
Acta Horticulturae Sinica ›› 2023, Vol. 50 ›› Issue (10): 2271-2287.doi: 10.16420/j.issn.0513-353x.2022-0835
• Plant Protection • Previous Articles Next Articles
WANG Yixi, YAN Shuangshuang, YU Bingwei, GAN Yuwei, QIU Zhengkun, ZHU Zhangsheng, CHEN Changming, CAO Bihao*( )
)
Received:2023-04-28
Revised:2023-05-09
Online:2023-10-25
Published:2023-10-31
WANG Yixi, YAN Shuangshuang, YU Bingwei, GAN Yuwei, QIU Zhengkun, ZHU Zhangsheng, CHEN Changming, CAO Bihao. Screening and Identification of E3 Ubiquitin Ligase Genes Relate to Bacterial Wilt Resistance in Eggplant[J]. Acta Horticulturae Sinica, 2023, 50(10): 2271-2287.
Add to citation manager EndNote|Ris|BibTeX
URL: https://www.ahs.ac.cn/EN/10.16420/j.issn.0513-353x.2022-0835
| 名称 Name | 序列(5′-3′) Sequence | 质粒 Plasmid | 酶切位点 Restriction site |
|---|---|---|---|
| SmSP1-GFP | F:ctgcccaaattcgcgaccggt ATGGCTCCATGGGGCGGA | pEAQ-EGFP | Age I |
| R:gcccttgctcaccataccggt ATGGCGAAAAGTTTTCACA | pEAQ-EGFP | Age I | |
| SmSPL2-GFP | F:ctgcccaaattcgcgaccggt ATGTCAATACACGACCAAGCGG | pEAQ-EGFP | Age I |
| R:gcccttgctcaccataccggt AGAATCATATATCCTTACGGAACT | pEAQ-EGFP | Age I | |
| SmDDA1a1-GFP | F:ctgcccaaattcgcgaccggtATGGGGTCAATTTTCGGTGAA | pEAQ-EGFP | Age I |
| R:gcccttgctcaccataccggt GGTGGACACCTTAAGATGCT | pEAQ-EGFP | Age I | |
| SmDDA1a2-GFP | F:ctgcccaaattcgcgaccggt ATGGGGTCAATTTTCGGTGAA | pEAQ-EGFP | Age I |
| R:gcccttgctcaccataccggt GGTGGACACCTTAAGATGCT | pEAQ-EGFP | Age I | |
| pEAQ-EGFP-Universal | F:AGAGTTTTCCCGTGGTTTTCGAACT | ||
| R:GGACACGCTGAACTTGTGGCCGTTT | |||
| pTRV2-SmSP1 | F:gtgagtaaggttaccgaattc ATGGCTCCATGGGGCGGA | pTRV2 | EcoR I |
| R:cgtgagctcggtaccggatcc TCAATGGCGAAAAGTTTTCACA | pTRV2 | BamH I | |
| pTRV2-SmSPL2 | F:gtgagtaaggttaccgaattc ATGTCAATACACGACCAAGCGG | pTRV2 | EcoR I |
| R:cgtgagctcggtaccggatcc TCAAGAATCATATATCCTTACGGA | pTRV2 | BamH I | |
| pTRV2-SmDDA1a1 | F:gtgagtaaggttaccgaattc ATGGGGTCAATTTTCGGTGAA | pTRV2 | EcoR I |
| R:cgtgagctcggtaccggatcc TCAGGTGGACACCTTAAGATGCT | pTRV2 | BamH I | |
| pTRV2-SmDDA1a2 | F:gtgagtaaggttaccgaattc ATGGGGTCAATTTTCGGTGAA | pTRV2 | EcoR I |
| R:cgtgagctcggtaccggatcc TCAGGTGGACACCTTAAGATGCT | pTRV2 | BamH I | |
| pTRV2-Universal | F:TGAGGGAAAAGTAGAGAACG | ||
| R:CCTATGGTAAGACAATGAGT |
Table 1 List of the primers for subcellular localization and VIGS assays
| 名称 Name | 序列(5′-3′) Sequence | 质粒 Plasmid | 酶切位点 Restriction site |
|---|---|---|---|
| SmSP1-GFP | F:ctgcccaaattcgcgaccggt ATGGCTCCATGGGGCGGA | pEAQ-EGFP | Age I |
| R:gcccttgctcaccataccggt ATGGCGAAAAGTTTTCACA | pEAQ-EGFP | Age I | |
| SmSPL2-GFP | F:ctgcccaaattcgcgaccggt ATGTCAATACACGACCAAGCGG | pEAQ-EGFP | Age I |
| R:gcccttgctcaccataccggt AGAATCATATATCCTTACGGAACT | pEAQ-EGFP | Age I | |
| SmDDA1a1-GFP | F:ctgcccaaattcgcgaccggtATGGGGTCAATTTTCGGTGAA | pEAQ-EGFP | Age I |
| R:gcccttgctcaccataccggt GGTGGACACCTTAAGATGCT | pEAQ-EGFP | Age I | |
| SmDDA1a2-GFP | F:ctgcccaaattcgcgaccggt ATGGGGTCAATTTTCGGTGAA | pEAQ-EGFP | Age I |
| R:gcccttgctcaccataccggt GGTGGACACCTTAAGATGCT | pEAQ-EGFP | Age I | |
| pEAQ-EGFP-Universal | F:AGAGTTTTCCCGTGGTTTTCGAACT | ||
| R:GGACACGCTGAACTTGTGGCCGTTT | |||
| pTRV2-SmSP1 | F:gtgagtaaggttaccgaattc ATGGCTCCATGGGGCGGA | pTRV2 | EcoR I |
| R:cgtgagctcggtaccggatcc TCAATGGCGAAAAGTTTTCACA | pTRV2 | BamH I | |
| pTRV2-SmSPL2 | F:gtgagtaaggttaccgaattc ATGTCAATACACGACCAAGCGG | pTRV2 | EcoR I |
| R:cgtgagctcggtaccggatcc TCAAGAATCATATATCCTTACGGA | pTRV2 | BamH I | |
| pTRV2-SmDDA1a1 | F:gtgagtaaggttaccgaattc ATGGGGTCAATTTTCGGTGAA | pTRV2 | EcoR I |
| R:cgtgagctcggtaccggatcc TCAGGTGGACACCTTAAGATGCT | pTRV2 | BamH I | |
| pTRV2-SmDDA1a2 | F:gtgagtaaggttaccgaattc ATGGGGTCAATTTTCGGTGAA | pTRV2 | EcoR I |
| R:cgtgagctcggtaccggatcc TCAGGTGGACACCTTAAGATGCT | pTRV2 | BamH I | |
| pTRV2-Universal | F:TGAGGGAAAAGTAGAGAACG | ||
| R:CCTATGGTAAGACAATGAGT |
| 用途 Application | 名称 Name | 序列(5′-3′) Sequence |
|---|---|---|
| 表达组织特异性分析 Expression tissue specificity analysis 青枯菌/激素处理 R. solanacearum/hormone treatment VIGS实验 VIGS assay | qPCR-SmSP1 | F:CAAGTATGCTTCAATGGGTT |
| R:GAGGTCCTTTCTATTATCTGCT | ||
| qPCR-SmSPL2 | F:CATCCCCTACCTCTTGTG | |
| R:AAATCCCATCCTTTGAGC | ||
| qPCR-SmDDA1a1 | F:ATCAAGGAAATGCAGGCA | |
| R:TGGACACCTTAAGATGCT | ||
| qPCR-SmDDA1a2 | F:CGCCTGTGACTTATCGCCCTAC | |
| R:CTCGCTTTGGTCTCAACTTCTC | ||
| SA途径信号基因表达 SA pathway signaling gene expression | qPCR-TAG | F:GCAAGTGACCCTGAACTACGAAG |
| R:GGGTTTTCCACATCCCTGACAAG | ||
| qPCR-NPR1 | F:CTTGGACTGGGTGTTGCTAATG | |
| R:TGCCCATCCAATGTAATGTCTG | ||
| qPCR-EDS1 | F:GTTTCGCAGACAAGTTGAGCC | |
| R:CTCTGTGTGAACCGATAACGC | ||
| qPCR-PAD4 | F:ACATCGGCTGAAACCTCCTTATT | |
| R:TTTGATAAGTGGTGGGGAAATGA | ||
| qPCR-SGT1 | F:TTCTCGGTTTTGAGGAAGGG | |
| R:GCAGATACCAAGTGATGTCTACCA | ||
| qPCR-GluA | F:GCCGACTGGGTGAGATGGTAA | |
| R:ACATTGTTGTGCCCGTGGAC | ||
| 内参基因 Internal reference gene | qPCR-SmActin | F:GTCGGAATGGGACAGAAGGATG |
| R:GTGCCTCAGTCAGGAGAACAGGGT |
Table 2 List of the primers of qRT-PCR
| 用途 Application | 名称 Name | 序列(5′-3′) Sequence |
|---|---|---|
| 表达组织特异性分析 Expression tissue specificity analysis 青枯菌/激素处理 R. solanacearum/hormone treatment VIGS实验 VIGS assay | qPCR-SmSP1 | F:CAAGTATGCTTCAATGGGTT |
| R:GAGGTCCTTTCTATTATCTGCT | ||
| qPCR-SmSPL2 | F:CATCCCCTACCTCTTGTG | |
| R:AAATCCCATCCTTTGAGC | ||
| qPCR-SmDDA1a1 | F:ATCAAGGAAATGCAGGCA | |
| R:TGGACACCTTAAGATGCT | ||
| qPCR-SmDDA1a2 | F:CGCCTGTGACTTATCGCCCTAC | |
| R:CTCGCTTTGGTCTCAACTTCTC | ||
| SA途径信号基因表达 SA pathway signaling gene expression | qPCR-TAG | F:GCAAGTGACCCTGAACTACGAAG |
| R:GGGTTTTCCACATCCCTGACAAG | ||
| qPCR-NPR1 | F:CTTGGACTGGGTGTTGCTAATG | |
| R:TGCCCATCCAATGTAATGTCTG | ||
| qPCR-EDS1 | F:GTTTCGCAGACAAGTTGAGCC | |
| R:CTCTGTGTGAACCGATAACGC | ||
| qPCR-PAD4 | F:ACATCGGCTGAAACCTCCTTATT | |
| R:TTTGATAAGTGGTGGGGAAATGA | ||
| qPCR-SGT1 | F:TTCTCGGTTTTGAGGAAGGG | |
| R:GCAGATACCAAGTGATGTCTACCA | ||
| qPCR-GluA | F:GCCGACTGGGTGAGATGGTAA | |
| R:ACATTGTTGTGCCCGTGGAC | ||
| 内参基因 Internal reference gene | qPCR-SmActin | F:GTCGGAATGGGACAGAAGGATG |
| R:GTGCCTCAGTCAGGAGAACAGGGT |

Fig. 1 The transcriptome heat map(A)and domain(B)of four genes of eggplant inbred lines with high resistance and high susceptibility(E-31 and E-32)inoculated with Ralstonia solanacearum(0 and 7 days)

Fig. 2 Subcellular localization analysis of SmSP1,SmSPL2,SmDDA1a1 and SmDDA1a2 GFP stands for green fluorescent protein,and NLS stands for nuclear localization signal(Sun et al.,2020).
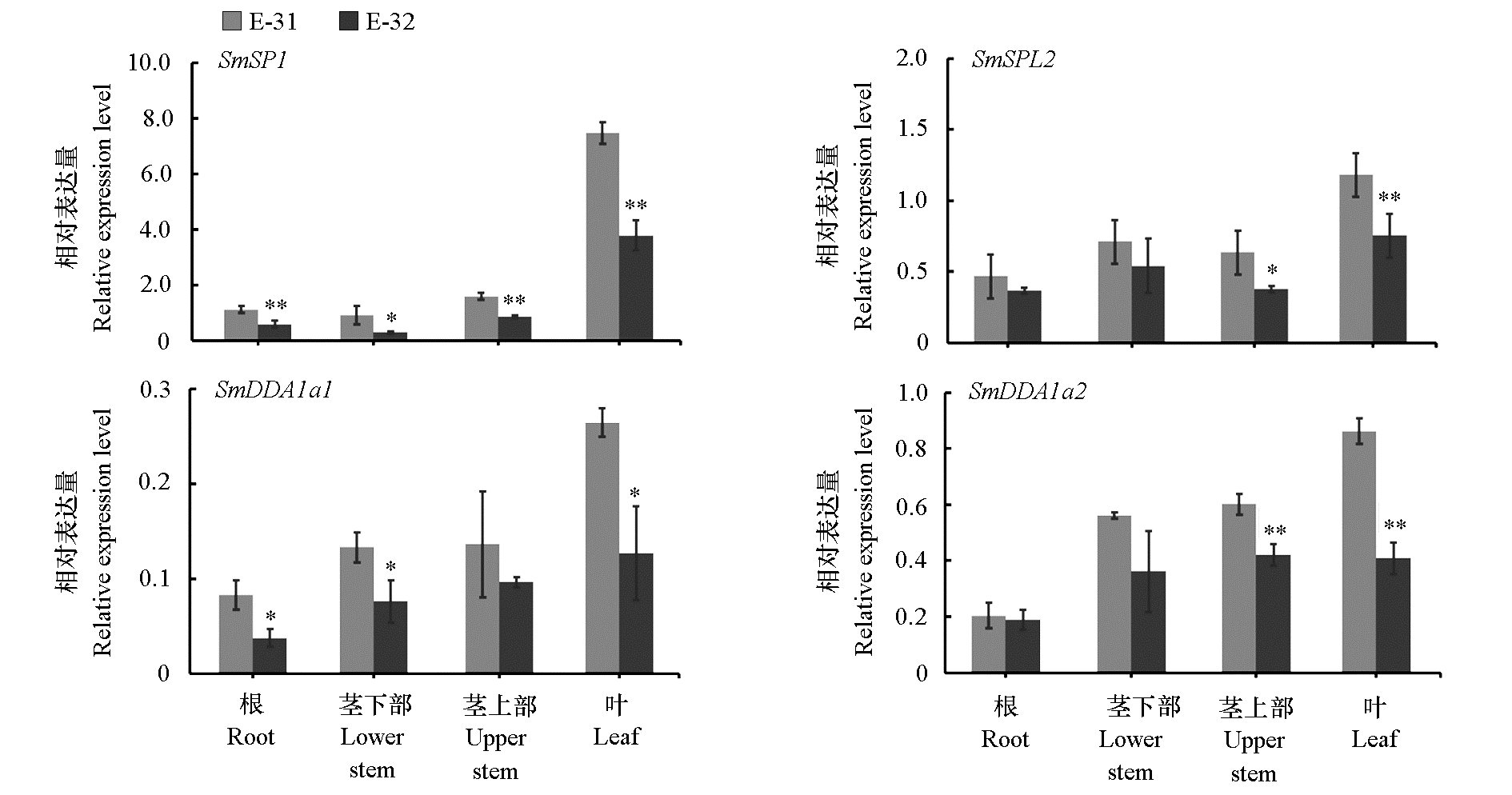
Fig. 3 The tissue-specific expression of four genes in eggplant seedlings of bacterial wilt highly resistant and highly susceptible inbred lines(E-31 and E-32) **P < 0.01;* P < 0.05.
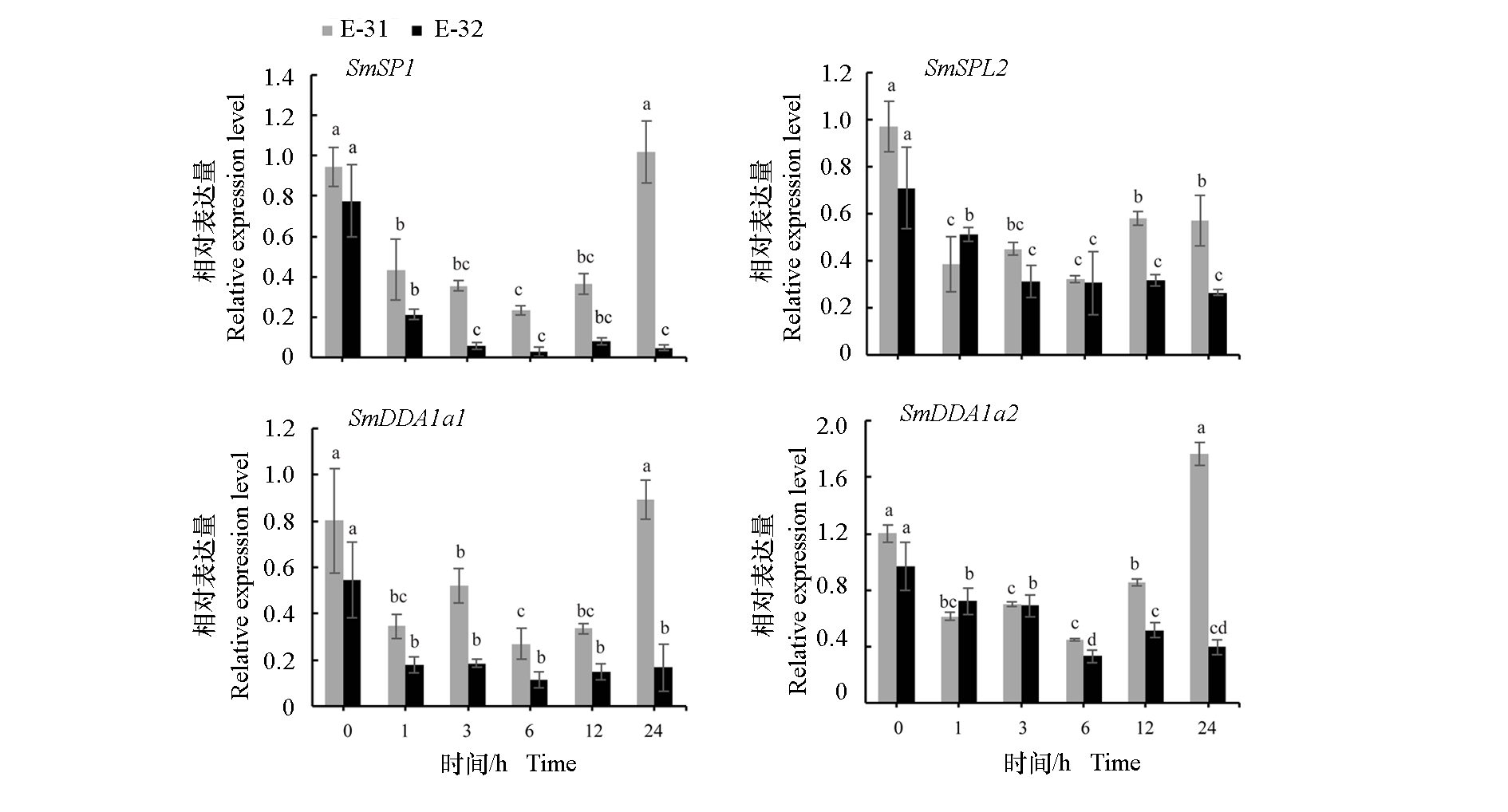
Fig. 4 The analysis of the expression levels of four genes in eggplant high resistance and high sensitivity inbred lines (E-31 and E-32)inoculated with Ralstonia solanacearum Significant difference analysis was conducted using LSD multiple comparisons,with different lowercase letters indicating significant differences in the same material at different times(P < 0.05).

Fig. 5 The expression analysis of four genes in eggplant inbred lines with high resistance to bacterial wilt after applying salicylic acid(SA),methyl jasmonic acid(MeJA)and ethrel(Eth) Significant difference analysis was conducted using LSD multiple comparisons,with different lowercase letters indicating significant differences at different times in the same treatment(P < 0.05).

Fig. 6 The expression levels and phenotypes of silenced plants after inoculation with Ralstonia solanacearum The control indicates that the plant is treated with H2O,while pTRV2 indicates that the plant injected with Agrobacterium infection solution carrying pTRV2 empty vector. The error lines of the control and pTRV2 treatment represent three biological repeated differences. Significant difference analysis was conducted using LSD multiple comparisons and represented by letter labeling(significance level 0.01),with different uppercase letters indicating significant differences in expression levels.
| 沉默基因 Silence gene | 总数/株 Total | 发病株数/株 Disease plants | 发病率/% Morbidity | 发病等级Evaluation of scale | 病情指数 Disease index | ||||
|---|---|---|---|---|---|---|---|---|---|
| 0 | 1 | 2 | 3 | 4 | |||||
| SmSP1 | 10 | 8 | 80 | 2 | 0 | 1 | 3 | 4 | 67.5 |
| SmSPL2 | 10 | 5 | 50 | 5 | 1 | 2 | 1 | 1 | 30.0 |
| SmDDA1a1 | 10 | 4 | 40 | 6 | 1 | 2 | 1 | 0 | 20.0 |
| SmDDA1a2 | 20 | 15 | 75 | 5 | 2 | 3 | 8 | 2 | 50.0 |
| 对照Control(H2O) | 10 | 0 | 0 | 10 | 0 | 0 | 0 | 0 | 0 |
| 对照Control(pTRV2) | 10 | 0 | 0 | 10 | 0 | 0 | 0 | 0 | 0 |
Table 3 The statistics list of morbidity and disease index of control and silent plants
| 沉默基因 Silence gene | 总数/株 Total | 发病株数/株 Disease plants | 发病率/% Morbidity | 发病等级Evaluation of scale | 病情指数 Disease index | ||||
|---|---|---|---|---|---|---|---|---|---|
| 0 | 1 | 2 | 3 | 4 | |||||
| SmSP1 | 10 | 8 | 80 | 2 | 0 | 1 | 3 | 4 | 67.5 |
| SmSPL2 | 10 | 5 | 50 | 5 | 1 | 2 | 1 | 1 | 30.0 |
| SmDDA1a1 | 10 | 4 | 40 | 6 | 1 | 2 | 1 | 0 | 20.0 |
| SmDDA1a2 | 20 | 15 | 75 | 5 | 2 | 3 | 8 | 2 | 50.0 |
| 对照Control(H2O) | 10 | 0 | 0 | 10 | 0 | 0 | 0 | 0 | 0 |
| 对照Control(pTRV2) | 10 | 0 | 0 | 10 | 0 | 0 | 0 | 0 | 0 |
| [1] |
|
| [2] |
doi: 10.1016/j.bbrc.2014.01.098 pmid: 24486490 |
| [3] |
doi: 10.1038/s41598-018-37186-2 |
| [4] |
doi: 10.1038/nsmb.2780 |
| [5] |
doi: 10.1105/tpc.106.048140 URL |
| [6] |
doi: 10.1104/pp.113.226837 URL |
| [7] |
|
| [8] |
doi: 10.1016/j.molp.2020.06.009 URL |
| [9] |
doi: 10.1038/srep31568 |
| [10] |
doi: 10.1016/j.gene.2017.11.003 URL |
| [11] |
doi: 10.1016/S0968-0004(02)02125-4 URL |
| [12] |
doi: 10.1111/mpp.2010.11.issue-2 URL |
| [13] |
doi: 10.3389/fpls.2015.00154 pmid: 25821454 |
| [14] |
doi: 10.1074/jbc.M413247200 pmid: 15749712 |
| [15] |
doi: 10.1007/s00425-018-03069-z |
| [16] |
|
| [17] |
doi: 10.3390/ijms19051426 URL |
| [18] |
doi: 10.1093/jxb/ers360 URL |
| [19] |
doi: 10.1007/s00709-015-0769-6 URL |
| [20] |
doi: 10.1093/jxb/eraa489 URL |
| [21] |
doi: 10.1105/tpc.107.055418 URL |
| [22] |
|
|
李可, 肖熙鸥, 林文秋, 李威, 吕玲玲, 曹必好. 2018. EDS1正调控茄子抗青枯病反应. 热带作物学报, 39 (2):332-337.
|
|
| [23] |
|
|
李明亮, 韩一凡. 2000. 乙烯在植物生长发育和抗病反应中的作用及其生物合成的反义抑制. 林业科学, 36 (4):77-84.
|
|
| [24] |
doi: 10.3389/fpls.2018.00521 URL |
| [25] |
|
|
李涛, 李植良, 夏碧波, 黎振兴, 孙保娟, 李颖, 徐小万, 王恒明. 2017. 茄子创新资源的耐热及抗青枯病评价研究. 中国农学通报, 33 (36):72-77.
doi: 10.11924/j.issn.1000-6850.casb17020118 |
|
| [26] |
doi: 10.1021/acs.jafc.1c04608 URL |
| [27] |
doi: 10.1038/s41477-021-00916-y pmid: 34007040 |
| [28] |
doi: 10.3389/fpls.2017.01710 URL |
| [29] |
doi: S0006-291X(18)32660-3 pmid: 30545635 |
| [30] |
doi: 10.3390/ijms22115507 URL |
| [31] |
doi: 10.1006/meth.2001.1262 pmid: 11846609 |
| [32] |
|
| [33] |
|
|
卢同. 1998. 我国作物细菌性青枯菌的研究进展. 福建农业学报, 13 (2):33-40.
|
|
| [34] |
|
| [35] |
doi: 10.1111/tpj.12035 pmid: 23013464 |
| [36] |
doi: 10.1111/tpj.2004.40.issue-6 URL |
| [37] |
|
| [38] |
doi: 10.1104/pp.17.01536 URL |
| [39] |
doi: 10.5352/JLS.2012.22.2.177 URL |
| [40] |
doi: 10.1104/pp.15.00479 pmid: 26320228 |
| [41] |
doi: 10.3390/ijms222413279 URL |
| [42] |
doi: 10.1038/nrm1547 pmid: 15688063 |
| [43] |
doi: 10.1186/s12870-014-0410-4 URL |
| [44] |
doi: 10.1046/j.1365-313X.2002.01461.x URL |
| [45] |
|
|
乔俊卿, 陈志谊, 刘邮洲, 刘永锋, 梁雪杰, 张磊. 2013. 茄科作物青枯病研究进展. 植物病理学报, 43 (1):1-10.
|
|
| [46] |
doi: 10.1007/s11103-019-00824-y |
| [47] |
doi: 10.1093/jxb/erz259 URL |
| [48] |
doi: 10.1038/373081a0 |
| [49] |
|
|
佘小漫, 何自福, 罗学梅, 罗方芳. 2011. 广东主要茄子品种对青枯病的抗性鉴定与评价. 广东农业科学, 14 (38):72-73.
|
|
| [50] |
doi: 10.1104/pp.104.052423 URL |
| [51] |
doi: 10.1038/s41438-019-0222-7 |
| [52] |
doi: 10.1016/j.ceb.2004.02.005 pmid: 15196553 |
| [53] |
doi: 10.1016/j.pbi.2010.04.002 pmid: 20471305 |
| [54] |
doi: 10.1038/nrm2688 pmid: 19424292 |
| [55] |
doi: 10.16420/j.issn.0513-353x.2015-0652 URL |
|
肖熙鸥, 蒋晶, 陈娜, 雷建军, 吕玲玲, 曹必好. 2016. 茄子调控抗青枯病反应信号基因的筛选和鉴定. 园艺学报, 43 (7):1295-1304.
doi: 10.16420/j.issn.0513-353x.2015-0652 URL |
|
| [56] |
|
|
肖熙鸥, 李冠男, 曹必好, 雷建军, 陈清华, 陈国菊. 2012. 青枯病病菌对茄子相关防卫信号基因表达及活性氧含量的影响. 植物生理学报, 48 (9):874-880.
|
|
| [57] |
|
| [58] |
doi: 10.1101/gad.1229504 URL |
| [59] |
|
|
杨虹伟, 翟先芝, 刘志斌, 王健美, 李旭锋, 杨毅. 2013. 两个高度同源的RING结构域蛋白功能初步研究. 中国农业科技导报, 15 (5):100-105.
|
|
| [60] |
|
|
杨伟, 简桂良, 赵磊, 陈大军. 2006. 乙烯代谢与植物抗病性//科技创新与绿色植保. 昆明:中国植物保护学会2006学术年会论文集.
|
|
| [61] |
|
|
翟先芝, 杨虹伟, 杨皓, 刘志斌, 王健美, 杨毅, 李旭锋. 2015. 拟南芥 AtRAC在盐胁迫应答中的功能研究. 四川大学学报(自然科学版), 52 (2):416-422.
|
|
| [62] |
doi: S0168-9452(16)30430-7 pmid: 28483055 |
| [63] |
|
|
周大祥, 熊书. 2015. 外源茉莉酸甲酯诱导竹根姜对青枯菌的抗性. 西北植物学报, 35 (7):1415-1420.
|
| [1] | WANG Hong and ZHANG Ya. A New Eggplant Cultivar‘Hangqie 2010’ [J]. Acta Horticulturae Sinica, 2023, 50(S1): 61-62. |
| [2] | LI Yuteng, CHEN Yao, REN Hengze, LI Congcong, WANG Haoqian, CAO Hongli, YUE Chuan, HAO Xinyuan, WANG Xinchao. Identification,Expression Analysis and Interaction Validation of CsIDM in Tea Plants [J]. Acta Horticulturae Sinica, 2023, 50(8): 1679-1696. |
| [3] | XUE Zhenzhen, GUAN Hongfa, LI Na, LI Lingfei, ZHONG Chunmei. Genome-wide Identification of GASA Family and Preliminary Exploration of Leaf Variegation Formation in Begonia masoniana [J]. Acta Horticulturae Sinica, 2023, 50(7): 1482-1494. |
| [4] | LI Shupei, ZHANG Ying, SHU Jinshuai, CHEN Yuhui, YANG Jinkun, CHEN Lulu, LIU Fuzhong. The Characteristics and Chloroplast Ultrastructure of Yellow Leaf Mutant chl234 in Eggplant [J]. Acta Horticulturae Sinica, 2023, 50(5): 1000-1008. |
| [5] | WANG Tonghuan, WU Yuxin, WU Yiyuan, LI Xinxin, LIU Mengyang, YANG Lianlian, LI Jiapeng, ZHANG Zhongshan, CAO Fang, ZHONG Xueting, WANG Zhanqi. Genome-wide Identification and Expression Analysis of the GRAS Gene Family in Response to Cold Stress in Chrysanthemum nankingense [J]. Acta Horticulturae Sinica, 2023, 50(4): 815-830. |
| [6] | LIU Hui, HUANG Ting, TAO Jianping, ZHANG Jiaqi, ZHANG Rongrong, SONG Liuxia, ZHAO Tongmin, YOU Xiong, XIONG Aisheng. Analysis of Circadian Clock Genes SlLNK1,SlEID1,and SlELF3 Response to Photoperiod in Tomato [J]. Acta Horticulturae Sinica, 2023, 50(10): 2079-2090. |
| [7] | ZHAO Xueyan, WANG Qi, WANG Li, WANG Fangyuan, WANG Qing, LI Yan. Comparative Transcriptome Analysis of Differential Expression in Different Tissues of Corydalis yanhusuo [J]. Acta Horticulturae Sinica, 2023, 50(1): 177-187. |
| [8] | LI Zhiliang, LI Zhenxing, LI Tao, SUN Baojuan, LI Ying, XU Xiaowan, WANG Hengming, HENG Zhou, GONG Chao. A New Eggplant Cultivar‘Xinfeng 3’ [J]. Acta Horticulturae Sinica, 2023, 50(1): 227-228. |
| [9] | CAI Peng, FANG Chao, LI Yuejian, LIU Duchen, LIU Xiaojun, LIANG Genyun. A New Eggplant Cultivar‘Tianjiao’ [J]. Acta Horticulturae Sinica, 2023, 50(1): 229-230. |
| [10] | TAN Jie, HUANG Shuping, CHEN Xia, ZHANG Hongyuan, LI Ye, WANG Benqi, CHEN Hao, WU Xuexia, and ZHANG Min, . A New Eggplant Cultivar‘Eqie 5’ [J]. Acta Horticulturae Sinica, 2022, 49(S2): 99-100. |
| [11] | HUANG Shuping, TAN Jie, Chen Xia, ZHANG Hongyuan, LI Ye, WANG Benqi, CHEN Hao, WU Xuexia, and ZHANG Min, . A New Eggplant Cultivar‘Eqie 6’ [J]. Acta Horticulturae Sinica, 2022, 49(S2): 101-102. |
| [12] | YANG Yang, TIAN Shibing, WANG Yongqing, JIANG Changchun, ZOU Min, TAO Tao, ZHOU Shanshan, WANG Zhijin, BAO Zhongxian, and TANG Xiaohua. A New Eggplant Cultivar‘Yuqie 5’ [J]. Acta Horticulturae Sinica, 2022, 49(S1): 77-78. |
| [13] | GAO Yanlong, WU Yuxia, ZHANG Zhongxing, WANG Shuangcheng, ZHANG Rui, ZHANG De, WANG Yanxiu. Bioinformatics Analysis of Apple ELO Gene Family and Its Expression Analysis Under Low Temperature Stress [J]. Acta Horticulturae Sinica, 2022, 49(8): 1621-1636. |
| [14] | LIN Yuanmi, ZHU Wenjiao, CHEN Min, XUE Chunmei, JIN Fangyu, ZHU Yuping, JIANG Xinyue, YE Lingfeng, NI Shunanling, YANG Qing. Mir396b Negatively Regulates Eggplant Defense Response to Verticillium Wilt [J]. Acta Horticulturae Sinica, 2022, 49(8): 1713-1722. |
| [15] | QIU Ziwen, LIU Linmin, LIN Yongsheng, LIN Xiaojie, LI Yongyu, WU Shaohua, YANG Chao. Cloning and Functional Analysis of the MbEGS Gene from Melaleuca bracteata [J]. Acta Horticulturae Sinica, 2022, 49(8): 1747-1760. |
| Viewed | ||||||
|
Full text |
|
|||||
|
Abstract |
|
|||||
Copyright © 2012 Acta Horticulturae Sinica 京ICP备10030308号-2 国际联网备案号 11010802023439
Tel: 010-82109523 E-Mail: yuanyixuebao@126.com
Support by: Beijing Magtech Co.Ltd