
园艺学报 ›› 2022, Vol. 49 ›› Issue (8): 1621-1636.doi: 10.16420/j.issn.0513-353x.2021-0545
• 研究论文 • 下一篇
高彦龙, 吴玉霞, 张仲兴, 王双成, 张瑞, 张德, 王延秀*( )
)
收稿日期:2022-04-27
修回日期:2022-06-06
出版日期:2022-08-25
发布日期:2022-09-05
通讯作者:
王延秀
E-mail:wangxy@gsau.edu.cn
基金资助:
GAO Yanlong, WU Yuxia, ZHANG Zhongxing, WANG Shuangcheng, ZHANG Rui, ZHANG De, WANG Yanxiu*( )
)
Received:2022-04-27
Revised:2022-06-06
Online:2022-08-25
Published:2022-09-05
Contact:
WANG Yanxiu
E-mail:wangxy@gsau.edu.cn
摘要:
为研究苹果ELO家族基因在蜡质合成及抗寒中的作用,利用生物信息的方法对苹果全基因组中的ELO家族成员进行鉴定与分析。结果表明,MdELO基因家族共包含7个成员,不均匀地分布在苹果的3条染色体(Chr2、Chr8和Chr15)上。MdELO蛋白包含211 ~ 305个不等的氨基酸残基,等电点在9.43 ~ 11.62之间。亚细胞定位结果表明,MdELO蛋白在细胞核、细胞膜和叶绿体中有分布。顺式作用元件分析表明,7个MdELO启动子上游2 000 bp序列分布有激素、环境适应性和逆境诱导等响应元件。进一步测定越冬期不同品种苹果枝条中的蜡质含量,结果显示,从初休眠期至萌芽期,蜡质含量呈逐渐下降趋势,且抗寒性强的‘俄矮元帅’蜡质含量显著高于‘宫崎短富’;从蜡质形态及枝条表皮形态来看,‘俄矮元帅’蜡质呈针芒状且分布紧密,其枝条表皮结构紧密,表面光滑,而‘宫崎短富’蜡质呈散块状,枝条表皮结构松散,表面粗糙;两种苹果枝条的脯氨酸含量、相对电导率和淀粉磷酸化酶活性随越冬期的延长呈先升后降的趋势,淀粉酶活性呈逐渐上升的趋势,其中‘俄矮元帅’各指标均高于‘宫崎短富’。综上表明‘俄矮元帅’抗寒性显著高于‘宫崎短富’。qRT-PCR分析发现,低温胁迫下苹果砧木垂丝海棠的叶和根中均能检测到ELO的表达,大部分基因表现出先升后降的趋势,在低温胁迫12或24 h表达量最高,说明该家族成员在植物抗寒中扮演着重要作用。
中图分类号:
高彦龙, 吴玉霞, 张仲兴, 王双成, 张瑞, 张德, 王延秀. 苹果ELO家族基因鉴定及其在低温胁迫下的表达分析[J]. 园艺学报, 2022, 49(8): 1621-1636.
GAO Yanlong, WU Yuxia, ZHANG Zhongxing, WANG Shuangcheng, ZHANG Rui, ZHANG De, WANG Yanxiu. Bioinformatics Analysis of Apple ELO Gene Family and Its Expression Analysis Under Low Temperature Stress[J]. Acta Horticulturae Sinica, 2022, 49(8): 1621-1636.
| 基因登录号 Accession No. | 基因名称 Gene name | 序列(5′-3′) Sequence | |
|---|---|---|---|
| MD02G1155100 | MdELO1 | F:TCCTACGTGTACTACCTGTCCAGATTC | R:AGAGGAACGAGATGACGGTGAGG |
| MD08G1036600 | MdELO2 | F:CAGTCTACCTCATCTTCATGTCGTTCC | R:AGAGCAGAGTGGTGGAGAGTATGG |
| MD08G1151400 | MdELO3 | F:CCGCCATCGCCGTCTACATAAC | R:GGCTGGAATAGGTCCGAGAGGTAC |
| MD08G1227900 | MdELO4 | F:ATTTGGCTCCACACATCGCAGTC | R:CGGTCACCCTTCTCTTCCACTTTG |
| MD15G1269600 | MdELO5 | F:CCGCCATCGCCGTCTACATAAC | R:GGCTGGAATAGGTCCGAGAGGTAC |
| MD15G1421700 | MdELO6 | F:CAACCTCAACCTCCTCCTCCTCTC | R:GCAAACAGATAATCCAGAAGGGGTAGG |
| MD15G1421900 | MdELO7 | F:CACAGCCGTCCACAACCTCATC | R:AGGTGTTGGGTGGGAAGCAGAG |
表1 qRT-PCR引物序列
Table 1 qRT-PCR primer sequence
| 基因登录号 Accession No. | 基因名称 Gene name | 序列(5′-3′) Sequence | |
|---|---|---|---|
| MD02G1155100 | MdELO1 | F:TCCTACGTGTACTACCTGTCCAGATTC | R:AGAGGAACGAGATGACGGTGAGG |
| MD08G1036600 | MdELO2 | F:CAGTCTACCTCATCTTCATGTCGTTCC | R:AGAGCAGAGTGGTGGAGAGTATGG |
| MD08G1151400 | MdELO3 | F:CCGCCATCGCCGTCTACATAAC | R:GGCTGGAATAGGTCCGAGAGGTAC |
| MD08G1227900 | MdELO4 | F:ATTTGGCTCCACACATCGCAGTC | R:CGGTCACCCTTCTCTTCCACTTTG |
| MD15G1269600 | MdELO5 | F:CCGCCATCGCCGTCTACATAAC | R:GGCTGGAATAGGTCCGAGAGGTAC |
| MD15G1421700 | MdELO6 | F:CAACCTCAACCTCCTCCTCCTCTC | R:GCAAACAGATAATCCAGAAGGGGTAGG |
| MD15G1421900 | MdELO7 | F:CACAGCCGTCCACAACCTCATC | R:AGGTGTTGGGTGGGAAGCAGAG |
| 基因登录号 Accession No. | 基因名称 Gene name | 大小/aa Size | 分子量 Molecular weight | 等电点 Isoelectric point | 亚细胞定位 Subcellular localization | 不稳定系数 Instability index |
|---|---|---|---|---|---|---|
| MD02G1155100 | MdELO1 | 305 | 34 129.36 | 10.18 | 质膜Plasma membrane | 43.13 |
| MD08G1036600 | MdELO2 | 211 | 24 143.86 | 11.62 | 质膜/细胞核Plasma membrane/nucleus | 75.39 |
| MD08G1151400 | MdELO3 | 302 | 33 869.14 | 10.23 | 质膜Plasma membrane | 41.39 |
| MD08G1227900 | MdELO4 | 265 | 30 759.69 | 9.43 | 叶绿体Chloroplast | 40.60 |
| MD15G1269600 | MdELO5 | 302 | 33 869.14 | 10.23 | 质膜Plasma membrane | 41.39 |
| MD15G1421700 | MdELO6 | 270 | 31 346.38 | 9.50 | 叶绿体Chloroplast | 38.86 |
| MD15G1421900 | MdELO7 | 273 | 31 159.93 | 9.53 | 叶绿体Chloroplast | 38.90 |
表2 苹果ELO基因家族理化性质分析
Table 2 Analysis of physicochemical properties of apple MdELO gene family
| 基因登录号 Accession No. | 基因名称 Gene name | 大小/aa Size | 分子量 Molecular weight | 等电点 Isoelectric point | 亚细胞定位 Subcellular localization | 不稳定系数 Instability index |
|---|---|---|---|---|---|---|
| MD02G1155100 | MdELO1 | 305 | 34 129.36 | 10.18 | 质膜Plasma membrane | 43.13 |
| MD08G1036600 | MdELO2 | 211 | 24 143.86 | 11.62 | 质膜/细胞核Plasma membrane/nucleus | 75.39 |
| MD08G1151400 | MdELO3 | 302 | 33 869.14 | 10.23 | 质膜Plasma membrane | 41.39 |
| MD08G1227900 | MdELO4 | 265 | 30 759.69 | 9.43 | 叶绿体Chloroplast | 40.60 |
| MD15G1269600 | MdELO5 | 302 | 33 869.14 | 10.23 | 质膜Plasma membrane | 41.39 |
| MD15G1421700 | MdELO6 | 270 | 31 346.38 | 9.50 | 叶绿体Chloroplast | 38.86 |
| MD15G1421900 | MdELO7 | 273 | 31 159.93 | 9.53 | 叶绿体Chloroplast | 38.90 |

图2 苹果ELO基因家族成员序列 序列一致性为100%用深蓝色标记,> 75%用粉色标记,> 50%用浅蓝色标记。红色下划线代表ELO结构域。
Fig. 2 Multiple sequence alignment of apple MdELO gene family members Sequence identity of 100% is marked in dark blue,> 75% in pink,and > 50% in light blue. The red underline represents the ELO domain.
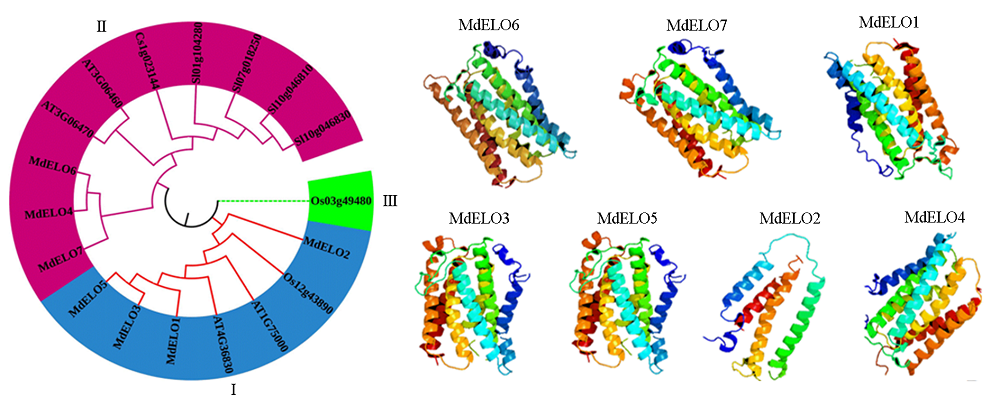
图3 苹果ELO基因家族进化及其三级结构 At:拟南芥;Sl:番茄;Cs:柑橘;Os:水稻。
Fig. 3 Phylogenetic tree and tertiary structure of apple MdELO gene family At:Arabidopsis thaliana;Sl:Solanum lycopersicum;Cs:Citrus reticulata Blanco;Os:Oryza sativa.
| 旁系同源基因对位点 Paralogous gene pair sites | Ka | Ks | Ka/Ks |
|---|---|---|---|
| MdELO1/MdELO2 | 0.427957 | 0.867576 | 0.493279 |
| MdELO1/MdELO3 | 0.026568 | 0.236089 | 0.112535 |
| MdELO1/MdELO4 | 0.755620 | 1.160104 | 0.651338 |
| MdELO1/MdELO5 | 0.026568 | 0.236089 | 0.112535 |
| MdELO1/MdELO6 | 0.745098 | 1.256016 | 0.593223 |
| MdELO1/MdELO7 | 0.671194 | 1.008214 | 0.665726 |
| MdELO2/MdELO3 | 0.368452 | 0.856895 | 0.429984 |
| MdELO2/MdELO4 | 0.701287 | 1.012445 | 0.692667 |
| MdELO2/MdELO5 | 0.368452 | 0.856895 | 0.429984 |
| MdELO2/MdELO6 | 0.700478 | 1.012911 | 0.691550 |
| MdELO2/MdELO7 | 0.743613 | 0.838943 | 0.886369 |
| MdELO3/MdELO4 | 0.718204 | 1.073463 | 0.669053 |
| MdELO3/MdELO5 | 0.701591 | 1.069425 | 0.656045 |
| MdELO3/MdELO6 | 0.706619 | 1.203640 | 0.587069 |
| MdELO3/MdELO7 | 0.636910 | 0.936570 | 0.680045 |
| MdELO4/MdELO5 | 0.718204 | 1.073463 | 0.669053 |
| MdELO4/MdELO6 | 0.018535 | 0.220872 | 0.083918 |
| MdELO4/MdELO7 | 0.244042 | 0.942442 | 0.258947 |
| MdELO5/MdELO6 | 0.706619 | 1.203640 | 0.587069 |
| MdELO5/MdELO7 | 0.636910 | 0.936570 | 0.680045 |
| MdELO6/MdELO7 | 0.246400 | 0.868520 | 0.283700 |
表3 MdELO基因的Ka/Ks
Table 3 Ka/Ks of MdELO gene
| 旁系同源基因对位点 Paralogous gene pair sites | Ka | Ks | Ka/Ks |
|---|---|---|---|
| MdELO1/MdELO2 | 0.427957 | 0.867576 | 0.493279 |
| MdELO1/MdELO3 | 0.026568 | 0.236089 | 0.112535 |
| MdELO1/MdELO4 | 0.755620 | 1.160104 | 0.651338 |
| MdELO1/MdELO5 | 0.026568 | 0.236089 | 0.112535 |
| MdELO1/MdELO6 | 0.745098 | 1.256016 | 0.593223 |
| MdELO1/MdELO7 | 0.671194 | 1.008214 | 0.665726 |
| MdELO2/MdELO3 | 0.368452 | 0.856895 | 0.429984 |
| MdELO2/MdELO4 | 0.701287 | 1.012445 | 0.692667 |
| MdELO2/MdELO5 | 0.368452 | 0.856895 | 0.429984 |
| MdELO2/MdELO6 | 0.700478 | 1.012911 | 0.691550 |
| MdELO2/MdELO7 | 0.743613 | 0.838943 | 0.886369 |
| MdELO3/MdELO4 | 0.718204 | 1.073463 | 0.669053 |
| MdELO3/MdELO5 | 0.701591 | 1.069425 | 0.656045 |
| MdELO3/MdELO6 | 0.706619 | 1.203640 | 0.587069 |
| MdELO3/MdELO7 | 0.636910 | 0.936570 | 0.680045 |
| MdELO4/MdELO5 | 0.718204 | 1.073463 | 0.669053 |
| MdELO4/MdELO6 | 0.018535 | 0.220872 | 0.083918 |
| MdELO4/MdELO7 | 0.244042 | 0.942442 | 0.258947 |
| MdELO5/MdELO6 | 0.706619 | 1.203640 | 0.587069 |
| MdELO5/MdELO7 | 0.636910 | 0.936570 | 0.680045 |
| MdELO6/MdELO7 | 0.246400 | 0.868520 | 0.283700 |
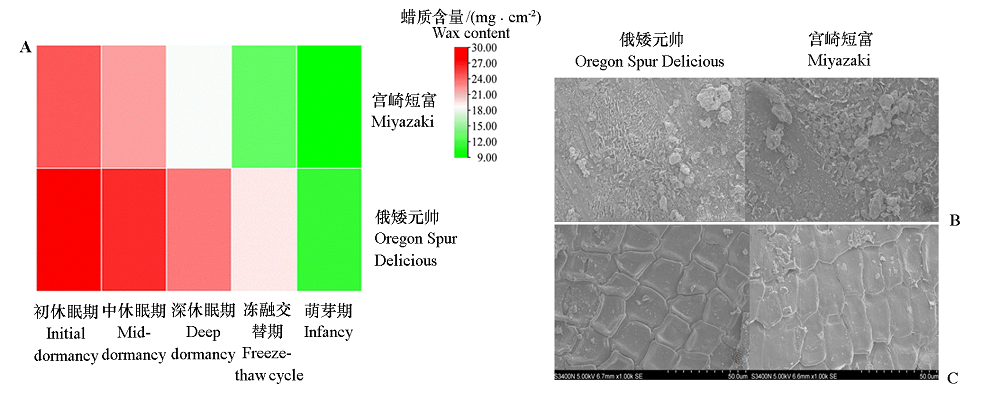
图7 ‘俄矮元帅’和‘宫崎短富’苹果越冬期蜡质含量(A)、蜡质形态(B)及枝条表皮形态(C)
Fig. 7 The wax content(A),wax morphology(B)and branch epidermis morphology(C)of‘Oregon Spur Delicious’and‘Miyazaki’during wintering
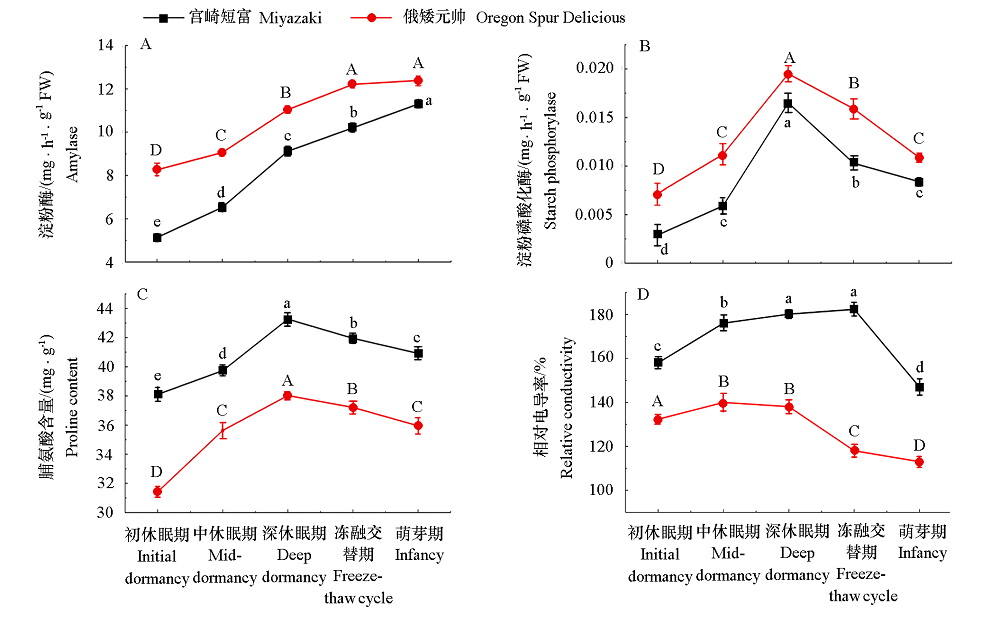
图8 ‘俄矮元帅’和‘宫崎短富’不同越冬期枝条淀粉酶活性、淀粉磷酸化酶活性、脯氨酸含量和相对电导率的变化 不同小写字母代表‘宫崎短富’苹果不同时期的差异显著性;不同大写字母代表‘俄矮元帅’苹果不同时期的差异显著性(P < 0.05)。
Fig. 8 Changes of amylase,starch phosphorylase,Proline and relative conductivity of‘Miyazaki’and ‘Oregon Spur Delicious’branches in different overwintering periods Different lowercase letters represent the significant differences of‘Miyazaki’at different times;Different capital letters represent the significant differences of‘Oregon Spur Delicious’at different times(P < 0.05).
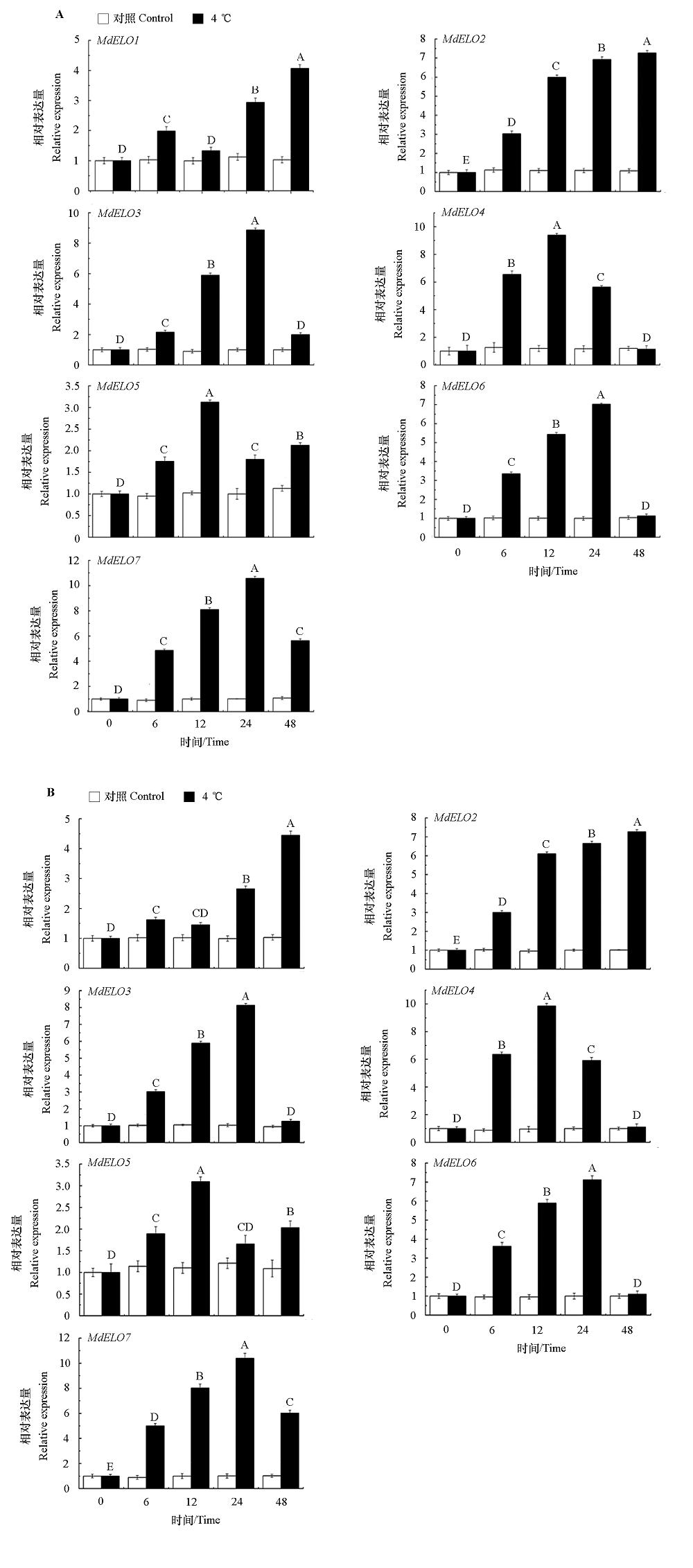
图9 低温胁迫下苹果砧木垂丝海棠根系(A)和叶片(B)中ELO基因的表达 图中不同大写字母表示不同处理组之间的显著差异(P < 0.01)。
Fig. 9 Expression analysis of apple MhELO gene family in roots(A)and leaves(B)under low temperature stress Different uppercase letters represent significant differences between treatment groups under different root treatment times(P < 0.01).
| [1] |
Achuba F I, Okoh P N. 2015. Effects of petroleum products in soil on α-amylase,starch phosphorylase and peroxidase activities in cowpea and maize seedlings. American Journal of Experimental Agriculture, 6 (6):112-120.
doi: 10.9734/AJEA/2015/9750 URL |
| [2] |
Adamski N M, Bush M S, Simmonds J, Turner A S, Mugford S G, Jones A, Findlay K, Pedentchouk N, Uauy C. 2013. The inhibitor of wax 1 locus(Iw1)prevents formation of β-and OH-β-diketones in wheat cuticular waxes and maps to a sub-cM interval on chromosome arm 2BS. The Plant Journal, 74 (6):989-1002.
doi: 10.1111/tpj.12185 URL |
| [3] |
Belding R D, Blankenship S M, Young E, Leidy R B. 1998. Composition and variability of epicuticular waxes in apple cultivars. Journal of the American Society for Horticultural Science, 123 (3):348-356.
doi: 10.21273/JASHS.123.3.348 URL |
| [4] |
Cahoon E B, Shockey J M, Dietrich C R, Gidda S K, Mullen R T, Dyer J M. 2007. Engineering oilseeds for sustainable production of industrial and nutritional feedstocks:solving bottlenecks in fatty acid flux. Current Opinion in Plant Biology, 10 (3):236-244.
pmid: 17434788 |
| [5] |
Cai Xiao-feng, Zhang Yu-yang. 2013. Genome-wide analysis of plant-specific dof transcription factor family in tomato. Journal of Integrative Plant Biology, 55 (6):552-566. (in Chinese)
doi: 10.1111/jipb.12043 URL |
| 蔡晓峰, 张玉阳. 2013. 番茄植物特异性多夫转录因子家族的全基因组分析. 植物生物学综合杂志, 55 (6):552-566. | |
| [6] |
Cinti D L, Cook L, Nagi M N, Suneja S K. 1992. The fatty acid chain elongation system of mammalian endoplasmic reticulum. Progress in Lipid Research, 31 (1):1-51.
pmid: 1641395 |
| [7] |
Dietbert E, Jorg J, Sauter. 2000. Seasonal changes of activity of a starch granule bound endoamylase and of a starch phosphorylase in poplar wood (Populus × canadensis Moench‘robusta’)and their possible regulation by temperature and phytohormones. Journal of Plant Physiology, 156 (5):731-740.
doi: 10.1016/S0176-1617(00)80239-4 URL |
| [8] |
Eigenbrode S D, Espelie K E. 1995. Effects of plant epicuticular lipids on insect herbivores. Annual Review of Entomology, 40 (1):171-194.
doi: 10.1146/annurev.en.40.010195.001131 URL |
| [9] | Fehling E, Mukherjee K D. 1991. Acyl-CoA elongase from a higher plant(Lunaria annua):metabolic intermediates of very-long chain acyl-CoA products and substrate specificity. Biochimica Biophysica Acta(BBA)-Lipids and Lipid Metabolism, 1082 (3):239-246. |
| [10] |
Jaganathan G K. 2017. Physiological mechanisms only tell half story:multiple biological processes are involved in regulating freezing tolerance of imbibed Lactuca sativa seeds. Scientific Reports, 7 (1):44166.
doi: 10.1038/srep44166 URL |
| [11] |
Jee E R, Peter M P, William W, Van O. 2007. Using the method of homogenization to calculate the effective diffusivity of the stratum corneum. Journal of Membrane Science, 293 (1):174-182.
doi: 10.1016/j.memsci.2007.02.018 URL |
| [12] |
Jin Z, Chandrasekaran U, Liu A. 2014. Genome-wide analysis of the dof transcription factors in castor bean(Ricinus communis L.). Genes and Genomics, 36 (4):527-537.
doi: 10.1007/s13258-014-0189-6 URL |
| [13] | Kazuyoshi T, Kiyohiro N, Ken S. 1978. Difference between the size of waxy and non-waxy kernel in the F1 rice plant.:unbalanced growth in floral glumes and caryopsis in rice. Japanese Journal of Breeding, 28 (3):225-233. |
| [14] |
Kohlwein S D, Eder S, Oh C S, Martin C E, Gable K, Bacikova D, Dunn T. 2001. Tsc13p is required for fatty acid elongation and localizes to a novel structure at the nuclear-vacuolar interface in SacChromyces cerevisiae. Molecular and Cellular Biology, 21 (1):109-125.
pmid: 11113186 |
| [15] | Krah C Y, Kumah P, Mardjan S, Njume A C. 2020. Effect of different threshing methods on the physical chracteristics of two varieties of paddy rice. IOP Conference Series: Earth and Environmental Science, 542 (1): 012022 (9pp). |
| [16] | Lassner M W, Lardizabal K, Metz J G. 1996. A jojoba beta-Ketoacyl-CoA synthase cDNA complements the canola fatty acid elongation mutation in transgenic plants. Th e Plant Cell, 8 (2):281-292. |
| [17] | Lescot M. 2002. Plant CARE,a database of plant cis-acting regulatory elements and a portal to tools for in silico analysis of promoter sequences. Nucleic Acids Research, 30 (1):325-327. |
| [18] | Makarenko S P, Konenkina T A, Salyaev R K. 2000. Chracteristics of fatty acid composition of lipids in higher plant vacuolar membranes. Membrane & Cell Biology, 13 (5):687-695. |
| [19] |
Millar A A, Clemens S, Zachgo S, Giblin E M, Taylor D C, Kunst L. 1999. CUT1,an arabidopsis gene required for cuticular wax biosynthesis and pollen fertility,encodes a very-long-chain fatty acid condensing enzyme. The Plant Cell, 11 (5):825-838.
doi: 10.1105/tpc.11.5.825 URL |
| [20] |
Millar A A, Kunst L. 1997. Very-long-chain fatty acid biosynthesis is controlled through the expression and specificity of the condensing enzyme. The Plant Journal:for Cell and Molecular Biology, 12 (1):121-31.
doi: 10.1046/j.1365-313X.1997.12010121.x URL |
| [21] |
Nijhawan A, Jain M, Tyagi A K, Khurana J P. 2008. Genomic survey and gene expression analysis of the basic leucine zipper transcription factor family in rice. Plant Physiology, 146 (2):333-350.
doi: 10.1104/pp.107.112821 pmid: 18065552 |
| [22] | Riederer M, Schneider G. 1990. The effect of the environment on the permeability and composition of citrus leaf cuticles:II. Composition of soluble cuticular lipids and correlation with transport properties. Planta, 180 (2):147-153. |
| [23] |
Shepherd T, Griffiths D W. 2006. The effects of stresson plant cuticular waxes. New Phytol, 171 (3):469-499.
pmid: 16866954 |
| [24] | Sun Bai-lin, Ma Li, Zeng Xiu-cun, Niu Zao-xia, Wang Wan-peng, Hu Fang-di, Lu Xiao-ming, Qi Wei-liang, Wu Jun-yan, Liu Li-jun, Li Xue-cai, Sun Wan-cang, Liao Chong-qing, Wang Xue-fang, Li Xiao-ze, Fang Yan. 2021. Differential expression analysis of fatty acid components and FAD 3 in winter rapeseed under low temperature stress. Agricultural Research in the Arid Areas, 39 (1):65-74. (in Chinese) |
| 孙柏林, 马骊, 曾秀存, 牛早霞, 王万鹏, 呼芳娣, 路晓明, 祁伟亮, 武军艳, 刘丽君, 李学才, 孙万仓, 缪纯庆, 王学芳, 李孝泽, 方彦. 2021. 低温胁迫下白菜型冬油菜脂肪酸组分及FAD3的差异表达分析. 干旱地区农业研究, 39 (1):65-74. | |
| [25] |
Tieman D M, Klee H J. 1999. Differential expression of two novel members of the tomato ethylene-receptor family. Plant Physiology, 120 (1):165-172.
pmid: 10318694 |
| [26] |
Tory A, Hendry, Joseph W, Brown, Geordan T. 2010. Angiosperm phylogeny inferred from sequences of four mitochondrial genes. Journal of Systematics and Evolution, 48 (6):391-425.
doi: 10.1111/j.1759-6831.2010.00097.x URL |
| [27] |
Velasco R, Zharkikh A, Affourtit J, Dhingra A, Cestaro A, Kalyanaraman A, Fontana P, Bhatnagar S K, Troggio M, Pruss D. 2010. The genome of the domesticated apple(Malus domestica Borkh.). Nature Genetics, 42:833-839.
doi: 10.1038/ng.654 URL |
| [28] |
Wang Q, Cheng T R, Yu X N, Jaime A, Teixeira D S, Byrne D H. 2014. Physiological and biochemical responses of six herbaceous peony cultivars to cold stress. South African Journal of Botany, 94:140-148.
doi: 10.1016/j.sajb.2014.05.012 URL |
| [29] | Wilhelm B, Christoph N, David C. 2008. Classification and terminology of plant epicuticular waxes. Botanical Journal of the Linnean Society,(3):237-260. |
| [30] | Xie Feng-lian. 2019. Comparison of expression differences of related genes in brown adipose tissue of yak in different seasons and comparison of fatty acid composition[M. D. Dissertation]. Xining: Qinghai University. (in Chinese) |
| 谢凤莲. 2019. 不同季节牦牛机体棕色脂肪组织相关基因表达差异及其脂肪酸组成成分的比较[硕士论文]. 西宁: 青海大学. | |
| [31] | Xu Guang-ping, Zhang Zhi-yong, Ren Zhong-hong, Zhang Zhi-wei. 2021. Bioinformatics analysis of the structure and function of black sea bream ELO gene-encoded protein. Advances in Biotechnology, 11 (3):344-352. (in Chinese) |
| 许广平, 张志勇, 任忠宏, 张志伟. 2021. 黑鲷ELO基因编码蛋白结构与功能的生物信息学分析. 生物技术进展, 11 (3):344-352. | |
| [32] | Xu J Y, Xue C C, Xue D, Zhao J M, Gai J Y, Guo N, Xing H, Swati P D. 2013a. Overexpression of GmHsp90s,a heat shock protein 90(HSP90)gene family cloning fromsoybean,decrease damage of abiotic stresses in Arabidopsis thaliana. PLoS ONE, 8 (7):e69810. |
| [33] |
Xu Q, Chen L L, Ruan X A, Chen D J, Zhu A D, Chen C L, Denis B, Jiao W B, Hao B H, Matthew P L, Chen J J, Gao S, Xing F, Lan H, Chang J W, Ge X H, Yang L, Hu Q, Miao Y, Wang L, Xiao S X, Manosh K B, Zeng W F, Guo F, Cao H B, Yang X M, Xu X W, Cheng Y J, Xu J, Liu J H, Oscar J L, Tang Z H, Guo W W, Kuang H H, Zhang H Y, Mikeal L R, Niranjan N, Deng X X, Ruan Y J. 2013b. The draft genome of sweet orange(Citrus sinensis). Nature genetics, 45 (1):59-66.
doi: 10.1038/ng.2472 URL |
| [34] | Yang Jie, Jiang Li, Zhang Jioajiao, Chen Ziping. 2013. Responses of the drought-resistent vem 1 mutant to NaCl and ABA stress in Arabidopsis. Plant Physiology Journal, 49 (4):337-342. (in Chinese) |
| 杨杰, 江力, 张娇娇, 陈子平. 2013. 拟南芥抗旱突变体vem1对NaCl和ABA胁迫的响应. 植物生理学报, 49 (4):337-342. | |
| [35] | Yang Ning-ning, Sun Wan-cang, Liu Zi-gang, Shi Peng-hui, Fang Yan, Wu Jun-yan, Zeng Xiu-cun, Kong De-jing, Lu Mei-hong, Wang Yue. 2014. Morphological and physiological mechanism of cold resistance in winter rape in northern China. Chinese Agricultural Sciences, 47 (3):452-461. (in Chinese) |
| 杨宁宁, 孙万仓, 刘自刚, 史鹏辉, 方彦, 武军艳, 曾秀存, 孔德晶, 鲁美宏, 王月. 2014. 北方冬油菜抗寒性的形态与生理机制. 中国农业科学, 47 (3):452-461. | |
| [36] | Yao Q M, Bai Y L, Kumar S, Au E, Orfao A, Chim C S. 2021. Minimal residual disease detection by next-generation sequencing in multiple myeloma:a comparison with real-time quantitative PCR. Frontiers in Oncology, 10:611021. |
| [37] | Yoshida K, Komae K. 2006. A rice family 9 glycoside hydrolase isozyme with broad substrate specificity for hemicelluloses in type II cell walls. Plant & Cell Physiology, 47 (11):1541-1554. |
| [38] |
Zhang K, Marina K, Han M, Li W, Yu Z Y, Yang Z L, Li Y, Michael L, Metzker, Rando A, Donald J Z, Laura E K, Pamela S L, Paul W W, Ian M M, Paul A S, David J F, Christopher P A, Robert J G, Radha A, Konstantin P. 2001. A 5-bp deletion in ELOVL 4 is associated with two related forms of autosomal dominant macular dystrophy. Nat Genet, 27 (1):89-93.
pmid: 11138005 |
| [39] | Zhou Jing-wen, Ding Yu-jiao, Zeng Xiao-rong, Ganesh K Jaganathan. 2020. Study on the relationship between freeze tolerance and fatty acid synthesis and metabolism of lettuce seeds after low temperature treatment. Subtropical Plant Science, 49 (1):9-14. (in Chinese) |
| 周婧雯, 丁玉娇, 曾晓蓉, Ganesh K Jaganathan. 2020. 生菜种子低温处理后耐冻性和脂肪酸合成代谢的关系研究. 亚热带植物科学, 49 (1):9-14. |
| [1] | 于婷婷, 李 欢, 宁源生, 宋建飞, 彭璐琳, 贾竣淇, 张玮玮, 杨洪强. 苹果GRAS全基因组鉴定及其对生长素的响应分析[J]. 园艺学报, 2023, 50(2): 397-409. |
| [2] | 赵雪艳, 王琪, 王莉, 王方圆, 王庆, 李艳. 基于比较转录组的延胡索组织差异性表达分析[J]. 园艺学报, 2023, 50(1): 177-187. |
| [3] | 韩晓蕾, 张彩霞, 刘 锴, 杨 安, 严家帝, 李武兴, 康立群, 丛佩华. 中熟苹果新品种‘中苹优蕾’[J]. 园艺学报, 2022, 49(S2): 1-2. |
| [4] | 韩晓蕾, 张彩霞, 刘 锴, 严家帝, 李武兴, 康立群, 丛佩华. 中熟苹果新品种‘苹优2号’[J]. 园艺学报, 2022, 49(S1): 1-2. |
| [5] | 王 强, 丛佩华, 刘肖烽. 晚熟苹果新品种‘华优甜娃’[J]. 园艺学报, 2022, 49(S1): 3-4. |
| [6] | 王 强, 丛佩华, 刘肖烽. 中熟苹果新品种‘华优宝蜜’[J]. 园艺学报, 2022, 49(S1): 5-6. |
| [7] | 杨 玲, 丛佩华, 王 强, 李武兴, 康立群. 中熟鲜食苹果新品种‘华丰’[J]. 园艺学报, 2022, 49(S1): 7-8. |
| [8] | 丁志杰, 包金波, 柔鲜古丽, 朱甜甜, 李雪丽, 苗浩宇, 田新民. 新疆野苹果与‘元帅’‘金冠’的叶绿体基因组比对研究[J]. 园艺学报, 2022, 49(9): 1977-1990. |
| [9] | 邱子文, 刘林敏, 林永盛, 林晓洁, 李永裕, 吴少华, 杨超. 千层金MbEGS基因的克隆与功能分析[J]. 园艺学报, 2022, 49(8): 1747-1760. |
| [10] | 郑晓东, 袭祥利, 李玉琪, 孙志娟, 马长青, 韩明三, 李少旋, 田义轲, 王彩虹. 油菜素内酯对盐碱胁迫下平邑甜茶幼苗生长的影响及调控机理研究[J]. 园艺学报, 2022, 49(7): 1401-1414. |
| [11] | 夏炎, 黄松, 武雪莉, 刘一琪, 王苗苗, 宋春晖, 白团辉, 宋尚伟, 庞宏光, 焦健, 郑先波. 基于宏病毒组测序技术的苹果病毒病鉴定与分析[J]. 园艺学报, 2022, 49(7): 1415-1428. |
| [12] | 郑林, 王帅, 刘语诺, 杜美霞, 彭爱红, 何永睿, 陈善春, 邹修平. 柑橘响应黄龙病菌侵染的NAC基因的克隆及表达分析[J]. 园艺学报, 2022, 49(7): 1441-1457. |
| [13] | 刘照霞, 张鑫, 王璐, 马玉婷, 陈倩, 朱占玲, 葛顺峰, 姜远茂. 肥料穴施位点对苹果细根生长、15N吸收利用及产量品质的影响[J]. 园艺学报, 2022, 49(7): 1545-1556. |
| [14] | 马维峰, 李艳梅, 马宗桓, 陈佰鸿, 毛娟. 苹果POD家族基因的鉴定与MdPOD15的功能分析[J]. 园艺学报, 2022, 49(6): 1181-1199. |
| [15] | 张凯, 麻明英, 王萍, 李益, 金燕, 盛玲, 邓子牛, 马先锋. 柑橘HSP20家族基因鉴定及其响应溃疡病菌侵染表达分析[J]. 园艺学报, 2022, 49(6): 1213-1232. |
| 阅读次数 | ||||||
|
全文 |
|
|||||
|
摘要 |
|
|||||
版权所有 © 2012 《园艺学报》编辑部 京ICP备10030308号-2 国际联网备案号 11010802023439
编辑部地址: 北京市海淀区中关村南大街12号中国农业科学院蔬菜花卉研究所 邮编: 100081
电话: 010-82109523 E-Mail: yuanyixuebao@126.com
技术支持:北京玛格泰克科技发展有限公司