
Acta Horticulturae Sinica ›› 2022, Vol. 49 ›› Issue (7): 1505-1518.doi: 10.16420/j.issn.0513-353x.2021-0474
• Research Papers • Previous Articles Next Articles
QIAN Jieyu, JIANG Lingli, ZHENG Gang, CHEN Jiahong, LAI Wuhao, XU Menghan, FU Jianxin, ZHANG Chao( )
)
Received:2022-02-27
Revised:2022-04-24
Online:2022-07-25
Published:2022-07-29
CLC Number:
QIAN Jieyu, JIANG Lingli, ZHENG Gang, CHEN Jiahong, LAI Wuhao, XU Menghan, FU Jianxin, ZHANG Chao. Identification and Expression Analysis of MYB Transcription Factors Regulating the Anthocyanin Biosynthesis in Zinnia elegans and Function Research of ZeMYB9[J]. Acta Horticulturae Sinica, 2022, 49(7): 1505-1518.
Add to citation manager EndNote|Ris|BibTeX
URL: https://www.ahs.ac.cn/EN/10.16420/j.issn.0513-353x.2021-0474
| 材料 Material | 基因名称 Gene name | 上游引物(5′-3′) Forward primer sequence | 下游引物(5′-3′) Reverse primer sequence |
|---|---|---|---|
| 百日草Zinnia elegans | ZeMYB3 | CGGGCCGTACAGATAACGAGAT | TGTTGGGATTTGTTGCGAGTG |
| ZeMYB6 | GACGGCTGAAGAAGATGAGAAGT | TCCGACCTCAAGTAGTTAATCCA | |
| ZeMYB9 | ACCAGGAAGAACTGCTAATGATGT | TGTGACCGCTCGTGGTTGTTA | |
| ZeMYB10 | ATGGTGCGGAAGGGTGTATG | GCTCGGAGTGGAACAAGGTG | |
| ZeMYB16 | AAAGCTCACGGTGAAGGTTGC | AAGGCTGTGGAGTTTGATGATAAGT | |
| ZeMYB20 | GTGGTCGTTGATTGCTGGAAG | TGAAGATCGCAGTCGTCGTAG | |
| ZeMYB22 | AGTTGTAGGTTGCGGTGGTTA | ATTGGCGGTTCTTCCTGGTAT | |
| ZeMYB23 | GGTCGTTGATTGCTGGAAGGT | TGAAGATCGCAGTCGTCGTAG | |
| ZeMYB32 | CGTCAAGGCTACCAGGAAGGA | ACTCAACGACCTGTGCGGAAT | |
| ZeMYB33 | TACCTGGACGAACAGACAACG | ACTCGACTGCTACGCCTGGAT | |
| ZeACT | TTGTGCTGGATTCTGGGGATGGT | GCAGTTTCAAGCTCTTGCTCGTAGTC | |
| 烟草Nicotiana tabacum | NtEF1α | TGGTTGTGACTTTTGGTCCCA | ACAAACCCACGCTTGAGATCC |
| NtCHS | AAGCAAGAGAAACTAAAGGCTACAAG | AAATCCAAAAAGCACACCCCAT | |
| NtCHI | CGGGTGCCTCCATTCTTTTTACT | CCTGACACTCTTTCGGCGATACTAC | |
| NtF3H | CCAGACAAACCAGATGGATGGATAG | CAAGGGTAAGGTCGGGCTGTG | |
| NtF3'H | TGGCTATTTCATTCCAAAAGGCTCA | CTTCAAAGTCATTTCCTCGCACATC | |
| NtDFR | GCAGTTGCTTCCCTTTTCTACC | TTCCCCATTGGTTGACTTTCC | |
| NtANS | GTGCCTGGGTTACAACTTTTCTATG | CATTGCTTAGGATTTCAAGGGTGTC | |
| NtUFGT | GAGTGCATTGGATGCCTTTT | CCAGCTCCATTAGGTCCTTG | |
| NtAN1a | ACCATTCTCGAACACCGAAG | TGCTAGGGCACAATGTGAAG | |
| NtAN1b | CTTGAACACTTCTCAAACCGA | TGCTAGGGCACAATGTGAAG |
Table 1 qRT-PCR primer sequences used in this study
| 材料 Material | 基因名称 Gene name | 上游引物(5′-3′) Forward primer sequence | 下游引物(5′-3′) Reverse primer sequence |
|---|---|---|---|
| 百日草Zinnia elegans | ZeMYB3 | CGGGCCGTACAGATAACGAGAT | TGTTGGGATTTGTTGCGAGTG |
| ZeMYB6 | GACGGCTGAAGAAGATGAGAAGT | TCCGACCTCAAGTAGTTAATCCA | |
| ZeMYB9 | ACCAGGAAGAACTGCTAATGATGT | TGTGACCGCTCGTGGTTGTTA | |
| ZeMYB10 | ATGGTGCGGAAGGGTGTATG | GCTCGGAGTGGAACAAGGTG | |
| ZeMYB16 | AAAGCTCACGGTGAAGGTTGC | AAGGCTGTGGAGTTTGATGATAAGT | |
| ZeMYB20 | GTGGTCGTTGATTGCTGGAAG | TGAAGATCGCAGTCGTCGTAG | |
| ZeMYB22 | AGTTGTAGGTTGCGGTGGTTA | ATTGGCGGTTCTTCCTGGTAT | |
| ZeMYB23 | GGTCGTTGATTGCTGGAAGGT | TGAAGATCGCAGTCGTCGTAG | |
| ZeMYB32 | CGTCAAGGCTACCAGGAAGGA | ACTCAACGACCTGTGCGGAAT | |
| ZeMYB33 | TACCTGGACGAACAGACAACG | ACTCGACTGCTACGCCTGGAT | |
| ZeACT | TTGTGCTGGATTCTGGGGATGGT | GCAGTTTCAAGCTCTTGCTCGTAGTC | |
| 烟草Nicotiana tabacum | NtEF1α | TGGTTGTGACTTTTGGTCCCA | ACAAACCCACGCTTGAGATCC |
| NtCHS | AAGCAAGAGAAACTAAAGGCTACAAG | AAATCCAAAAAGCACACCCCAT | |
| NtCHI | CGGGTGCCTCCATTCTTTTTACT | CCTGACACTCTTTCGGCGATACTAC | |
| NtF3H | CCAGACAAACCAGATGGATGGATAG | CAAGGGTAAGGTCGGGCTGTG | |
| NtF3'H | TGGCTATTTCATTCCAAAAGGCTCA | CTTCAAAGTCATTTCCTCGCACATC | |
| NtDFR | GCAGTTGCTTCCCTTTTCTACC | TTCCCCATTGGTTGACTTTCC | |
| NtANS | GTGCCTGGGTTACAACTTTTCTATG | CATTGCTTAGGATTTCAAGGGTGTC | |
| NtUFGT | GAGTGCATTGGATGCCTTTT | CCAGCTCCATTAGGTCCTTG | |
| NtAN1a | ACCATTCTCGAACACCGAAG | TGCTAGGGCACAATGTGAAG | |
| NtAN1b | CTTGAACACTTCTCAAACCGA | TGCTAGGGCACAATGTGAAG |
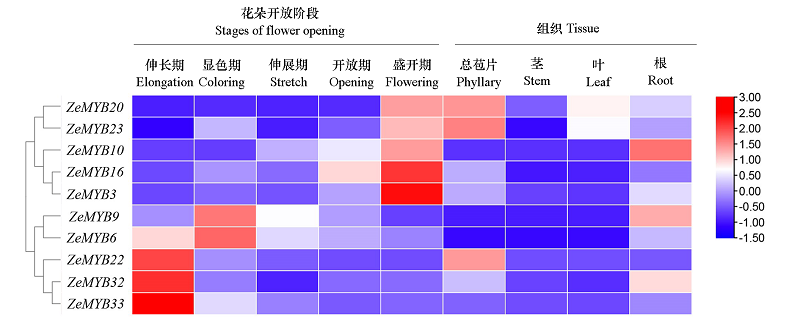
Fig. 5 Heatmap clustering analysis of R2R3-MYB expression in during flower development stages flowers of Zinnia elegansThe gene expression data was converted into the base-2 logarithm for calculation and hierarchical clustering. Red and blue boxes indicated high and low expression levels correspondingly.
| 基因 Gene | S4 | S6 | S7 | |||||||
|---|---|---|---|---|---|---|---|---|---|---|
| ZeMYB3 | ZeMYB16 | ZeMYB20 | ZeMYB23 | ZeMYB32 | ZeMYB9 | ZeMYB10 | ZeMYB22 | ZeMYB6 | ZeMYB33 | |
| ZeCHS | -0.529* | -0.614* | -0.427 | -0.584* | 0.940** | 0.084 | -0.687** | 0.987** | 0.420 | 0.983** |
| ZeCHI | -0.684** | -0.727** | -0.628* | -0.594* | 0.835** | 0.425 | -0.883** | 0.911** | 0.620* | 0.935** |
| ZeF3H | -0.616* | -0.563* | -0.520* | -0.229 | 0.170 | 0.877** | -0.747** | 0.277 | 0.673** | 0.363 |
| ZeF3'H | -0.814** | -0.880** | -0.896** | -0.693** | 0.050 | 0.785** | -0.820** | 0.215 | 0.522 | 0.262 |
| ZeDFR | -0.312 | -0.346 | -0.451 | -0.264 | -0.686** | 0.511 | -0.128 | -0.564* | 0.047 | -0.532* |
| ZeANS | -0.677** | -0.610* | -0.812** | -0.487 | -0.185 | 0.806** | -0.633* | -0.069 | 0.459 | -0.035 |
| Ze3GT | 0.207 | 0.350 | 0.610* | 0.574* | 0.410 | -0.045 | 0.078 | 0.346 | 0.147 | 0.391 |
| Ze5GT | -0.622* | -0.634* | -0.671** | -0.463 | 0.274 | 0.680** | -0.703** | 0.349 | 0.413 | 0.397 |
| ZeAT1 | -0.553* | -0.632* | -0.529* | -0.622* | 0.556* | 0.258 | -0.548* | 0.607* | 0.315 | 0.617* |
| ZeAT2 | -0.281 | -0.336 | -0.217 | -0.332 | 0.708** | 0.062 | -0.379 | 0.654** | 0.440 | 0.641* |
| ZeAT3 | 0.105 | 0.241 | 0.423 | 0.531* | -0.102 | 0.263 | 0.077 | -0.098 | 0.056 | -0.008 |
Table 2 Correlation analysis of expression level of ZeMYB genes with expression level of structural genes in Zinnia elegans
| 基因 Gene | S4 | S6 | S7 | |||||||
|---|---|---|---|---|---|---|---|---|---|---|
| ZeMYB3 | ZeMYB16 | ZeMYB20 | ZeMYB23 | ZeMYB32 | ZeMYB9 | ZeMYB10 | ZeMYB22 | ZeMYB6 | ZeMYB33 | |
| ZeCHS | -0.529* | -0.614* | -0.427 | -0.584* | 0.940** | 0.084 | -0.687** | 0.987** | 0.420 | 0.983** |
| ZeCHI | -0.684** | -0.727** | -0.628* | -0.594* | 0.835** | 0.425 | -0.883** | 0.911** | 0.620* | 0.935** |
| ZeF3H | -0.616* | -0.563* | -0.520* | -0.229 | 0.170 | 0.877** | -0.747** | 0.277 | 0.673** | 0.363 |
| ZeF3'H | -0.814** | -0.880** | -0.896** | -0.693** | 0.050 | 0.785** | -0.820** | 0.215 | 0.522 | 0.262 |
| ZeDFR | -0.312 | -0.346 | -0.451 | -0.264 | -0.686** | 0.511 | -0.128 | -0.564* | 0.047 | -0.532* |
| ZeANS | -0.677** | -0.610* | -0.812** | -0.487 | -0.185 | 0.806** | -0.633* | -0.069 | 0.459 | -0.035 |
| Ze3GT | 0.207 | 0.350 | 0.610* | 0.574* | 0.410 | -0.045 | 0.078 | 0.346 | 0.147 | 0.391 |
| Ze5GT | -0.622* | -0.634* | -0.671** | -0.463 | 0.274 | 0.680** | -0.703** | 0.349 | 0.413 | 0.397 |
| ZeAT1 | -0.553* | -0.632* | -0.529* | -0.622* | 0.556* | 0.258 | -0.548* | 0.607* | 0.315 | 0.617* |
| ZeAT2 | -0.281 | -0.336 | -0.217 | -0.332 | 0.708** | 0.062 | -0.379 | 0.654** | 0.440 | 0.641* |
| ZeAT3 | 0.105 | 0.241 | 0.423 | 0.531* | -0.102 | 0.263 | 0.077 | -0.098 | 0.056 | -0.008 |
| [1] |
Aharoni A, Vos C, Wein M, Sun Z, O'Connell A P. 2010. The strawberry FaMYB1 transcription factor suppresses anthocyanin and flavonol accumulation in transgenic tobacco. The Plant Journal, 28 (3):319-332.
doi: 10.1046/j.1365-313X.2001.01154.x URL |
| [2] |
Boyle T H, Stimart D P. 1989. Anatomical and biochemical factors determining ray floret color of Zinnia angustifolia,Z. elegans,and their interspecific hybrids. Journal of the American Society for Horticultural Science, 114:499-505.
doi: 10.21273/JASHS.114.3.499 URL |
| [3] |
Dubos C, Stracke R, Grotewold E, Weisshaar B, Martin C, Lepiniec L. 2010. MYB transcription factors in Arabidopsis. Trends in Plant Science, 15:573-581.
doi: 10.1016/j.tplants.2010.06.005 URL |
| [4] |
Gerats A G M, Farcy E, Wallroth M, Groot S P C, Schram A. 1984. Control of anthocyanin biosynthesis in Petunia hybrida by multiple allelic series of the genes An1and An2. Genetics, 106 (3):501-508.
doi: 10.1093/genetics/106.3.501 pmid: 17246198 |
| [5] |
Gonzalez A, Zhao M, Leavitt J M, Lloyd A M. 2010. Regulation of the anthocyanin biosynthetic pathway by the TTG1/bHLH/MYB transcriptional complex in Arabidopsis seedlings. The Plant Journal, 53:814-827.
doi: 10.1111/j.1365-313X.2007.03373.x URL |
| [6] |
Grotewold E. 2006. The genetics and biochemistry of floral pigments. Annual Review of Plant Biology, 57:761-780.
pmid: 16669781 |
| [7] |
Heppel S C, Jaffé F W, Takos A M, Schellmann S, Bogs J. 2013. Identification of key amino acids for the evolution of promoter target specificity of anthocyanin and proanthocyanidin regulating MYB factors. Plant Molecular Biology, 82:457-471.
doi: 10.1007/s11103-013-0074-8 pmid: 23689818 |
| [8] | Hong Yanhong, Ye Qinghua, Li Zekun, Wang Wei, Xie Qian, Chen Qingxi, Chen Jianqing. 2021. Accumulation of anthocyanins in red-flowered strawberry‘Meihong’petals and expression analysis of MYB gene. Acta Horticulturae Sinica, 48 (8):1470-1484. |
| 洪燕红, 叶清华, 李泽坤, 王威, 谢倩, 陈清西, 陈建清. 2021. 红花草莓‘莓红’花瓣花色苷积累及其MYB基因的表达分析. 园艺学报, 48 (8):1470-1484. | |
| [9] |
Jin H, Cominelli E, Bailey P, Parr A, Mehrtens F, Jones J, Tonelli C, Weisshaar B, Martin C. 2000. Transcriptional repression by AtMYB4 controls production of UV-protecting sunscreens in Arabidopsis. The EMBO Journal, 19:6150-6161.
doi: 10.1093/emboj/19.22.6150 URL |
| [10] | Ke Yujie, Chen Mingkun, Ma Shanhu, Ou Yue, Wang Yi, Zheng Qingdong, Liu Zhongjian, Ai Ye. 2021. Research progress of MYB transcription factors in Orchidaceae. Acta Horticulturae Sinica, 48 (11):2311-2320. (in Chinese) |
| 柯玉洁, 陈明堃, 马山虎, 欧悦, 王艺, 郑清冬, 刘仲健, 艾叶. 2021. 兰科植物MYB 转录因子研究进展. 园艺学报, 48 (11):2311-2320. | |
| [11] |
Li C, Qiu J, Yang G, Huang S, Yin J. 2016. Isolation and characterization of a R2R3-MYB transcription factor gene related to anthocyanin biosynthesis in the spathes of Anthurium andraeanum(Hort.). Plant Cell Reports, 35 (10):2151-2165.
doi: 10.1007/s00299-016-2025-8 URL |
| [12] | Li Chonghui, Yang Guangsui, Zhang Zhiqun, Yin Junmei. 2021. A novel R2R3-MYB transcription factor gene AaMYB6involved in anthocyanin biosynthesis in Anthurium andraeanum. Acta Horticulturae Sinica, 48 (10):1859-1872. (in Chinese) |
| 李崇晖, 杨光穗, 张志群, 尹俊梅. 2021. 红掌R2R3-MYB转录因子基因AaMYB6调控花青素苷合成. 园艺学报, 48 (10):1859-1872. | |
| [13] | Li Guisheng. 2021. Comparative analysis of‘Jinyan’and‘Hongyang’kiwifruit transcriptomes. Acta Horticulturae Sinica, 48 (6):1183-1196. (in Chinese) |
| 李贵生. 2021. 猕猴桃‘金艳’和‘红阳’果实转录组的比较分析. 园艺学报, 48 (6):1183-1196. | |
| [14] |
Li M, Li Y, Guo L, Gong N, Pang Y, Jiang W, Liu Y, Jiang X, Zhao L, Wang Y, Xie D-Y, Gao L, Xia T. 2017. Functional characterization of tea(Camellia sinensis)MYB4a transcription factor using an integrative approach. Frontiers in Plant Science, 8:943-960.
doi: 10.3389/fpls.2017.00943 URL |
| [15] | Li Mou-liang, Zhang Xiao-ni, Lin Sheng-nan, Wu Quan-shu, Bao Man-zhu, Fu Xiao-peng. 2020. Genome-wide Identification and Analysis of R2R3-MYB Transcription Factor in Rosa chinensis. Molecular Plant Breeding(online), 18 (33):1-11. (in Chinese) |
| 李谋亮, 张晓妮, 林胜男, 吴全淑, 包满珠, 傅小鹏. 2020. 月季R2R3-MYB转录因子全基因组鉴定与分析. 分子植物育种(网络版), 18 (33):1-11. | |
| [16] |
Li Y, Liang J, Zeng X, Guo H, Luo Y, Kear P, Zhang S, Zhu G. 2021. Genome-wide analysis of MYB gene family in potato provides insights into tissue-specific regulation of anthocyanin biosynthesis. Horticultural Plant Journal, 7 (2):129-141.
doi: 10.1016/j.hpj.2020.12.001 URL |
| [17] |
Li Y, Shan X, Tong L, Wei C, Lu K, Li S, Kimani S, Wang S, Wang L, Gao X. 2020. The conserved and particular roles of the R2R3-MYB regulator FhPAP1 from Freesia hybrida in flower anthocyanin biosynthesis. Plant and Cell Physiology, 61:1365-1380.
doi: 10.1093/pcp/pcaa065 URL |
| [18] |
Matus J T, Aquea F, Arce-Johnson P. 2008. Analysis of the grape MYB R2R3 subfamily reveals expanded wine quality-related clades and conserved gene structure organization across Vitis and Arabidopsis genomes. BMC Plant Biology, 8 (1):83.
doi: 10.1186/1471-2229-8-83 URL |
| [19] |
Qian J, Lai W, Jiang L, Zhan H, Zhai M, Fu J, Zhang C. 2021. Association between differential gene expression and anthocyanin biosynthesis underlying the diverse array of petal colors in Zinnia elegans. Scientia Horticulturae, 277:109809.
doi: 10.1016/j.scienta.2020.109809 URL |
| [20] |
Shen H, He X, Poovaiah C R, Wuddineh W A, Ma J, Mann D G, Wang H, Jackson L, Tang Y, Fang C, Richard A D. 2012. Functional characterization of the switchgrass(Panicum virgatum)R2R3-MYB transcription factor PvMYB4for improvement of lignocellulosic feedstocks. New Phytologist, 193:121-136.
doi: 10.1111/j.1469-8137.2011.03922.x pmid: 21988539 |
| [21] |
Stevenson C C, Harrington G N. 2009. The impact of supplemental carbon sources on Arabidopsis thaliana growth,chlorophyll content and anthocyanin accumulation. Plant Growth Regulation, 59 (3):255.
doi: 10.1007/s10725-009-9412-x URL |
| [22] |
Stracke R, Ishihara H, Huep G, Barsch A, Mehrtens F, Niehaus K, Weisshaar B. 2007. Differential regulation of closely related R2R3-MYB transcription factors controls flavonol accumulation in different parts of the Arabidopsis thaliana seedling. The Plant Journal, 50:660-677.
doi: 10.1111/j.1365-313X.2007.03078.x URL |
| [23] |
Stracke R, Werber M, Weisshaar B. 2001. The R2R3-MYB gene family in Arabidopsis thaliana. Current Opinion in Plant Biology, 4 (5):447-456.
pmid: 11597504 |
| [24] |
Sun B, Zhu Z, Cao P, Chen H, Chen C, Zhou X, Mao Y, Lei J, Jiang Y, Meng W, Wang Y, Liu S. 2016. Purple foliage coloration in tea (Camellia sinensis L.)arises from activation of the R2R3-MYB transcription factor CsAN1. Scientific Reports, 6:32534.
doi: 10.1038/srep32534 URL |
| [25] |
Tanaka Y, Sasaki N, Ohmiya A. 2008. Biosynthesis of plant pigments: anthocyanins,betalains and carotenoids. The Plant Journal, 54:733-749.
doi: 10.1111/j.1365-313X.2008.03447.x URL |
| [26] |
Viegas P, Mathews H, Bhatia C R, Notani N K. 1987. Monohybrid and dihybrid segregations in the progenies of tobacco transformed for kanamycin resistance with a Ti-vector system. Journal of Genetics, 66 (1):25-31.
doi: 10.1007/BF02934453 URL |
| [27] |
Vimolmangkang S, Han Y, Wei G, Korban S S. 2013. An apple MYB transcription factor,MdMYB3,is involved in regulation of anthocyanin biosynthesis and flower development. BMC Plant Biology, 13 (1):176-188.
doi: 10.1186/1471-2229-13-176 URL |
| [28] |
Xu H, Wang N, Liu J, Qu C, Wang Y, Jiang S, Lu N, Wang D, Zhang Z, Chen X. 2017. The molecular mechanism underlying anthocyanin metabolism in apple using the MdMYB16 and MdbHLH33genes. Plant Molecular Biology, 94:149-165.
doi: 10.1007/s11103-017-0601-0 URL |
| [29] |
Yan H, Pei X, Zhang H, Li X, Zhang X, Zhao M, Chiang V, Sederoff R R, Zhao X. 2021. MYB-mediated regulation of anthocyanin biosynthesis. International Journal of Molecular Sciences, 22 (6):3103.
doi: 10.3390/ijms22063103 URL |
| [30] |
Yang C Q, Sha G Y, Wei T, Ma B Q, Li C Y, Li P M, Zou Y J, Xu L F, Ma F W. 2021. Linkage map and QTL mapping of red flesh locus in apple using a R1R1 × R6R6 population. Horticultural Plant Journal, 7 (5):393-400.
doi: 10.1016/j.hpj.2020.12.008 URL |
| [31] | Yao Yifan, Dong Bin, Feng Chengyong, Yang Liyuan, Zhao Hongbo. 2020. Identification of R2R3-MYB family genes in Osmanthus fragrans and analysis of their expression during flower opening. Acta Horticulturae Sinica, 47 (10):164-176. (in Chinese) |
| 姚亦凡, 董彬, 冯成庸, 杨丽媛, 赵宏波. 2020. 桂花R2R3-MYB家族基因鉴定及其在花开放过程中的表达分析. 园艺学报, 47 (10):2027-2039. | |
| [32] | Zhao Jia, Liu Rong, Yang Fan, Li Xin, Liu Housheng, Yan Qian, Xiao Yuehua. 2015. Cloning and expression analysis of anthocyanidin-related R2R3-MYB protein gene from Rosa chinensis. Scientia Agricultura Sinica, 48 (7):1392-1404. (in Chinese) |
| 赵佳, 刘荣, 杨帆, 李鑫, 刘厚生, 严倩, 肖月华. 2015. 月季花青素苷相关R2R3-MYB蛋白基因的克隆和表达分析. 中国农业科学, 48 (7):1392-1404. | |
| [33] | Zheng Qingdong, Wang Yi, Ou Yue, Ke Yujie, Yao Yahe, Wang Mengjie, Chen Jiayi, Ai Ye. 2021. Research advances of genes responsible for flower colors in Orchidaceae. Acta Horticulturae Sinica, 48 (10):2057-2072. (in Chinese) |
| 郑清冬, 王艺, 欧悦, 柯玉洁, 姚亚合, 王梦洁, 陈嘉忆, 艾叶. 2021. 兰科植物花色相关基因研究进展. 园艺学报, 48 (10):2057-2072. | |
| [34] |
Zhong C, Tang Y, Pang B, Li X, Yang Y, Deng J, Feng C, Li L, Ren G, Wang Y, Peng J, Sun S, Liang S, Wang X. 2020. The R2R3-MYB transcription factor GhMYB1a regulates flavonol and anthocyanin accumulation in Gerbera hybrida. Horticulture Research, 7:78.
doi: 10.1038/s41438-020-0296-2 URL |
| [35] | Zhou Yang-Li, Hou Shuo, Zheng Zhengquan, Gao Yan-hui, Tong Zaikang. 2020. Functional study of LsMYBs gene in VIGS gene silencing system. Chinese Journal of Agricultural Biotechnology, 28 (6):974-983. (in Chinese) |
| 周洋丽, 侯朔, 郑正权, 高燕会, 童再康. 2020. 基于VIGS基因沉默体系的换锦花LsMYBs基因功能研究. 农业生物技术学报, 28 (6):974-983. |
| [1] | ZOU Xue, DING Fan, LIU Lifang, YU Hankaizong, CHEN Nianwei, and RAO Liping. A New Purple Potato Cultivar‘Mianziyu 1’ [J]. Acta Horticulturae Sinica, 2022, 49(S1): 93-94. |
| [2] | WANG Sha, ZHANG Xinhui, ZHAO Yujie, LI Bianbian, ZHAO Xueqing, SHEN Yu, DONG Jianmei, YUAN Zhaohe. Cloning and Functional Analysis of PgMYB111 Related to Anthocyanin Synthesis in Pomegranate [J]. Acta Horticulturae Sinica, 2022, 49(9): 1883-1894. |
| [3] | HUANG Ling, HU Xianmei, LIANG Zehui, WANG Yanping, CHAN Zhulong, XIANG Lin. Cloning and Function Identification of Anthocyanidin Synthase Gene TgANS in Tulipa gesneriana [J]. Acta Horticulturae Sinica, 2022, 49(9): 1935-1944. |
| [4] | LI Maofu, YANG Yuan, WANG Hua, FAN Youwei, SUN Pei, JIN Wanmei. Analysis the Function of R2R3 MYB Transcription Factor RhMYB113c on Regulating Anthocyanin Synthesis in Rosa hybrida [J]. Acta Horticulturae Sinica, 2022, 49(9): 1957-1966. |
| [5] | GAO Yanlong, WU Yuxia, ZHANG Zhongxing, WANG Shuangcheng, ZHANG Rui, ZHANG De, WANG Yanxiu. Bioinformatics Analysis of Apple ELO Gene Family and Its Expression Analysis Under Low Temperature Stress [J]. Acta Horticulturae Sinica, 2022, 49(8): 1621-1636. |
| [6] | XU Haifeng, WANG Zhongtang, CHEN Xin, LIU Zhiguo, WANG Lihu, LIU Ping, LIU Mengjun, ZHANG Qiong. The Analyses of Target Metabolomics in Flavonoid and Its Potential MYB Regulation Factors During Coloring Period of Winter Jujube [J]. Acta Horticulturae Sinica, 2022, 49(8): 1761-1771. |
| [7] | ZHANG Lugang, LU Qianqian, HE Qiong, XUE Yihua, MA Xiaomin, MA Shuai, NIE Shanshan, YANG Wenjing. Creation of Novel Germplasm of Purple-orange Heading Chinese Cabbage [J]. Acta Horticulturae Sinica, 2022, 49(7): 1582-1588. |
| [8] | CHEN Daozong, LIU Yi, SHEN Wenjie, ZHU Bo, TAN Chen. Identification and Analysis of PAP1/2 Homologous Genes in Brassica rapa,B. oleracea and B. napus [J]. Acta Horticulturae Sinica, 2022, 49(6): 1301-1312. |
| [9] | LI Xiaoming, YU Junchi, WANG Chunxia. Comparison of Growth and Secondary Metabolites of Purple and White Flower Dracocephalum moldavica Under Field,Greenhouse and Greenhouse Shading Conditions [J]. Acta Horticulturae Sinica, 2022, 49(6): 1363-1370. |
| [10] | LI Lixian, WANG Shuo, CHEN Ying, WU Yingtao, WANG Yaqian, FANG Yue, CHEN Xuesen, TIAN Changping, FENG Shouqian. PavMYB10.1 Promotes Anthocyanin Accumulation by Positively Regulating PavRiant in Sweet Cherry [J]. Acta Horticulturae Sinica, 2022, 49(5): 1023-1030. |
| [11] | JIANG Cuicui, FANG Zhizhen, ZHOU Danrong, LIN Yanjuan, YE Xinfu. Identification and Expression Analysis of Sugar Transporter Family Genes in‘Furongli’(Prunus salicina) [J]. Acta Horticulturae Sinica, 2022, 49(2): 252-264. |
| [12] | DENG Jiao, SU Mengyue, LIU Xuelian, OU Kefang, HU Zhengrong, YANG Pingfang. Transcriptome Analysis Revealed the Formation Mechanism of Floral Color of Lotus‘Dasajin’with Bicolor Petal [J]. Acta Horticulturae Sinica, 2022, 49(2): 365-377. |
| [13] | WANG Zhiyu, CHANG Beibei, LIU Qi, CHENG Xiaofan, DU Xiaoyun, YU Xiaoli, SONG Laiqing, ZHAO Lingling. Study on Expression and Anthocyanin Accumulation of Solute Carrier Gene MdSLC35F2-like in Apple [J]. Acta Horticulturae Sinica, 2022, 49(11): 2293-2303. |
| [14] | SUN Wei, SUN Shiyu, CHEN Yiran, WANG Yuhan, ZHANG Yan, JU Zhigang, YI Yin. Cloning and Function Analysis of Chalcone Isomerase Gene RdCHI1 in Rhododendron delavayi [J]. Acta Horticulturae Sinica, 2022, 49(11): 2407-2418. |
| [15] | HONG Yanhong, YE Qinghua, LI Zekun, WANG Wei, XIE Qian, CHEN Qingxi, CHEN Jianqing. Accumulation of Anthocyanins in Red-flowered Strawberry‘Meihong’Petals and Expression Analysis of MYB Gene [J]. Acta Horticulturae Sinica, 2021, 48(8): 1470-1484. |
| Viewed | ||||||
|
Full text |
|
|||||
|
Abstract |
|
|||||
Copyright © 2012 Acta Horticulturae Sinica 京ICP备10030308号-2 国际联网备案号 11010802023439
Tel: 010-82109523 E-Mail: yuanyixuebao@126.com
Support by: Beijing Magtech Co.Ltd