):6396-6405. | |
| [8] |
Chen Y J, Perera V, Christiansen M W, Holme I B, Gregersen P L, Grant M R, Collinge D B, Lyngkjar M F. 2013. The barley HvNAC6 transcription factor affects ABA accumulation and promotes basal resistance against powdery mildew. Plant Molecular Biology, 83 (6):577-590.
doi: 10.1007/s11103-013-0109-1 URL |
| [9] | Cheng Bao-ping, Zhao Hong-wei, Peng Ai-tian, Song Xiao-bing, Ling Jin-feng, Chen Xia. 2016. Investigation on the occurrence of citrus psyllid,a transmission agent of citrus greening disease in Guangdong orchards. Plant Protection, 42 (1):189-192,196. (in Chinese) |
| 程保平, 赵弘巍, 彭埃天, 宋晓兵, 凌金锋, 陈霞. 2016. 柑橘黄龙病的传播介体-柑橘木虱在广东果园的发生调查. 植物保护, 42 (1):189-192,196. | |
| [10] |
da Graca J V, Douhan G W, Halbert S E, Keremane M L, Lee R F, Vidalakis G, Zhao H W. 2016. Huanglongbing:an overview of a complex pathosystem ravaging the world’s citrus. Journal of Integrative Plant Biology, 58 (4):373-387.
doi: 10.1111/jipb.12437 URL |
| [11] |
Damsteegt V D, Postnikovae N, Stone A L, Kuhlmann M, Wilson C, Sechler A, Schaad N W, Brlansky R H, Schneider W L. 2010. Murraya paniculata and related species as potential hosts and inoculum reservoirs of‘Candidatus Liberibacter asiaticus’causal agent of Huanglongbing. Plant Disease, 94 (5):528-533.
doi: 10.1094/PDIS-94-5-0528 pmid: 30754478 |
| [12] |
Delessert C, Kazan K, Wilson I W, Straeten D V D, Manners J, Dennis E S, Dolferus R. 2005. The transcription factor ATAF2 represses the expression of pathogenesis-related genes in Arabidopsis. The Plant Journal, 43 (5):745-757.
doi: 10.1111/j.1365-313X.2005.02488.x URL |
| [13] |
Duval M, Hsieh T F, Kim S Y, Thomas T L. 2002. Molecular characterization of AtNAM:a member of the Arabidopsis NAC domain superfamily. Plant Mol Biol, 50 (2):237-248.
doi: 10.1023/A:1016028530943 URL |
| [14] |
Ernst H A, Olsen A N, Skriver K, Larsen S, Leggio L L. 2004. Structure of the conserved domain of ANAC,a member of the NAC family of transcription factors. Embo Reports, 5 (3):297-303.
doi: 10.1038/sj.embor.7400093 URL |
| [15] | Fan Jing. 2007. Molecular cloning and expression pattern of CsCP and CsNAC from‘Navel’orange related to citrus peel pitting[Ph. D. Dissertation]. Chongqing: Chongqing University. (in Chinese) |
| 祝光涛. 2015. 番茄重要农艺性状的全基因组关联分析及野生种质在栽培种的渐渗分析[博士论文]. 重庆: 重庆大学. | |
| [16] | Guo Wen-fang, Liu De-chun, Yang Li, Zhuang Xia, Zhang Juan-juan, Wang Shu-sheng, Liu Yong. 2015. Cloning and expression analysis of new stress-resistant NAC83 gene from citrus. Acta Horticulturae Sinica, 42 (3):445-454. (in Chinese) |
| 郭文芳, 刘德春, 杨莉, 庄霞, 张涓涓, 王书胜, 刘勇. 2015. 柑橘抗逆基因NAC83的克隆与表达分析. 园艺学报, 42 (3):445-454. | |
| [17] |
Halbert S E, Manjunath K L. 2004. Asian citrus psyllids(Sternor-rhyncha:Psyllidae)and greening disease of citrus:a literature review and assessment of risk in Florida. Florida Entomologist, 87 (3):330-353.
doi: 10.1653/0015-4040(2004)087[0330:ACPSPA]2.0.CO;2 URL |
| [18] |
Hu R B, Qi G, Kong Y Z, Kong D J, Gao Q, Zhou G K. 2010. Comprehensive analysis of NAC domain transcription factor gene family in Populus trichocarpa. BMC Plant Biology, 10 (1):145.
doi: 10.1186/1471-2229-10-145 URL |
| [19] |
Johnson E G, Wu J, Bright D B, Graham J H. 2014. Association of‘Candidatus Liberibacter asiaticus’root infection,but not phloem plugging with root loss on Huanglongbing-affected trees prior to appearance of foliar symptoms. Plant Pathology, 63 (5):290-298.
doi: 10.1111/ppa.12109 URL |
| [20] |
Kumar S, Stecher G, Tamura K. 2016. MEGA7:molecular evolutionary genetics analysis version 7.0 for bigger datasets. Molecular Biology and Evolution, 33 (7):1870-1874.
doi: 10.1093/molbev/msw054 URL |
| [21] |
Le D T, Nishiyama R, Watanabe Y, Mochida K, Yamaguchi S K, Shinozaki K, Tran L S. 2011. Genome-wide survey and expression analysis of the plant-specific NAC transcription factor family in soybean during development and dehydration stress. DNA Research, 18 (4):263-276.
doi: 10.1093/dnares/dsr015 URL |
| [22] |
Lee M H, Jeon H S, Kim H G, Park O K. 2017. An Arabidopsis NAC transcription factor NAC4 promotes pathogen-induced cell death under negative regulation by microRNA164. New Phytologist, 214 (1):343-360.
doi: 10.1111/nph.14371 URL |
| [23] | Li Cheng-hui, Cai Bin. 2013. Genome-wide analysis of NAC transcription factor gene family in grape. Acta Agriculturae Shanghai, 29 (6):7-13. (in Chinese) |
| 李成慧, 蔡斌. 2013. 葡萄NAC转录因子家族全基因组分析. 上海农业学报, 29 (6):7-13. | |
| [24] |
Lin R M, Zhao W S, Meng X B, Wang M, Peng Y L. 2007. Rice gene OsNAC19 encodes a novel NAC-domain transcription factor and responds to infection by Magnaporthe grisea. Plant Science, 172 (1):120-130.
doi: 10.1016/j.plantsci.2006.07.019 URL |
| [25] |
Liu Y Z, Baig M N R, Fan R, Ye J L, Cao Y C, Deng X X. 2009. Identification and expression pattern of a novel NAM,ATAF,and CUC-like gene from Citrus sinensis osbeck. Plant Molecular Biology Reporter, 27 (3):292-297.
doi: 10.1007/s11105-008-0082-z URL |
| [26] | Liu Zhan-ji, Shao Feng-xia, Tang Gui-ying. 2007. The research progress of structure function and regulation of plant NAC transcription factors. Acta Bot Boreal-Occident Sin, 27 (9):1915-1920. (in Chinese) |
| 柳展基, 邵凤霞, 唐桂英. 2007. 植物NAC转录因子的结构功能及其表达调控研究进展. 西北植物学报, 27 (9):1915-1920. | |
| [27] |
Livak K J, Schmittgen T D. 2001. Analysis of relative gene expression data using real-time quantitative PCR and the 2-ΔΔCt method. Methods, 25 (4):402-408.
doi: 10.1006/meth.2001.1262 pmid: 11846609 |
| [28] |
Nakashima K, Tran L P, Nguyen D V, Fujita M, Maruyama K, Todaka D, Ito Y, Hayashi N, Shinozaki K, Yamaguchi-Shinozaki K. 2007. Functional analysis of a NAC-type transcription factor OsNAC6 involved in abiotic and biotic stress-responsive gene expression in rice. The Plant Journal, 51 (4):617-630.
doi: 10.1111/j.1365-313X.2007.03168.x URL |
| [29] |
Nuruzzaman M, Manimekalai R, Sharoni A M, Satoh K, Kondoh K, Ooka H, Kikuchi S. 2010. Genome-wide analysis of NAC transcription factor family in rice. Gene, 465 (1):30-44.
doi: 10.1016/j.gene.2010.06.008 URL |
| [30] |
Nuruzzaman M, Sharoni A M, Kikuchi S. 2013. Roles of NAC transcription factors in the regulation of biotic and abiotic stress responses in plants. Frontiers in Microbiology, 4:248.
doi: 10.3389/fmicb.2013.00248 pmid: 24058359 |
| [31] |
Olsen A N, Ernst H A, Leggio L L, Skriver K. 2005. NAC transcription factors:structurally distinct,functionally diverse. Trends Plant Sci, 10 (2):79-87.
doi: 10.1016/j.tplants.2004.12.010 URL |
| [32] |
Ooka H, Satoh K, Doi K, Nagata T, Otomo Y, Murakami K, Matsubara K, Osato N, Kawai J, Carninci P, Hayashizaki Y, Suzuki K, Kojima K, Takahara Y, Yamatomo K, Kikuchi S. 2003. Comprehensive analysis of NAC family genes in Oryza sativa and Arabidopsis thaliana. DNA Research, 10 (6):239-247.
doi: 10.1093/dnares/10.6.239 URL |
| [33] |
Peng A H, Zou X P, He Y R, Chen S C, Liu X F, Zhang J Y, Zhang Q W, Xie Z, Long J H, Zhao X C. 2021. Overexpressing a NPR1-like gene from Citrus paradisi enhanced Huanglongbing resistance in C. sinensis. Plant Cell Reports, 40 (3):529-541.
doi: 10.1007/s00299-020-02648-3 URL |
| [34] | Peng Hui, Yu Xing-wang, Cheng Hui-ying, Zhang Hua, Shi Qing-hua, Li Jian-gui, Ma Hao. 2010. Review on the family of plant NAC transcription factors. Chinese Bulletin of Botany, 45 (2):236-248. (in Chinese) |
| 彭辉, 于兴旺, 成慧颖, 张桦, 石庆华, 李建贵, 麻浩. 2010. 植物NAC转录因子家族研究概况. 植物学报, 45 (2):236-248. | |
| [35] | Qiu Zhi-yu, Huang Hong-lan, Shu Chang, Qiu Jun-jie. 2015. Citrus yellow dragon disease pathogenesis,symptoms and prevention measure. Biological Disaster Science, 38 (3):193-200. (in Chinese) |
| 邱志燏, 黄红兰, 舒畅, 邱俊洁. 2015. 柑橘黄龙病发病机理、症状及防控措施. 生物灾害科学, 38 (3):193-200. | |
| [36] |
Ren T, Qu F, Morris T J. 2000. HRT gene function requires interaction between a NAC protein and viral capsid protein to confer resistance to turnip crinkle virus. Plant Cell, 12 (10):1917-1926.
pmid: 11041886 |
| [37] | Rong Huan, Ren Shi-jie, Wang Zi-ping, Wang Fei, Zhou Yong. 2020. Advances in the structure and function of plant NAC transcription factors. Jiangsu Agricultural Sciences, 48 (18):44-53. (in Chinese) |
| 荣欢, 任师杰, 汪梓坪, 王飞, 周勇. 2020. 植物NAC转录因子的结构及功能研究进展. 江苏农业科学, 48 (18):44-53. | |
| [38] |
Rushton P J, Bokowiec M T, Han S, Zhang H, Brannock J F, Chen X, Laudeman T W, Timko M P. 2008. Tobacco transcription factors:novel insights into transcriptional regulation in the Solanaceae. Plant Physiology, 147 (1):280-295.
doi: 10.1104/pp.107.114041 pmid: 18337489 |
| [39] |
Seo P J, Kim M J, Park J Y, Kim S Y, Jeon J, Lee Y H, Kim J, Park C M. 2010. Cold activation of a plasma membrane-tethered NAC transcription factor induces a pathogen resistance response in Arabidopsis. The Plant Journal, 61 (4):661-671.
doi: 10.1111/j.1365-313X.2009.04091.x URL |
| [40] |
Su H Y, Zhang S Z, Yin Y L, Zhu D Z, Han L Y. 2015. Genome-wide analysis of NAM-ATAF1,2-CUC2 transcription factor family in Solanum lycopersicum. Journal of Plant Biochemistry and Biotechnology, 24 (2):176-183.
doi: 10.1007/s13562-014-0255-9 URL |
| [41] |
Sun H, Hu M L, Li J Y, Chen L, Li M, Zhang S Q, Zhang X L, Yang X Y. 2018. Comprehensive analysis of NAC transcription factors uncovers their roles during fiber development and stress response in cotton. BMC Plant Biology, 18 (1):1-15.
doi: 10.1186/s12870-017-1213-1 URL |
| [42] |
Sun L J, Zhang H J, Li A Y, Huang L, Hong Y B, Ding X S, Nelson R S, Zhou X P, Song F M. 2013. Functions of rice NAC transcriptional factors,ONAC122 and ONAC131,in defense responses against Magnaporthe grisea. Plant Mol Biol, 81 (1):41-56.
doi: 10.1007/s11103-012-9981-3 URL |
| [43] | Sun Li-jun, Li Da-yong, Zhang Hui-juan, Song Feng-ming. 2012. Functions of NAC transcription factors in biotic and abiotic stress responses in plants. Hereditas, 34 (8):993-1002. (in Chinese) |
| 孙利军, 李大勇, 张慧娟, 宋凤鸣. 2012. NAC转录因子在植物抗病和抗非生物胁迫反应中的作用. 遗传, 34 (8):993-1002. | |
| [44] |
Vand S H, Abdullah T L, Sijam k, Abdullah S N, Abdullah N A. 2009. Differential reaction of citrus species in Malaysia to Huanglongbing(HLB) disease using grafting method. American Journal of Agricultural and Biological Sciences, 4 (1):32-38.
doi: 10.3844/ajabssp.2009.32.38 URL |
| [45] |
Wang X E, Basnayake B M, Vindhya S, Zhang H J, Li G J, Li W, Virk N, Mengiste T, Song F M. 2009. The Arabidopsis ATAF1,a NAC transcription factor,is a negative regulator of defense responses against necrotrophic fungal and bacterial pathogens. Molecular Plant-microbe Interactions, 22 (10):1227-1238.
doi: 10.1094/MPMI-22-10-1227 URL |
| [46] |
Wu Y R, Deng Z Y, Lai J B, Zhang Y Y, Yang C P, Yin B J, Zhao Q Z, Zhang L, Li Y, Yang C W, Xie Q. 2009. Dual function of Arabidopsis ATAF1 in abiotic and biotic stress responses. Cell Research, 19 (11):1279-1290.
doi: 10.1038/cr.2009.108 URL |
| [47] |
Xie Z, Zhao K, Long J H, Zhen L, Zou X P, Chen S C. 2021. Comparative analysis of Wanjincheng orange leaf and root responses to‘Candidatus liberibacter asiaticus’infection using leaf-disc grafting. Horticultural Plant Journal, 7 (5):401-410.
doi: 10.1016/j.hpj.2021.01.007 URL |
| [48] | Xie Zhu, Zhao Ke, Zheng Lin, Long Junhong, Du Meixia, He Yongrui, Chen Shanchun, Zou Xiuping. 2020. Cloning and expression analysis of alcohol dehydrogenase CsADH1gene responding to Huanglongbing infection in Citrus. Acta Horticulturae Sinica, 47 (3):445-454. (in Chinese) |
| 谢竹, 赵珂, 郑林, 龙俊宏, 杜美霞, 何永睿, 陈善春, 邹修平. 2020. 响应黄龙病侵染的柑橘乙醇脱氢酶基因CsADH1的克隆与表达分析. 园艺学报, 47 (3):445-454. | |
| [49] |
Xu Q, Chen L L, Ruan X A, Chen D J, Zhu A D, Chen C L, Bertrand D, Jiao W B, Hao B H, Lyon M P, Chen J J, Gao S, Xing F, Lan H, Chang J W, Ge X H, Lei Y, Hu Q, Miao Y, Wang L, Xiao S X, Biswas M K, Zeng W F, Guo F, Cao H B, Yang X M, Xu X W, Cheng Y J, Xu J, Liu J H, Luo J H, Tang Z H, Guo W W, Kuang H H, Zhang H Y, Roose M L, Nagarajan N, Deng X X, Ruan Y J. 2013. The draft genome of sweet orange(Citrus sinensis). Nature Genetics, 45 (1):59-66.
doi: 10.1038/ng.2472 URL |
| [50] |
Yoshii M, Shimizu T, Yamazaki M, Higashi T, Miyao A, Hirochika H, Omura T. 2009. Disruption of a novel gene for a NAC-domain protein in rice confers resistance to rice dwarf virus. The Plant Journal, 57 (4):615-625.
doi: 10.1111/j.1365-313X.2008.03712.x URL |
| [51] |
Yoshii M, Yamazaki M, Rakwal R, KIshi-kaboshi M, Miyaol A, Hirohiko H. 2010. The NAC transcription factor RIM1 of rice is a new regulator of jasmonate signaling. The Plant Journal, 61 (5):804-815.
doi: 10.1111/j.1365-313X.2009.04107.x URL |
| [52] |
Yu C S, Lin C J, Kwang J K. 2004. Predicting subcellular localization of proteins for Gram-negative bacteria by support vector machines based on n-peptide compositions. Protein Science, 13 (5):1402-1406.
doi: 10.1110/ps.03479604 URL |
| [53] |
Yuan X, Wang H, Cai J T, Li D Y, Song F M. 2019. NAC transcription factors in plant immunity. Phytopathology Research, 1 (1):1-13.
doi: 10.1186/s42483-018-0011-5 URL |
| [54] |
Zou X P, Bai X J, Wen Q L, Xie Z, Wu L, Peng A H, He Y R, Xu L Z, Chen S C. 2019. Comparative analysis of tolerant and susceptible citrus reveals the role of methyl salicylate signaling in the response to Huanglongbing. Journal of Plant Growth Regulation, 38 (4):1516-1528.
doi: 10.1007/s00344-019-09953-6 URL |
| [55] |
Zou X P, Jiang X Y, Xu L Z, Lei T G, Peng A H, He Y R, Yao L X, Chen S C. 2017. Transgenic citrus expressing synthesized cecropin B genes in the phloem exhibits decreased susceptibility to Huanglongbing. Plant Molecular Biology, 93 (4):341-353.
doi: 10.1007/s11103-016-0565-5 URL |
| [56] |
Zou X P, Zhao K, Liu Y N, Du M X, Zheng L, Wang S, Xu L Z, Peng A H, He Y R, Long Q, Chen S C. 2021. Overexpression of salicylic acid carboxyl methyltransferase(CsSAMT1)enhances tolerance to Huanglongbing disease in Wanjincheng orange[Citrus sinensis(L.)Osbeck]. International Journal of Molecular Sciences, 22 (6):2803.
doi: 10.3390/ijms22062803 URL |
| [1] | 叶子茂, 申晚霞, 刘梦雨, 王 彤, 张晓楠, 余 歆, 刘小丰, 赵晓春, . R2R3-MYB转录因子CitMYB21对柑橘类黄酮生物合成的影响[J]. 园艺学报, 2023, 50(2): 250-264. |
| [2] | 蒋靖东, 韦壮敏, 王楠, 朱晨桥, 叶俊丽, 谢宗周, 邓秀新, 柴利军. 山金柑四倍体资源的发掘与鉴定[J]. 园艺学报, 2023, 50(1): 27-35. |
| [3] | 杜玉玲, 杨凡, 赵娟, 刘书琪, 龙超安. 新鱼腥草素钠对柑橘指状青霉的抑菌作用[J]. 园艺学报, 2023, 50(1): 145-152. |
| [4] | 赵雪艳, 王琪, 王莉, 王方圆, 王庆, 李艳. 基于比较转录组的延胡索组织差异性表达分析[J]. 园艺学报, 2023, 50(1): 177-187. |
| [5] | 李镇希, 潘睿翾, 许美容, 郑正, 邓晓玲. 柑橘黄龙病菌双重实时荧光PCR检测方法的建立[J]. 园艺学报, 2023, 50(1): 188-196. |
| [6] | 朱凯杰, 张哲惠, 曹立新, 向舜德, 叶俊丽, 谢宗周, 柴利军, 邓秀新, . 棕色晚熟脐橙新品种‘宗橙’[J]. 园艺学报, 2022, 49(S1): 41-42. |
| [7] | 朱世平, 文荣中, 王媛媛, 曾 杨. 特晚熟柑橘新品种‘金乐柑’[J]. 园艺学报, 2022, 49(S1): 43-44. |
| [8] | 王沙, 张心慧, 赵玉洁, 李变变, 招雪晴, 沈雨, 董建梅, 苑兆和. 石榴花青苷合成相关基因PgMYB111的克隆与功能分析[J]. 园艺学报, 2022, 49(9): 1883-1894. |
| [9] | 高彦龙, 吴玉霞, 张仲兴, 王双成, 张瑞, 张德, 王延秀. 苹果ELO家族基因鉴定及其在低温胁迫下的表达分析[J]. 园艺学报, 2022, 49(8): 1621-1636. |
| [10] | 邱子文, 刘林敏, 林永盛, 林晓洁, 李永裕, 吴少华, 杨超. 千层金MbEGS基因的克隆与功能分析[J]. 园艺学报, 2022, 49(8): 1747-1760. |
| [11] | 杨海健, 张云贵, 周心智. 柑橘新品种‘云贵脆橙’[J]. 园艺学报, 2022, 49(7): 1611-1612. |
| [12] | 马维峰, 李艳梅, 马宗桓, 陈佰鸿, 毛娟. 苹果POD家族基因的鉴定与MdPOD15的功能分析[J]. 园艺学报, 2022, 49(6): 1181-1199. |
| [13] | 张凯, 麻明英, 王萍, 李益, 金燕, 盛玲, 邓子牛, 马先锋. 柑橘HSP20家族基因鉴定及其响应溃疡病菌侵染表达分析[J]. 园艺学报, 2022, 49(6): 1213-1232. |
| [14] | 李文婷, 李翠晓, 林小清, 郑永钦, 郑正, 邓晓玲. 基于STR位点对广东省柑橘溃疡病菌种群遗传结构的分析[J]. 园艺学报, 2022, 49(6): 1233-1246. |
| [15] | 麻明英, 郝晨星, 张凯, 肖桂华, 苏翰英, 文康, 邓子牛, 马先锋. 甜橙SWEET2a促进柑橘溃疡病菌侵染[J]. 园艺学报, 2022, 49(6): 1247-1260. |
| 阅读次数 | ||||||
|
全文 |
|
|||||
|
摘要 |
|
|||||
版权所有 © 2012 《园艺学报》编辑部 京ICP备10030308号-2 国际联网备案号 11010802023439
编辑部地址: 北京市海淀区中关村南大街12号中国农业科学院蔬菜花卉研究所 邮编: 100081
电话: 010-82109523 E-Mail: yuanyixuebao@126.com
技术支持:北京玛格泰克科技发展有限公司