
Acta Horticulturae Sinica ›› 2024, Vol. 51 ›› Issue (2): 266-280.doi: 10.16420/j.issn.0513-353x.2023-0833
• Genetic&Breeding?Germplasm Resources?Molecular Biology • Previous Articles Next Articles
YAN Chaofan, SUN Xuemei, ZHONG Qiwen, SHAO Dengkui, DENG Changrong, WEN Junqin*( )
)
Received:2023-10-20
Revised:2023-12-25
Online:2024-02-25
Published:2024-02-26
Contact:
WEN Junqin
YAN Chaofan, SUN Xuemei, ZHONG Qiwen, SHAO Dengkui, DENG Changrong, WEN Junqin. Identification and Bioinformatics Analysis of 20S Proteasome Gene Family in Tomato[J]. Acta Horticulturae Sinica, 2024, 51(2): 266-280.
Add to citation manager EndNote|Ris|BibTeX
URL: https://www.ahs.ac.cn/EN/10.16420/j.issn.0513-353x.2023-0833
| 基因 Gene | 基因ID Gene ID | 染色体定位 Chromosome location | 基因位置/ bp Gene location | |
|---|---|---|---|---|
| 起始Start | 终止End | |||
| SlPRA1-01 | Solyc01g111450.3.1 | Chr01:97599172..97602337(+) | 97 599 172 | 97 602 337 |
| SlPRA2-01 | Solyc01g111460.3.1 | Chr01:97605332..97606609(-) | 97 605 332 | 97 606 609 |
| SlPRA3-02 | Solyc02g070510.3.1 | Chr02:40805105..40810898(+) | 40 805 105 | 40 810 898 |
| SlPRA4-02 | Solyc02g081700.1.1 | Chr02:46112367..46113113(+) | 46 112 367 | 46 113 113 |
| SlPRA5-02 | Solyc02g084920.3.1 | Chr02:48607717..48616195(-) | 48 607 717 | 48 616 195 |
| SlPRA6-03 | Solyc03g033745.1.1 | Chr03:5362233..5373114(-) | 5 362 233 | 5 373 114 |
| SlPRA7-03 | Solyc03g063420.2.1 | Chr03:36767871..36777655(-) | 36 767 871 | 36 777 655 |
| SlPRA8-04 | Solyc04g009410.3.1 | Chr04:2830248..2840325(+) | 2 830 248 | 2 840 325 |
| SlPRA9-04 | Solyc04g024420.3.1 | Chr04:31500067..31505977(+) | 31 500 067 | 31 505 977 |
| SlPRA10-04 | Solyc04g080590.3.1 | Chr04:64787899..64791729(-) | 64 787 899 | 64 791 729 |
| SlPRA11-05 | Solyc05g013820.3.1 | Chr05:7123832..7132561(-) | 7 123 832 | 7 132 561 |
| SlPRA12-05 | Solyc05g056160.3.1 | Chr05:66400334..66404473(-) | 66 400 334 | 66 404 473 |
| SlPRA13-07 | Solyc07g016200.3.1 | Chr07:6438876..6447830(-) | 6 438 876 | 6 447 830 |
| SlPRA14-07 | Solyc07g055080.3.1 | Chr07:63355928..63361757(+) | 63 355 928 | 63 361 757 |
| SlPRA15-08 | Solyc08g016510.3.1 | Chr08:7711628..7717929(-) | 7 711 628 | 7 717 929 |
| SlPRA16-09 | Solyc09g082320.3.1 | Chr09:68500118..68505978(-) | 68 500 118 | 68 505 978 |
| SlPRA17-10 | Solyc10g008010.3.1 | Chr10:2165246..2178357(+) | 2 165 246 | 2 178 357 |
| SlPRA18-10 | Solyc10g077030.2.1 | Chr10:60039871..60049610(-) | 60 039 871 | 60 049 610 |
| SlPRA19-10 | Solyc10g081130.2.1 | Chr10:62399725..62404172(+) | 62 399 725 | 62 404 172 |
| SlPRA20-11 | Solyc11g069150.2.1 | Chr11:54042842..54049437(-) | 54 042 842 | 54 049 437 |
| SlPRA21-12 | Solyc12g009140.2.1 | Chr12:2452941..2457449(-) | 2 452 941 | 2 457 449 |
Table 1 20S proteasome gene family of tomato
| 基因 Gene | 基因ID Gene ID | 染色体定位 Chromosome location | 基因位置/ bp Gene location | |
|---|---|---|---|---|
| 起始Start | 终止End | |||
| SlPRA1-01 | Solyc01g111450.3.1 | Chr01:97599172..97602337(+) | 97 599 172 | 97 602 337 |
| SlPRA2-01 | Solyc01g111460.3.1 | Chr01:97605332..97606609(-) | 97 605 332 | 97 606 609 |
| SlPRA3-02 | Solyc02g070510.3.1 | Chr02:40805105..40810898(+) | 40 805 105 | 40 810 898 |
| SlPRA4-02 | Solyc02g081700.1.1 | Chr02:46112367..46113113(+) | 46 112 367 | 46 113 113 |
| SlPRA5-02 | Solyc02g084920.3.1 | Chr02:48607717..48616195(-) | 48 607 717 | 48 616 195 |
| SlPRA6-03 | Solyc03g033745.1.1 | Chr03:5362233..5373114(-) | 5 362 233 | 5 373 114 |
| SlPRA7-03 | Solyc03g063420.2.1 | Chr03:36767871..36777655(-) | 36 767 871 | 36 777 655 |
| SlPRA8-04 | Solyc04g009410.3.1 | Chr04:2830248..2840325(+) | 2 830 248 | 2 840 325 |
| SlPRA9-04 | Solyc04g024420.3.1 | Chr04:31500067..31505977(+) | 31 500 067 | 31 505 977 |
| SlPRA10-04 | Solyc04g080590.3.1 | Chr04:64787899..64791729(-) | 64 787 899 | 64 791 729 |
| SlPRA11-05 | Solyc05g013820.3.1 | Chr05:7123832..7132561(-) | 7 123 832 | 7 132 561 |
| SlPRA12-05 | Solyc05g056160.3.1 | Chr05:66400334..66404473(-) | 66 400 334 | 66 404 473 |
| SlPRA13-07 | Solyc07g016200.3.1 | Chr07:6438876..6447830(-) | 6 438 876 | 6 447 830 |
| SlPRA14-07 | Solyc07g055080.3.1 | Chr07:63355928..63361757(+) | 63 355 928 | 63 361 757 |
| SlPRA15-08 | Solyc08g016510.3.1 | Chr08:7711628..7717929(-) | 7 711 628 | 7 717 929 |
| SlPRA16-09 | Solyc09g082320.3.1 | Chr09:68500118..68505978(-) | 68 500 118 | 68 505 978 |
| SlPRA17-10 | Solyc10g008010.3.1 | Chr10:2165246..2178357(+) | 2 165 246 | 2 178 357 |
| SlPRA18-10 | Solyc10g077030.2.1 | Chr10:60039871..60049610(-) | 60 039 871 | 60 049 610 |
| SlPRA19-10 | Solyc10g081130.2.1 | Chr10:62399725..62404172(+) | 62 399 725 | 62 404 172 |
| SlPRA20-11 | Solyc11g069150.2.1 | Chr11:54042842..54049437(-) | 54 042 842 | 54 049 437 |
| SlPRA21-12 | Solyc12g009140.2.1 | Chr12:2452941..2457449(-) | 2 452 941 | 2 457 449 |

Fig. 2 Chromosomal localization and replication relationships of the 20S proteasome gene family The outer box represents the chromosome skeleton,the middle and inner boxes indicate gene density,and the approximate distribution of each tomato 20S proteasome gene is marked on the chromosome skeleton by a short black line. Coloured lines refer to the linkage groups with more than 0.95 similarity,red lines 0.98-0.99,green lines 0.97-0.98,and light grey lines refer to the linkage groups with less than 0.95 similarity and the linkage groups of other genes.

Fig. 6 Covariance analysis of 20S proteasome genes in different species Gray lines indicate blocks of covariance between tomato and other species,and red lines highlight 20S proteasome gene pairs.
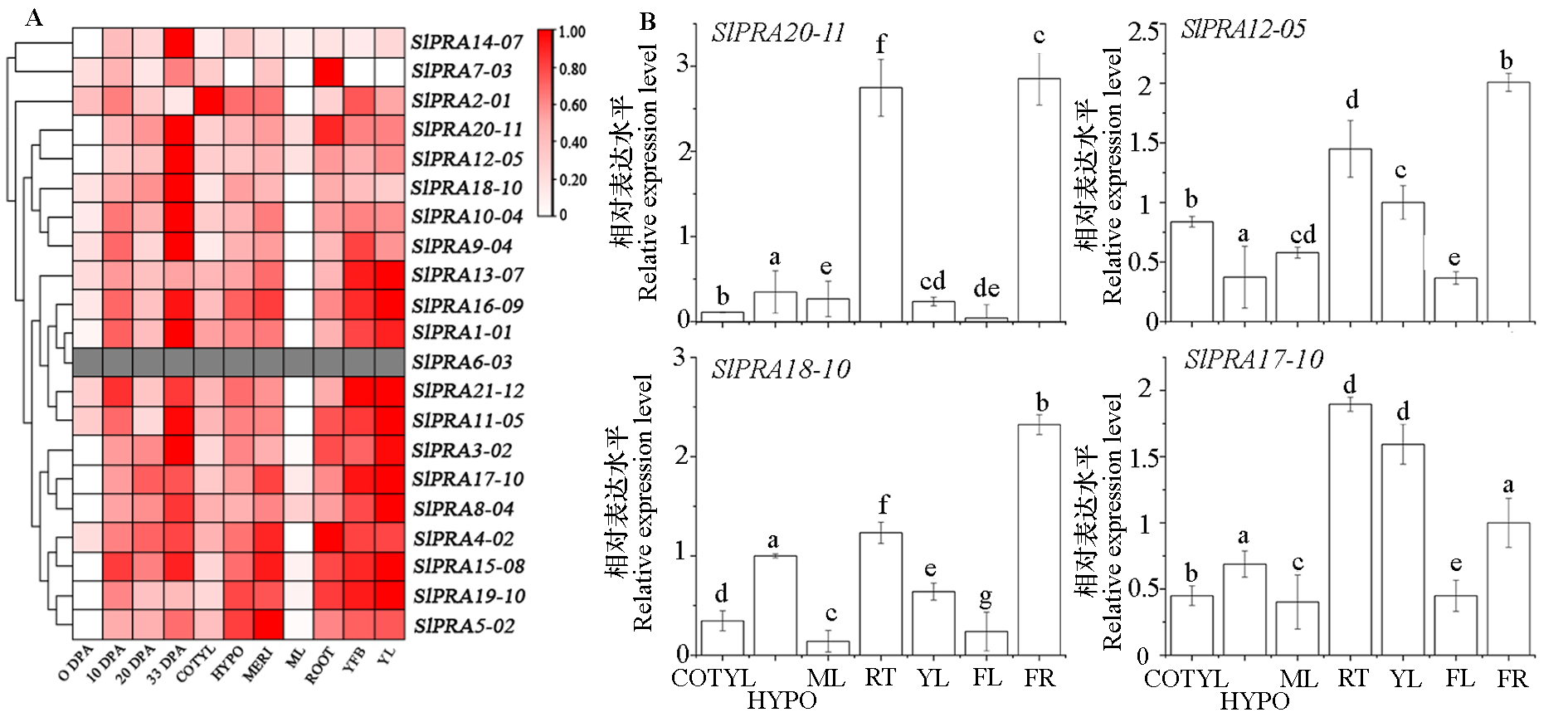
Fig. 8 Tissue-specific expression of 20S proteasome gene in tomato A shows the relative expression of tomato 20S proteasome genes in different tissues and different developmental stages(TFGD),and figure B shows qPCR analysis of the expression levels of some genes. 0 DPA:Anthesis flowers;10 DPA:10 days post anthesis fruit;20 DPA:20 days post anthesis fruit;33 DPA:Rippening fruit. COTYL:Cotyledons;HYPO:Hypocotyl;MERI:Vegitative meristems;ML:Mature leaves;RT:Root;YFB:Young flower buds,YL:Young flower buds;FL:Flower;FR:Fruit.
| [1] |
|
| [2] |
doi: 10.3390/biom4030862 pmid: 25250704 |
| [3] |
doi: 10.1105/tpc.021345 pmid: 15208399 |
| [4] |
|
| [5] |
doi: S0022-2836(17)30270-X pmid: 28583440 |
| [6] |
|
|
陈文奕, 陈惠萍. 2023. 水稻糊粉层蛋白酶体参与种子萌发的研究. 热带作物学报, http://kns.cnki.net/kcms/detail/46.1019.S.20230403.0936.002.html.
|
|
| [7] |
|
|
杜国华, 张立军, 樊金娟, 阮燕晔, 刘淳, 许红梅. 2010. 高等植物蛋白质的特异性降解系统. 分子植物育种, 8 (3):567-576.
|
|
| [8] |
doi: 10.1093/genetics/149.2.677 pmid: 9611183 |
| [9] |
|
|
韩晔, 种康. 2004. 泛素降解途径与生长素的调节. 植物生理学通讯,(6):653-658.
|
|
| [10] |
|
| [11] |
doi: 10.1016/j.str.2014.11.017 pmid: 25599644 |
| [12] |
|
|
何浩, 王志明, 张立强, 唐艺真, 夏琪, 李忠源, 张怀渝. 2023. 小麦PDR基因家族的鉴定与表达分析. 农业生物技术学报,(3):445-459.
|
|
| [13] |
|
| [14] |
|
| [15] |
|
| [16] |
|
| [17] |
doi: 10.1038/s41477-020-0721-4 pmid: 32690892 |
| [18] |
|
| [19] |
|
| [20] |
doi: 10.1158/1078-0432.CCR-07-2218 pmid: 18347166 |
| [21] |
|
| [22] |
|
| [23] |
|
| [24] |
|
|
孙鹏, 刘淼, 冯利兴, 刘璇. 2015. 蛋白酶体结构和活性调节机制的研究进展. 生物化学与生物物理进展, 42 (12):1084-1093.
|
|
| [25] |
|
| [26] |
doi: 10.1039/c1mb05283g pmid: 22027891 |
| [27] |
|
| [28] |
|
| [29] |
|
|
王丰, 施一公. 2014. 26S蛋白酶体的结构生物学研究进展. 中国科学:生命科学, 44 (10):965-974.
|
|
| [30] |
|
|
许传俊, 李玲. 2007. 泛素/26S蛋白酶体途径与植物的生长发育. 西北植物学报,(3):635-643.
|
|
| [31] |
|
|
余婷, 关洪鑫, 欧阳松应. 2021. 蛋白酶体调节颗粒的结构生物学特征及其功能. 中国生物化学与分子生物学报, 37 (3):270-288.
doi: 10.13865/j.cnki.cjbmb.2021.01.1523 |
|
| [32] |
|
|
张亚飞, 曹丽娜, 樊昊驹, 严海川, 刘关君. 2021. 毛果杨20S蛋白酶体基因家族成员预测及生物信息学分析. 分子植物育种,
|
| [1] | HAN Ying, DUAN Ying, NIU Yijie, LI Yansu, HE Chaoxing, SUN Mintao, WANG Jun, LI Qiang, CHEN Shuangchen, YAN Yan. Transcription Metabolic Mechanism of Humic Acid Biodegradable Plastic Film to Improving the Fruit Quality of Tomato in the Greenhouse [J]. Acta Horticulturae Sinica, 2024, 51(8): 1758-1772. |
| [2] | GONG Xiaoya, LI Xian, ZHOU Xingang, WU Fengzhi. Effect of Rhizosphere Microorganisms Induced by Potato-Onion on Tomato Root-Knot Nematode Disease [J]. Acta Horticulturae Sinica, 2024, 51(8): 1913-1926. |
| [3] | MENG Sida, HAN Leilei, XIANG Hengzuo, ZHU Meiyu, FENG Zhen, YE Yunzhu, SUN Meihua, LI Yanbing, ZHAO Liping, TAN Changhua, QI Mingfang, and LI Tianlai. Research Progress on the Mechanism of Regulating the Number of Tomato Locules [J]. Acta Horticulturae Sinica, 2024, 51(7): 1649-1664. |
| [4] | MA Xingyun, FAN Bingli, TANG Guangcai, JIA Zhiqi, LI Ying, XUE Dongqi, ZHANG Shiwen. Preliminary Study on the Mechanism of DXR Regulating Chloroplast Development Flower Color and Fruit Coloring in Tomato [J]. Acta Horticulturae Sinica, 2024, 51(6): 1241-1255. |
| [5] | ZHANG Wenjing, XU Dayong, WU Qianlin, YANG Fo, XIN Bingyue, ZENG Xin, LI Feng. Genome Analysis of Bacillus velezensis XDY66,an Antagonist of Tomato Botrytis cinerea [J]. Acta Horticulturae Sinica, 2024, 51(6): 1413-1425. |
| [6] | WANG Yongzhen, ZHANG Jianguo, LIU Caihong, LI Sibei, LÜ Tiantian. A New Tomato Hybrid Cultivar‘Yuanhong 212’ [J]. Acta Horticulturae Sinica, 2024, 51(6): 1435-1436. |
| [7] | LIU Zeying, SUN Shuai, LIU Zhiqiang, CUI Xia, LI Ren. Mapping of the Sharp Blossom-end Gene and Screening of Candidate Gene in Tomato [J]. Acta Horticulturae Sinica, 2024, 51(5): 982-992. |
| [8] | LI Pin, GAN Ning, CHEN Jiawei, XIANG Sixiang, SHEN Jingyi, OUYANG Bo, LU Yong’en. Analysis of Phosphorus Utilization Efficiency in Natural Population of Tomato and Screening of Low Phosphorus Tolerant Germplasm [J]. Acta Horticulturae Sinica, 2024, 51(5): 993-1004. |
| [9] | YANG Ting, XI Dehui, XIA Ming, LI Jianan. Studies on the Mechanism of α-Momordicin Gene Enhancing Tomato Resistance to Tobacco Mosaic Virus [J]. Acta Horticulturae Sinica, 2024, 51(5): 1126-1136. |
| [10] | HU Zhifeng, SHAO Jingcheng, ZHANG Li. A New Tomato Cultivar‘Longfan 15’ [J]. Acta Horticulturae Sinica, 2024, 51(4): 917-918. |
| [11] | LIU Genzhong, LI Fangman, GE Pingfei, TAO Jinbao, ZHANG Xingyu, YE Zhibiao, ZHANG Yuyang. QTL Mapping and Candidate Gene Identification Related to Ascorbic Acid Content in Tomato [J]. Acta Horticulturae Sinica, 2024, 51(2): 219-228. |
| [12] | DONG Shuchao, HONG Jun, LING Jiayi, XIE Zixin, ZHANG Shengjun, ZHAO Liping, SONG Liuxia, WANG Yinlei, ZHAO Tongmin. Genome-wide Association Studies of Drought Tolerance in Tomato [J]. Acta Horticulturae Sinica, 2024, 51(2): 229-238. |
| [13] | XU Qin, WANG Jiaying, ZHANG Mannan, XIAO Zhihao, ZHENG Hankai, LU Yong'en, WANG Taotao, ZHANG Yuyang, ZHANG Junhong, YE Zhibiao, YE Jie. Identification of Genetic Loci and Molecular Marker Development of Salt Tolerance in Tomato Seedlings [J]. Acta Horticulturae Sinica, 2024, 51(2): 239-252. |
| [14] | YANG Liang, LIU Huan, MA Yanqin, LI Ju, WANG Hai'e, ZHOU Yujie, LONG Haicheng, MIAO Mingjun, LI Zhi, CHANG Wei. Creating High Lycopene Fruit Using CRISPR/Cas9 Technology in Tomato [J]. Acta Horticulturae Sinica, 2024, 51(2): 253-265. |
| [15] | ZHANG Wenhao, ZHANG Hui, LIU Yuting, WANG Yan, ZHANG Yingying, WANG Xinman, WANG Quanhua, ZHU Weimin, YANG Xuedong. Preliminary Transcriptome Analysis in Two Tomato Fruits Materials with Different Sugar Content [J]. Acta Horticulturae Sinica, 2024, 51(2): 281-294. |
| Viewed | ||||||
|
Full text |
|
|||||
|
Abstract |
|
|||||
Copyright © 2012 Acta Horticulturae Sinica 京ICP备10030308号-2 国际联网备案号 11010802023439
Tel: 010-82109523 E-Mail: yuanyixuebao@126.com
Support by: Beijing Magtech Co.Ltd