
Acta Horticulturae Sinica ›› 2025, Vol. 52 ›› Issue (3): 603-622.doi: 10.16420/j.issn.0513-353x.2023-0974
• Genetic & Breeding·Germplasm Resources·Molecular Biology • Previous Articles Next Articles
SUN Tingzhen1, SHU Qin1, MA Wei1, SHI Yuzi1, ZHANG Meng1, XIANG Chenggang2, BO Kailiang1, DUAN Ying1,*( ), WANG Changlin1,*(
), WANG Changlin1,*( )
)
Received:2024-10-10
Revised:2025-01-09
Online:2025-03-25
Published:2025-03-25
Contact:
DUAN Ying, WANG Changlin
SUN Tingzhen, SHU Qin, MA Wei, SHI Yuzi, ZHANG Meng, XIANG Chenggang, BO Kailiang, DUAN Ying, WANG Changlin. Identification and Phylogenetic Analysis of Gibberellin Oxidase Gene Family and Its Expression Pattern in Main Vine Growth in Cucurbita maxima[J]. Acta Horticulturae Sinica, 2025, 52(3): 603-622.
Add to citation manager EndNote|Ris|BibTeX
URL: https://www.ahs.ac.cn/EN/10.16420/j.issn.0513-353x.2023-0974
| 引物名称 Primer name | 上游引物(5′-3′) Forward primer | 下游引物(5′-3′) Reverse primer |
|---|---|---|
| CmaGA2ox1 | GGCGATAAGGGCTGGATCGAATATCTC | ACCACTTCGCCGCTCAATTTCTTC |
| CmaGA2ox2 | CTACAGGCTTGGAGCAACGA | GGCAACTCCCGATTCCAGAA |
| CmaGA2ox3 | TGGCGTTCCCATGGAA TTCA | TATTCAACCCAGCCCACGTC |
| CmaGA2ox4 | AGCACGGAATTGGGGTTTCT | CACCGGTAAGTTCCACCCAA |
| CmaGA2ox5 | TGAAGAAGCTGAGCTGCGAA | AGCTTGGAGTTCCGGACATG |
| CmaGA2ox6 | GTTGTGGACACCTGTTTGCC | ATTCAATGGTGGTGCCCCAA |
| CmaGA2ox7 | TCCCGAGATTTTCCGGTGTG | ACAGCCTACTCAATGCGCTT |
| CmaGA2ox8 | ACAGCCGTTTGAGCGTAAGA | ATCATCGGAGCCGTTGGA TC |
| CmaGA2ox9 | ATGGCTGAGGGGTTAGGGAT | ACCCAAGCTTATTGGAGGGC |
| CmaGA2ox10 | AAAACAGAGAGCTGGCCCTC | ATGGAGAGGAACCCATCGGA |
| CmaGA2ox11 | TGGCTGGAACTTACCGATGG | GTGGGTCTGAGGGTGACATG |
| CmaGA2ox12 | TTTGGGGAGCATTCTGACCC | ACGTAGAATGCAGACGGGTC |
| CmaGA2ox13 | CCAGCCGACCCATTTGGATA | TCCACCGCACACCTGAAAAT |
| CmaGA20ox1 | GTGTTCGTCGACAACCAATGGAGAC | TCTGAGTGAACCGGAGGAGCATC |
| CmaGA20ox2 | CGACCCAACATCCCTCACCATTCT | ATCAGGGTACTGTCTTGGCCCTT |
| CmaGA20ox5 | GTAGACAACCAATGGCGTCTCATTGC | ATGTCAGCTCTGTAATGGTTGAGAGTG |
| CmaGA20ox10 | GGCTTTCAAGTGTGCGTCGATGATC | TCAAGCAACGTTCCCCATGTGTAATC |
| CmaGA20ox12 | CATCGCATGACAAAGTGGTGAGAGC | ATGGCAAGATCTCGGAAACTGGGTTC |
| CmaGA3ox1 | GATCCTCCGCAGAACATGCTTCTCA | TGCTGCTTTGGTGGAGGATTGTC |
| CmaGA3ox4 | GATTCAACGTCATAGGTTCTCCGGTTG | GTGTCTGGTGAAGCATGGTGAAGA |
| CmEF1a | ACGGTGATGCTGGTATGGTTA | CATTGTTGTTGGTTGGCTTATT |
Table 1 Primers of qRT-PCR
| 引物名称 Primer name | 上游引物(5′-3′) Forward primer | 下游引物(5′-3′) Reverse primer |
|---|---|---|
| CmaGA2ox1 | GGCGATAAGGGCTGGATCGAATATCTC | ACCACTTCGCCGCTCAATTTCTTC |
| CmaGA2ox2 | CTACAGGCTTGGAGCAACGA | GGCAACTCCCGATTCCAGAA |
| CmaGA2ox3 | TGGCGTTCCCATGGAA TTCA | TATTCAACCCAGCCCACGTC |
| CmaGA2ox4 | AGCACGGAATTGGGGTTTCT | CACCGGTAAGTTCCACCCAA |
| CmaGA2ox5 | TGAAGAAGCTGAGCTGCGAA | AGCTTGGAGTTCCGGACATG |
| CmaGA2ox6 | GTTGTGGACACCTGTTTGCC | ATTCAATGGTGGTGCCCCAA |
| CmaGA2ox7 | TCCCGAGATTTTCCGGTGTG | ACAGCCTACTCAATGCGCTT |
| CmaGA2ox8 | ACAGCCGTTTGAGCGTAAGA | ATCATCGGAGCCGTTGGA TC |
| CmaGA2ox9 | ATGGCTGAGGGGTTAGGGAT | ACCCAAGCTTATTGGAGGGC |
| CmaGA2ox10 | AAAACAGAGAGCTGGCCCTC | ATGGAGAGGAACCCATCGGA |
| CmaGA2ox11 | TGGCTGGAACTTACCGATGG | GTGGGTCTGAGGGTGACATG |
| CmaGA2ox12 | TTTGGGGAGCATTCTGACCC | ACGTAGAATGCAGACGGGTC |
| CmaGA2ox13 | CCAGCCGACCCATTTGGATA | TCCACCGCACACCTGAAAAT |
| CmaGA20ox1 | GTGTTCGTCGACAACCAATGGAGAC | TCTGAGTGAACCGGAGGAGCATC |
| CmaGA20ox2 | CGACCCAACATCCCTCACCATTCT | ATCAGGGTACTGTCTTGGCCCTT |
| CmaGA20ox5 | GTAGACAACCAATGGCGTCTCATTGC | ATGTCAGCTCTGTAATGGTTGAGAGTG |
| CmaGA20ox10 | GGCTTTCAAGTGTGCGTCGATGATC | TCAAGCAACGTTCCCCATGTGTAATC |
| CmaGA20ox12 | CATCGCATGACAAAGTGGTGAGAGC | ATGGCAAGATCTCGGAAACTGGGTTC |
| CmaGA3ox1 | GATCCTCCGCAGAACATGCTTCTCA | TGCTGCTTTGGTGGAGGATTGTC |
| CmaGA3ox4 | GATTCAACGTCATAGGTTCTCCGGTTG | GTGTCTGGTGAAGCATGGTGAAGA |
| CmEF1a | ACGGTGATGCTGGTATGGTTA | CATTGTTGTTGGTTGGCTTATT |
| 基因名称 Gene name | 基因ID Gene ID | 位置 Location | 氨基酸长 Number of amino acids | 正负链 Strand | 等电点 pI | 分子量/kD Molecular weight | 疏水性 Hydro- phobicity | 亚细胞定位 Subcellular localization |
|---|---|---|---|---|---|---|---|---|
| CmaGA20ox1 | CmaCh02G010210 | 6058004 ~ 6060196 | 378 | - | 6.17 | 43.18 | -0.42 | Mb |
| CmaGA20ox2 | CmaCh03G011090 | 7574527 ~ 7576330 | 364 | - | 7.10 | 41.07 | -0.31 | Mb |
| CmaGA20ox3 | CmaCh07G008110 | 3735328 ~ 3737605 | 369 | - | 6.57 | 42.00 | -0.41 | Mb |
| CmaGA20ox4 | CmaCh15G006420 | 3035007 ~ 3036820 | 351 | + | 6.73 | 39.48 | -0.20 | Mb |
| CmaGA20ox5 | CmaCh15G013930 | 8764141 ~ 8766571 | 361 | - | 5.85 | 40.96 | -0.28 | Mb |
| CmaGA20ox6 | CmaCh16G001620 | 703878 ~ 705410 | 386 | - | 6.25 | 44.19 | -0.43 | Nu |
| CmaGA20ox7 | CmaCh17G000020 | 9087 ~ 10692 | 336 | + | 5.45 | 38.17 | -0.36 | Mb |
| CmaGA20ox8 | CmaCh18G012060 | 9561682 ~ 9563098 | 385 | + | 5.36 | 43.50 | -0.32 | Chl |
| CmaGA20ox9 | CmaCh18G012070 | 9565859 ~ 9567931 | 295 | + | 6.12 | 33.49 | -0.43 | Chl |
| CmaGA20ox10 | CmaCh18G012080 | 9570626 ~ 9572255 | 384 | + | 5.33 | 43.27 | -0.42 | Cy |
| CmaGA20ox11 | CmaCh18G012090 | 9574376 ~ 9575714 | 386 | + | 5.47 | 43.32 | -0.39 | Cy |
| CmaGA20ox12 | CmaCh18G012100 | 9579614 ~ 9583282 | 427 | + | 5.82 | 48.00 | -0.31 | Cy |
| CmaGA3ox1 | CmaCh05G006480 | 3289026 ~ 3290688 | 370 | + | 6.75 | 41.32 | -0.10 | Er |
| CmaGA3ox2 | CmaCh05G006490 | 3293066 ~ 3294672 | 367 | + | 6.03 | 41.37 | -0.13 | Cy |
| CmaGA3ox3 | CmaCh05G006500 | 3297430 ~ 3298596 | 346 | + | 6.67 | 38.94 | -0.20 | Mb |
| CmaGA3ox4 | CmaCh05G006510 | 3304268 ~ 3307142 | 383 | + | 6.13 | 43.31 | -0.19 | Cy |
| CmaGA3ox5 | CmaCh05G006520 | 3320084 ~ 3321250 | 344 | + | 6.50 | 38.77 | -0.21 | Cy |
| CmaGA3ox6 | CmaCh05G006530 | 3325577 ~ 3326736 | 348 | + | 6.22 | 39.53 | -0.22 | Cy |
| CmaGA3ox7 | CmaCh08G006320 | 3573502 ~ 3574908 | 377 | + | 6.62 | 41.73 | -0.21 | Mb |
| CmaGA3ox8 | CmaCh12G003100 | 1581698 ~ 1584654 | 348 | + | 5.72 | 39.17 | -0.23 | Cy |
| CmaGA3ox9 | CmaCh17G008310 | 6527065 ~ 6528927 | 342 | + | 7.27 | 38.55 | -0.26 | Mb |
| CmaGA2ox1 | CmaCh03G013990 | 8817543 ~ 8818816 | 331 | + | 6.27 | 36.85 | -0.31 | Cy |
| CmaGA2ox2 | CmaCh04G000330 | 191407 ~ 194668 | 374 | - | 6.70 | 41.93 | -0.30 | Cy |
| CmaGA2ox3 | CmaCh04G006490 | 3321297 ~ 3322998 | 340 | - | 8.31 | 37.75 | -0.20 | Chl |
| CmaGA2ox4 | CmaCh04G009310 | 4804076 ~ 4807014 | 360 | + | 5.70 | 41.35 | -0.31 | Cy |
| CmaGA2ox5 | CmaCh07G001050 | 581638 ~ 582804 | 332 | - | 5.76 | 36.82 | -0.21 | Mit |
| CmaGA2ox6 | CmaCh08G004850 | 2762847 ~ 2764115 | 331 | + | 8.75 | 36.59 | -0.22 | Mb |
| CmaGA2ox7 | CmaCh13G005360 | 5582531 ~ 5584247 | 336 | - | 5.96 | 37.73 | -0.16 | Cy |
| CmaGA2ox8 | CmaCh13G007180 | 6423868 ~ 6426404 | 341 | - | 5.96 | 38.51 | -0.28 | Mb |
| CmaGA2ox9 | CmaCh14G009480 | 4909702 ~ 4911273 | 335 | + | 5.89 | 37.09 | -0.10 | Chl |
| CmaGA2ox10 | CmaCh16G004530 | 2269857 ~ 2271606 | 327 | + | 7.02 | 36.21 | -0.23 | Chl |
| CmaGA2ox11 | CmaCh16G008420 | 4911602 ~ 4915537 | 349 | + | 6.07 | 40.10 | -0.44 | Cy |
| CmaGA2ox12 | CmaCh17G010160 | 7383663 ~ 7385904 | 329 | - | 8.70 | 36.45 | -0.26 | Mb |
| CmaGA2ox13 | CmaCh18G004710 | 2653034 ~ 2655073 | 336 | + | 6.14 | 37.85 | -0.20 | Cy |
Table 2 Information of GA2ox、GA3ox and GA20ox gene families in Cucurbita maxima genome
| 基因名称 Gene name | 基因ID Gene ID | 位置 Location | 氨基酸长 Number of amino acids | 正负链 Strand | 等电点 pI | 分子量/kD Molecular weight | 疏水性 Hydro- phobicity | 亚细胞定位 Subcellular localization |
|---|---|---|---|---|---|---|---|---|
| CmaGA20ox1 | CmaCh02G010210 | 6058004 ~ 6060196 | 378 | - | 6.17 | 43.18 | -0.42 | Mb |
| CmaGA20ox2 | CmaCh03G011090 | 7574527 ~ 7576330 | 364 | - | 7.10 | 41.07 | -0.31 | Mb |
| CmaGA20ox3 | CmaCh07G008110 | 3735328 ~ 3737605 | 369 | - | 6.57 | 42.00 | -0.41 | Mb |
| CmaGA20ox4 | CmaCh15G006420 | 3035007 ~ 3036820 | 351 | + | 6.73 | 39.48 | -0.20 | Mb |
| CmaGA20ox5 | CmaCh15G013930 | 8764141 ~ 8766571 | 361 | - | 5.85 | 40.96 | -0.28 | Mb |
| CmaGA20ox6 | CmaCh16G001620 | 703878 ~ 705410 | 386 | - | 6.25 | 44.19 | -0.43 | Nu |
| CmaGA20ox7 | CmaCh17G000020 | 9087 ~ 10692 | 336 | + | 5.45 | 38.17 | -0.36 | Mb |
| CmaGA20ox8 | CmaCh18G012060 | 9561682 ~ 9563098 | 385 | + | 5.36 | 43.50 | -0.32 | Chl |
| CmaGA20ox9 | CmaCh18G012070 | 9565859 ~ 9567931 | 295 | + | 6.12 | 33.49 | -0.43 | Chl |
| CmaGA20ox10 | CmaCh18G012080 | 9570626 ~ 9572255 | 384 | + | 5.33 | 43.27 | -0.42 | Cy |
| CmaGA20ox11 | CmaCh18G012090 | 9574376 ~ 9575714 | 386 | + | 5.47 | 43.32 | -0.39 | Cy |
| CmaGA20ox12 | CmaCh18G012100 | 9579614 ~ 9583282 | 427 | + | 5.82 | 48.00 | -0.31 | Cy |
| CmaGA3ox1 | CmaCh05G006480 | 3289026 ~ 3290688 | 370 | + | 6.75 | 41.32 | -0.10 | Er |
| CmaGA3ox2 | CmaCh05G006490 | 3293066 ~ 3294672 | 367 | + | 6.03 | 41.37 | -0.13 | Cy |
| CmaGA3ox3 | CmaCh05G006500 | 3297430 ~ 3298596 | 346 | + | 6.67 | 38.94 | -0.20 | Mb |
| CmaGA3ox4 | CmaCh05G006510 | 3304268 ~ 3307142 | 383 | + | 6.13 | 43.31 | -0.19 | Cy |
| CmaGA3ox5 | CmaCh05G006520 | 3320084 ~ 3321250 | 344 | + | 6.50 | 38.77 | -0.21 | Cy |
| CmaGA3ox6 | CmaCh05G006530 | 3325577 ~ 3326736 | 348 | + | 6.22 | 39.53 | -0.22 | Cy |
| CmaGA3ox7 | CmaCh08G006320 | 3573502 ~ 3574908 | 377 | + | 6.62 | 41.73 | -0.21 | Mb |
| CmaGA3ox8 | CmaCh12G003100 | 1581698 ~ 1584654 | 348 | + | 5.72 | 39.17 | -0.23 | Cy |
| CmaGA3ox9 | CmaCh17G008310 | 6527065 ~ 6528927 | 342 | + | 7.27 | 38.55 | -0.26 | Mb |
| CmaGA2ox1 | CmaCh03G013990 | 8817543 ~ 8818816 | 331 | + | 6.27 | 36.85 | -0.31 | Cy |
| CmaGA2ox2 | CmaCh04G000330 | 191407 ~ 194668 | 374 | - | 6.70 | 41.93 | -0.30 | Cy |
| CmaGA2ox3 | CmaCh04G006490 | 3321297 ~ 3322998 | 340 | - | 8.31 | 37.75 | -0.20 | Chl |
| CmaGA2ox4 | CmaCh04G009310 | 4804076 ~ 4807014 | 360 | + | 5.70 | 41.35 | -0.31 | Cy |
| CmaGA2ox5 | CmaCh07G001050 | 581638 ~ 582804 | 332 | - | 5.76 | 36.82 | -0.21 | Mit |
| CmaGA2ox6 | CmaCh08G004850 | 2762847 ~ 2764115 | 331 | + | 8.75 | 36.59 | -0.22 | Mb |
| CmaGA2ox7 | CmaCh13G005360 | 5582531 ~ 5584247 | 336 | - | 5.96 | 37.73 | -0.16 | Cy |
| CmaGA2ox8 | CmaCh13G007180 | 6423868 ~ 6426404 | 341 | - | 5.96 | 38.51 | -0.28 | Mb |
| CmaGA2ox9 | CmaCh14G009480 | 4909702 ~ 4911273 | 335 | + | 5.89 | 37.09 | -0.10 | Chl |
| CmaGA2ox10 | CmaCh16G004530 | 2269857 ~ 2271606 | 327 | + | 7.02 | 36.21 | -0.23 | Chl |
| CmaGA2ox11 | CmaCh16G008420 | 4911602 ~ 4915537 | 349 | + | 6.07 | 40.10 | -0.44 | Cy |
| CmaGA2ox12 | CmaCh17G010160 | 7383663 ~ 7385904 | 329 | - | 8.70 | 36.45 | -0.26 | Mb |
| CmaGA2ox13 | CmaCh18G004710 | 2653034 ~ 2655073 | 336 | + | 6.14 | 37.85 | -0.20 | Cy |

Fig. 4 Phylogenetic tree of GA2ox,GA3ox and GA20ox in different plants At:Arabidopsis;Cma:Cucurbita maxima;Pt:Populus trichocarpa;Cs:Camellia sinensis;Md:Malus × domestica;Vv:Vitis vinifera;Citrus sinensis:Sweet orange;Gm:Glycine max;Mt:Medicago truncatula;Cucumis sativus:Cucumber;Cla:Citrullus lanatus;Sb:Sorghum bicolor;Zm:Zea mays;Os:Oryza sativa;Pe:Phyllostachys edulis
| 串联复制基因对 Tandem duplication | Ka | Ks | Ka/Ks | 纯化选择 Purifying selection | 分化时间/Mya Divergence time |
|---|---|---|---|---|---|
| CmaGA20ox8/CmaGA20ox9 | 0.092 | 0.231 | 0.402 | 是 Yes | 7.70 |
| CmaGA20ox9/CmaGA20ox10 | 0.101 | 0.171 | 0.594 | 是 Yes | 5.69 |
| CmaGA20ox10/CmaGA20ox11 | 0.177 | 0.210 | 0.846 | 是 Yes | 6.99 |
| CmaGA20ox11/CmaGA20ox12 | 0.024 | 0.056 | 0.424 | 是 Yes | 1.87 |
| CmaGA3ox1/CmaGA3ox2 | 0.516 | 1.760 | 0.293 | 是 Yes | 58.65 |
| CmaGA3ox2/CmaGA3ox3 | 0.120 | 0.165 | 0.727 | 是 Yes | 5.50 |
| CmaGA3ox3/CmaGA3ox4 | 0.051 | 0.073 | 0.700 | 是 Yes | 2.45 |
| CmaGA3ox4/CmaGA3ox5 | 0.054 | 0.079 | 0.693 | 是 Yes | 2.62 |
| CmaGA3ox5/CmaGA3ox6 | 0.117 | 0.107 | 1.086 | 否 No | 3.58 |
Table 3 The Ka/Ks ratios and estimated divergence time for tandem duplication in Cucurbita maxima
| 串联复制基因对 Tandem duplication | Ka | Ks | Ka/Ks | 纯化选择 Purifying selection | 分化时间/Mya Divergence time |
|---|---|---|---|---|---|
| CmaGA20ox8/CmaGA20ox9 | 0.092 | 0.231 | 0.402 | 是 Yes | 7.70 |
| CmaGA20ox9/CmaGA20ox10 | 0.101 | 0.171 | 0.594 | 是 Yes | 5.69 |
| CmaGA20ox10/CmaGA20ox11 | 0.177 | 0.210 | 0.846 | 是 Yes | 6.99 |
| CmaGA20ox11/CmaGA20ox12 | 0.024 | 0.056 | 0.424 | 是 Yes | 1.87 |
| CmaGA3ox1/CmaGA3ox2 | 0.516 | 1.760 | 0.293 | 是 Yes | 58.65 |
| CmaGA3ox2/CmaGA3ox3 | 0.120 | 0.165 | 0.727 | 是 Yes | 5.50 |
| CmaGA3ox3/CmaGA3ox4 | 0.051 | 0.073 | 0.700 | 是 Yes | 2.45 |
| CmaGA3ox4/CmaGA3ox5 | 0.054 | 0.079 | 0.693 | 是 Yes | 2.62 |
| CmaGA3ox5/CmaGA3ox6 | 0.117 | 0.107 | 1.086 | 否 No | 3.58 |
| 大片段复制基因对 Segmental duplication | Ka | Ks | Ka/Ks | 纯化选择 Purifying selection | 分化时间/Mya Divergence ime |
|---|---|---|---|---|---|
| CmaGA20ox1/CmaGA20ox5 | 0.085 | 0.553 | 0.154 | 是 Yes | 18.45 |
| CmaGA20ox2/CmaGA20ox3 | 0.082 | 0.872 | 0.094 | 是 Yes | 29.08 |
| CmaGA20ox6/CmaGA20ox8 | 0.185 | 0.462 | 0.400 | 是 Yes | 15.40 |
| CmaGA2ox1/CmaGA2ox13 | 0.262 | 1.876 | 0.140 | 是 Yes | 62.54 |
| CmaGA2ox1/CmaGA2ox6 | 0.515 | NaN | NaN | 否 No | — |
| CmaGA2ox6/CmaGA2ox13 | 0.448 | 2.994 | 0.149 | 是 Yes | 99.80 |
| CmaGA2ox3/CmaGA2ox10 | 0.047 | 0.533 | 0.089 | 是 Yes | 17.77 |
| CmaGA2ox4/CmaGA2ox11 | 0.116 | 0.416 | 0.280 | 是 Yes | 13.87 |
| CmaGA2ox12/CmaGA2ox5 | 0.538 | NaN | NaN | 否 No | — |
| CmaGA3ox1/CmaGA3ox8 | 0.429 | 1.563 | 0.274 | 是 Yes | 52.10 |
Table 4 The Ka/Ks ratios and estimated divergence time for segmental duplication in Cucurbita maxima
| 大片段复制基因对 Segmental duplication | Ka | Ks | Ka/Ks | 纯化选择 Purifying selection | 分化时间/Mya Divergence ime |
|---|---|---|---|---|---|
| CmaGA20ox1/CmaGA20ox5 | 0.085 | 0.553 | 0.154 | 是 Yes | 18.45 |
| CmaGA20ox2/CmaGA20ox3 | 0.082 | 0.872 | 0.094 | 是 Yes | 29.08 |
| CmaGA20ox6/CmaGA20ox8 | 0.185 | 0.462 | 0.400 | 是 Yes | 15.40 |
| CmaGA2ox1/CmaGA2ox13 | 0.262 | 1.876 | 0.140 | 是 Yes | 62.54 |
| CmaGA2ox1/CmaGA2ox6 | 0.515 | NaN | NaN | 否 No | — |
| CmaGA2ox6/CmaGA2ox13 | 0.448 | 2.994 | 0.149 | 是 Yes | 99.80 |
| CmaGA2ox3/CmaGA2ox10 | 0.047 | 0.533 | 0.089 | 是 Yes | 17.77 |
| CmaGA2ox4/CmaGA2ox11 | 0.116 | 0.416 | 0.280 | 是 Yes | 13.87 |
| CmaGA2ox12/CmaGA2ox5 | 0.538 | NaN | NaN | 否 No | — |
| CmaGA3ox1/CmaGA3ox8 | 0.429 | 1.563 | 0.274 | 是 Yes | 52.10 |
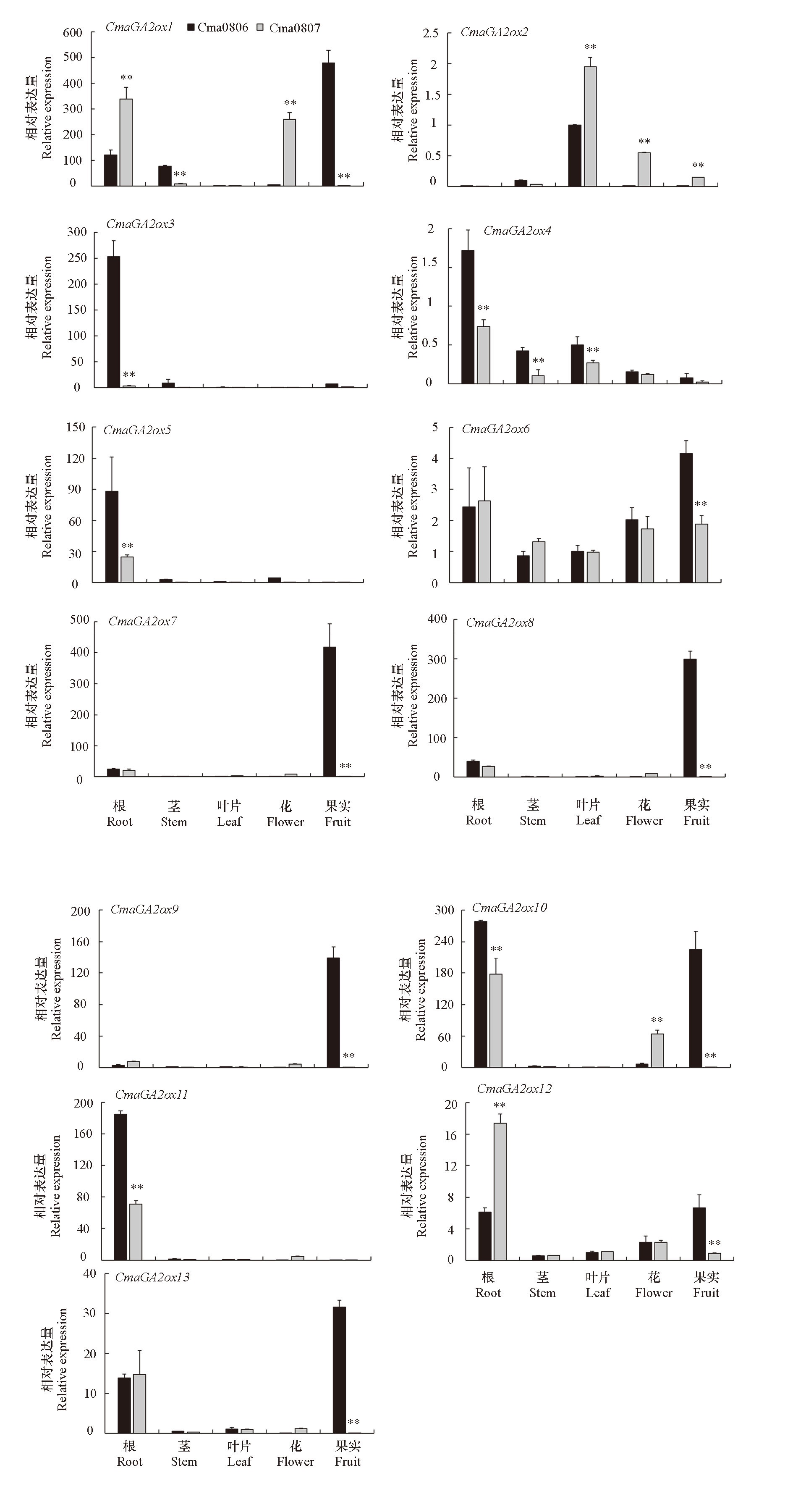
Fig. 9 Tissue-specific expression analysis of GA2ox gene family members in dwarf-vine germplasm Cma0806 and long-vine germplasm Cma0807 in Cucurbita maxima Student’s t-test was used to perform the significance analysis of gene expression data of different germplasm in the same tissue

Fig. 10 Tissue-specific expression analysis of GA3ox and GA20ox gene family members in dwarf-vine germplasm Cma0806 and long-vine germplasm Cma0807 in Cucurbita maxima Student’s t test was used to perform the significance analysis of gene expression data of different germplasm in the same tissue
| [1] |
|
| [2] |
|
| [3] |
|
| [4] |
|
| [5] |
|
| [6] |
|
|
董凤, 樊胜, 马小龙, 孟媛, 左希亚, 刘小杰, 李珂, 刘桢, 韩明玉, 张东. 2018. 苹果赤霉素氧化酶基因GA2ox、GA3ox和GA20ox家族全基因组鉴定及表达分析. 园艺学报, 45 (4):613-626.
doi: 10.16420/j.issn.0513-353x.2017-0452 |
|
| [7] |
doi: 10.1038/s41598-018-21293-1 pmid: 29440685 |
| [8] |
|
| [9] |
|
| [10] |
|
| [11] |
|
| [12] |
|
| [13] |
doi: 10.1016/s1360-1385(00)01790-8 pmid: 11120474 |
| [14] |
|
| [15] |
|
| [16] |
|
| [17] |
|
| [18] |
pmid: 8078921 |
| [19] |
|
| [20] |
|
| [21] |
|
| [22] |
doi: 10.1006/meth.2001.1262 pmid: 11846609 |
| [23] |
|
|
刘港运, 段世享, 许娜娜, 郭姚淼, 豆峻岭, 杨森, 牛欢欢, 刘东明, 杨路明, 胡建斌, 朱华玉. 2024. 分子标记辅助构建甜瓜矮化基因Cmerecta近等基因系. 园艺学报, 51 (9):2048-2062.
|
|
| [24] |
|
| [25] |
doi: 10.1093/jxb/erv300 pmid: 26093022 |
| [26] |
|
| [27] |
doi: 10.1111/pbi.12860 pmid: 29112324 |
| [28] |
|
| [29] |
doi: 10.1038/nbt847 pmid: 12858182 |
| [30] |
doi: 10.7505/j.issn.1007-9084.2019.03.012 |
|
盛晨, 张艳欣, 于景印, 高媛, 黎冬华, 周瑢, 张秀荣, 王林海. 2019. 芝麻赤霉素合成相关基因鉴定与表达分析. 中国油料作物学报, 41 (3):399-408.
|
|
| [31] |
|
| [32] |
|
| [33] |
|
| [34] |
|
| [35] |
|
| [36] |
doi: 10.1073/pnas.96.8.4698 pmid: 10200325 |
| [37] |
|
| [38] |
doi: 10.1038/s41477-022-01297-6 pmid: 36509843 |
| [39] |
|
| [40] |
|
| [41] |
|
| [42] |
|
|
向成钢. 2019. 印度南瓜矮生基因CmDw-1图位克隆及美洲南瓜矮生基因定位[博士论文]. 北京: 中国农业大学.
|
|
| [43] |
|
| [44] |
|
| [45] |
|
|
杨意宏, 王思宁, 徐浩, 孙化雨, 赵韩生, 陈段芬, 高志民. 2018. 毛竹赤霉素合成相关酶基因的全基因组鉴定和表达分析. 基因组学与应用生物学, 37 (9):3966-3977.
|
|
| [46] |
|
| [47] |
|
| [48] |
doi: 10.11869/j.issn.100-8551.2019.06.1096 |
|
朱海生, 刘建汀, 温文旭, 李永平, 王彬, 陈敏氡, 温庆放. 2019. 印度南瓜延伸因子基因CmEF1a的克隆与分析. 核农学报,(6):1096-1104.
doi: 10.11869/j.issn.100-8551.2019.06.1096 |
| [1] | WANG Shanshan, GUO Rui, HE Ling, WU Chunhong, CHEN Chanyou, WAN Heping, ZHAO Huixia. Identification of Long Cowpea Lhc Gene Family and Its Expression Analysis Under Salt Stress [J]. Acta Horticulturae Sinica, 2025, 52(1): 111-122. |
| [2] | LIU Jianhao, JING Yanfu, LIU Yuexin, XU Yaoguang, YU Yang, GE Xiuxiu, XIE Hua. Identification of Peach NAC Gene Family and Role of PpNAC050 in Promoting Fruit Fructose Accumulation [J]. Acta Horticulturae Sinica, 2024, 51(9): 1983-1996. |
| [3] | ZHAO Jiaying, ZENG Zhouting, CEN Xinying, SHI Jiaoqi, LI Xiaoxian, SHEN Xiaoxia, YU Zhenming. Genome-Wide Identification and Expression Analysis of CCO Gene Family in Dendrobium officinale During Flower Development [J]. Acta Horticulturae Sinica, 2024, 51(9): 2075-2088. |
| [4] | WANG Yanhong, XI Keyong, TIAN Ye, LIU Deqi, ZHOU Kegui, YIN Junliang, LIU Yiqing, ZHU Yongxing. Identification and Expression Pattern Analysis of GST in Zingiber officinale [J]. Acta Horticulturae Sinica, 2024, 51(8): 1803-1822. |
| [5] | CHEN Jiayue, DUAN Yingming, ZHOU Yan, XIAO Yang, BIAN Yinbing, GONG Yuhua. Identification,Expression and Function Analysis of ALDH Gene Family in Lentinula edodes [J]. Acta Horticulturae Sinica, 2024, 51(5): 1033-1046. |
| [6] | LIU Jinhong, WANG Zheng, YU Hao, XIN Yirui, QI Guoning, LIU Shenkui, REN Huimin. Identification of SLAC Gene Family in Phyllostachys edulis and Characterization of PheSLAC1 [J]. Acta Horticulturae Sinica, 2024, 51(3): 545-559. |
| [7] | LUO Xinrui, ZHANG Xiaoxu, WANG Yuping, WANG Zhi, MA Yuanyuan, ZHOU Bingyue. Identification and Expression Analysis of Trihelix Gene Family in Common Bean [J]. Acta Horticulturae Sinica, 2024, 51(12): 2775-2790. |
| [8] | HAN Shiwen, LIU Tao, WANG Liping, LI Nanyang, WANG Suna, WANG Xing. Genome-Wide Identification and Stress-Responsive Expression Analysis of the Cucumber SRS Gene Family [J]. Acta Horticulturae Sinica, 2024, 51(10): 2281-2296. |
| [9] | HUANG Zhihao, LIU Tingting, DONG Xujie, YAN Mingli, LIUZhixiang , ZENG Chaozhen. Identification and Expression Analysis of HMA Gene Family in Brassica juncea Under Cadmium Stress [J]. Acta Horticulturae Sinica, 2023, 50(6): 1230-1242. |
| [10] | HE Weizhi, LEI Weiqi, GUO Xiangxin, LI Ruilian, CHEN Guanqun. Identification of the MYB Gene Family and Functional Analysis of Key Genes Related to Blue Flower Coloration in Agapanthus praecox [J]. Acta Horticulturae Sinica, 2023, 50(6): 1255-1268. |
| [11] | YU Tingting, LI Huan, NING Yuansheng, SONG Jianfei, PENG Lulin, JIA Junqi, ZHANG Weiwei, YANG Hongqiang. Genome-wide Identification of GRAS Gene Family in Apple and Expression Analysis of Its Response to Auxin [J]. Acta Horticulturae Sinica, 2023, 50(2): 397-409. |
| [12] | SHEN Yuxiao, ZOU Jinyu, LUO Ping, SHANG Wenqian, LI Yonghua, HE Songlin, WANG Zheng, SHI Liyun. Genome-wide Identification and Abiotic Stress Response Analysis of PP2C Family Genes in Rosa chinensis‘Old Blush’ [J]. Acta Horticulturae Sinica, 2023, 50(10): 2139-2156. |
| [13] | ZHAI Hanhan, ZHAI Yujie, TIAN Yi, ZHANG Ye, YANG Li, WEN Zhiliang, CHEN Haijiang. Genome-wide Identification of Peach SAUR Gene Family and Characterization of PpSAUR5 Gene [J]. Acta Horticulturae Sinica, 2023, 50(1): 1-14. |
| [14] | GAO Yanlong, WU Yuxia, ZHANG Zhongxing, WANG Shuangcheng, ZHANG Rui, ZHANG De, WANG Yanxiu. Bioinformatics Analysis of Apple ELO Gene Family and Its Expression Analysis Under Low Temperature Stress [J]. Acta Horticulturae Sinica, 2022, 49(8): 1621-1636. |
| [15] | LIU Jinming, GUO Caihua, YUAN Xing, KANG Chao, QUAN Shaowen, NIU Jianxin. Genome-wide Identification of Dof Family Genes and Expression Analysis Sepal Persistent and Abscission in Pear [J]. Acta Horticulturae Sinica, 2022, 49(8): 1637-1649. |
| Viewed | ||||||
|
Full text |
|
|||||
|
Abstract |
|
|||||
Copyright © 2012 Acta Horticulturae Sinica 京ICP备10030308号-2 国际联网备案号 11010802023439
Tel: 010-82109523 E-Mail: yuanyixuebao@126.com
Support by: Beijing Magtech Co.Ltd