
Acta Horticulturae Sinica ›› 2023, Vol. 50 ›› Issue (3): 523-533.doi: 10.16420/j.issn.0513-353x.2021-1196
• Research Papers • Previous Articles Next Articles
MA Shuaihui, HE Guangqi, CHENG Yizhe, GUO Dalong*( )
)
Received:2022-04-18
Revised:2022-10-03
Online:2023-03-25
Published:2023-04-03
Contact:
*(E-mail:guodalong@haust.edu.cn)
CLC Number:
MA Shuaihui, HE Guangqi, CHENG Yizhe, GUO Dalong. Analysis of Alternative Splicing at Different Developmental Stages of Kyoho Grapes with 5-azaC Treatment[J]. Acta Horticulturae Sinica, 2023, 50(3): 523-533.
Add to citation manager EndNote|Ris|BibTeX
URL: https://www.ahs.ac.cn/EN/10.16420/j.issn.0513-353x.2021-1196
| 处理 Treatment | 花后天数 Days post-anthesis | 原始序列 Raw data | 高质量序列 Clean data | 唯一比对率/% Unique mapping rate |
|---|---|---|---|---|
| 5-azaC | 25 | 46 079 024 | 44 673 747 | 96.81 |
| 35 | 47 832 989 | 46 379 318 | 96.78 | |
| 45 | 48 174 649 | 46 431 546 | 96.76 | |
| 55 | 46 588 342 | 45 124 854 | 96.71 | |
| 65 | 46 919 474 | 45 539 185 | 96.25 | |
| H2O(对照Control) | 25 | 47 422 767 | 45 914 784 | 96.89 |
| 35 | 48 921 036 | 47 464 829 | 96.81 | |
| 45 | 43 858 148 | 42 261 038 | 95.99 | |
| 55 | 47 506 354 | 45 916 451 | 96.42 | |
| 65 | 48 515 002 | 46 692 047 | 95.91 |
Table 1 Fruit sample collection and RNA-seq data mapping information
| 处理 Treatment | 花后天数 Days post-anthesis | 原始序列 Raw data | 高质量序列 Clean data | 唯一比对率/% Unique mapping rate |
|---|---|---|---|---|
| 5-azaC | 25 | 46 079 024 | 44 673 747 | 96.81 |
| 35 | 47 832 989 | 46 379 318 | 96.78 | |
| 45 | 48 174 649 | 46 431 546 | 96.76 | |
| 55 | 46 588 342 | 45 124 854 | 96.71 | |
| 65 | 46 919 474 | 45 539 185 | 96.25 | |
| H2O(对照Control) | 25 | 47 422 767 | 45 914 784 | 96.89 |
| 35 | 48 921 036 | 47 464 829 | 96.81 | |
| 45 | 43 858 148 | 42 261 038 | 95.99 | |
| 55 | 47 506 354 | 45 916 451 | 96.42 | |
| 65 | 48 515 002 | 46 692 047 | 95.91 |
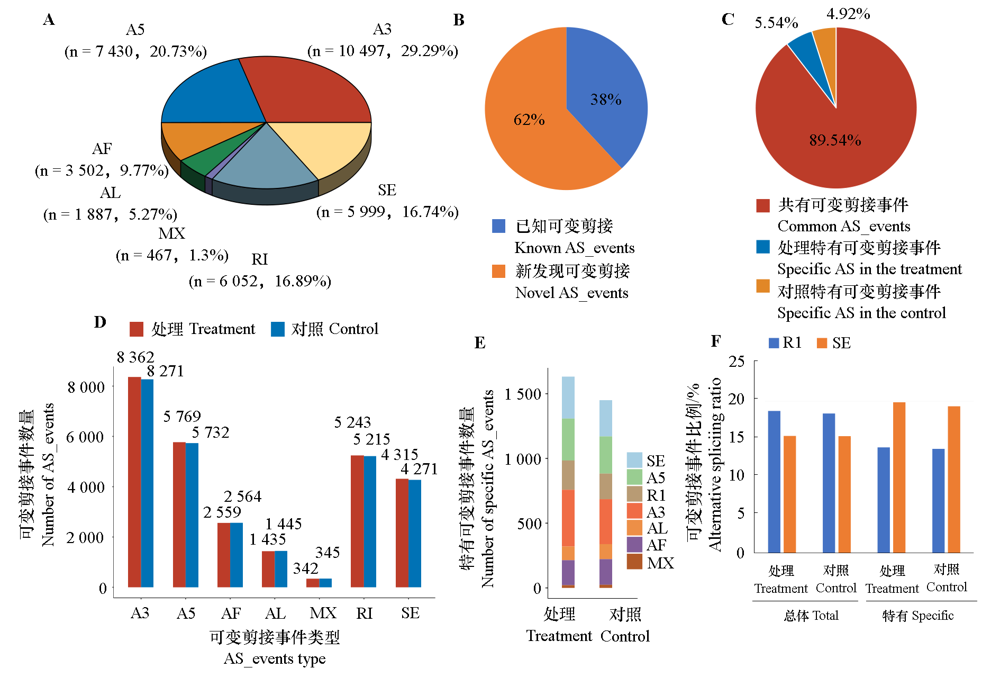
Fig. 1 Identification of alternative splicing(AS)events in Kyoho A:Overall proportion ofseven types of AS;B:Overall proportion of known and novel AS events;C:Proportion of common and unique AS to effective AS events;D:Proportion of each type of effective AS events in the treatment and control groups;E:Number of each type of unique AS events in the treatment and control groups;F:Change in proportion of both RI and SE types of events in the overall and unique AS events.

Fig. 3 Comparison of the number of AS events in Kyoho 5-azaC treatment and control A:Number of Number of AS types;B and C:Comparison of AS events at five sampling points in the control group(C1-C5)and treatment group(A1-A5);D:Comparison of conservative AS events in the treatment and control groups.
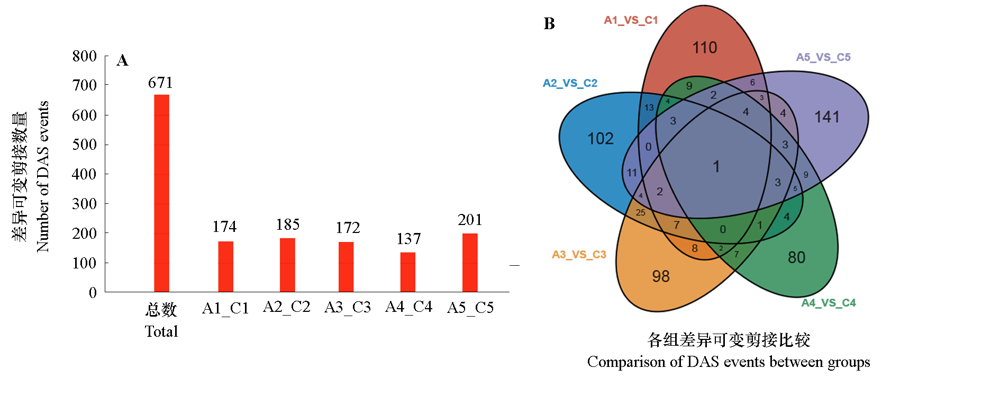
Fig. 4 Statistics on the number of DAS events between the 5-azaC treatment and control in kyoho C1-C5 and A1-A5 are samples from 25,35,45,55 and 65 d after flowering for control and treatment,respectively.
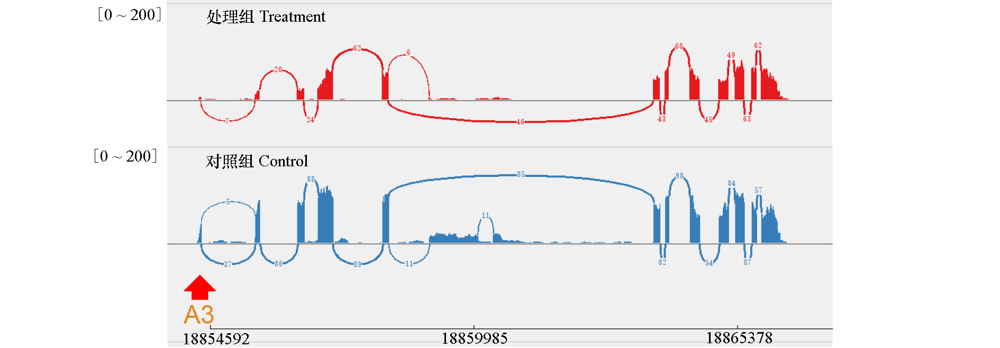
Fig. 5 Visualization of sequencing results for the Vitvi11g01240 locus Red arrows indicate alternative splicing positions.“A3”character indicate the alternative splicing type,the lower numbers indicated the location site in the genome,the upper numbers showed the value range of the reads.
| 差异组 Difference group | 可变剪接事件ID AS_events ID | 参考基因组ID Reference genome ID | Swissprot注释 Swissprot annotation | 剪接率变化 ΔPSI | P |
|---|---|---|---|---|---|
| A1 vs. C1 | MSTRG.15527;A3:chr11:18854416-18855518:18854416-18855521:+ | Vitvi11g01240 | Q803K4|N-lysine methyltransferase setd6 | -0.385000 | 0.016983 |
| MSTRG.24077;A3:chr18:4489688-4490608:4488547-4490608:- | Vitvi18g00414 | Q9C9Q8|Probable pectin methylransferase QUA2 | -0.954810 | 0.013986 | |
| MSTRG.27205;A5:chr19:23341818-23342980:23341818-23342984:- | Vitvi19g01686 | O34614|Putative rRNA methylase YtqB | 0.238771 | 0.042957 | |
| MSTRG.42;A3:chr00:3483728-3498435:3483728-3498440:+ | Vitvi07g02774 | O49354|Ubiquinone biosynthesis O-methyltransferase,mitochondrial | 0.397344 | 0.003996 | |
| A2 vs. C2 | MSTRG.14725;SE:chr11:2089410-2089537:2089678-2089991:+ | Vitvi11g00218 | Q9C5D7|Probable caffeoyl-CoA O-methyltransferase At4g26220 | -0.218300 | 0.011988 |
| MSTRG.22755;A3:chr17:1079931-1080008:1079931-1080110:+ | Vitvi17g00106 | Q6NMK1|Glucuronoxylan 4-O-methyltransferase 1 | 0.436952 | 0.036963 | |
| MSTRG.4536;AL:chr03:17733301-17735190:17735272:17733301-17736694:17736812:+ | Vitvi03g01141 | Q8VXV7|Lysine-specific histone demethylase 1 homolog 1 | 0.953023 | 0.003996 | |
| MSTRG.6764;A5:chr05:9227636-9227771:9227636-9227788:- | Vitvi05g00821 | Q8W595|Histone-lysine N-methyltransferase SUVR4 | 0.340451 | 0.036963 | |
| MSTRG.9695;A5:chr07:16439920-16440048:16439920-16440056:- | Vitvi07g01181 | P19672|Putative rRNA methyltransferase YqxC | -0.214630 | 0.032967 | |
| A3 vs. C3 | MSTRG.3140;A3:chr02:6450274-6450465:6450274-6450502:+ | Vitvi02g01481 | Q8GT41|PLA1_PLAAC(Putative invertase inhibitor | -0.298220 | 0.047952 |
| MSTRG.5600;RI:chr04:19046492:19046896-19046989:19047215:- | Vitvi04g01339 | Q9FYZ9|Benzoate carboxyl methyltransferase | -0.425830 | 0.018482 | |
| A4 vs. C4 | MSTRG.4755;A5:chr04:1209673-1211033:1209641-1211033:+ | Vitvi04g00134 | Q96P11|Probable 28S rRNA(cytosine-C(5))- methyltransferase | 0.922624 | 0.044955 |
| A5 vs. C5 | MSTRG.17335;A3:chr13:1446344-1446866:1446344-1446940:+ | Vitvi13g00159 | B6YUU9|tRNA(guanine(26)- N(2))-dimethyltransferase | 0.482021 | 0.040460 |
| MSTRG.17774;A3:chr13:6735007-6735041:6734974-6735041:- | Vitvi13g00674 | F4JW79|Protein RNA-directed DNA methylation 3 | 0.638740 | 0.016983 |
Table 2 DAS events associated with methylation
| 差异组 Difference group | 可变剪接事件ID AS_events ID | 参考基因组ID Reference genome ID | Swissprot注释 Swissprot annotation | 剪接率变化 ΔPSI | P |
|---|---|---|---|---|---|
| A1 vs. C1 | MSTRG.15527;A3:chr11:18854416-18855518:18854416-18855521:+ | Vitvi11g01240 | Q803K4|N-lysine methyltransferase setd6 | -0.385000 | 0.016983 |
| MSTRG.24077;A3:chr18:4489688-4490608:4488547-4490608:- | Vitvi18g00414 | Q9C9Q8|Probable pectin methylransferase QUA2 | -0.954810 | 0.013986 | |
| MSTRG.27205;A5:chr19:23341818-23342980:23341818-23342984:- | Vitvi19g01686 | O34614|Putative rRNA methylase YtqB | 0.238771 | 0.042957 | |
| MSTRG.42;A3:chr00:3483728-3498435:3483728-3498440:+ | Vitvi07g02774 | O49354|Ubiquinone biosynthesis O-methyltransferase,mitochondrial | 0.397344 | 0.003996 | |
| A2 vs. C2 | MSTRG.14725;SE:chr11:2089410-2089537:2089678-2089991:+ | Vitvi11g00218 | Q9C5D7|Probable caffeoyl-CoA O-methyltransferase At4g26220 | -0.218300 | 0.011988 |
| MSTRG.22755;A3:chr17:1079931-1080008:1079931-1080110:+ | Vitvi17g00106 | Q6NMK1|Glucuronoxylan 4-O-methyltransferase 1 | 0.436952 | 0.036963 | |
| MSTRG.4536;AL:chr03:17733301-17735190:17735272:17733301-17736694:17736812:+ | Vitvi03g01141 | Q8VXV7|Lysine-specific histone demethylase 1 homolog 1 | 0.953023 | 0.003996 | |
| MSTRG.6764;A5:chr05:9227636-9227771:9227636-9227788:- | Vitvi05g00821 | Q8W595|Histone-lysine N-methyltransferase SUVR4 | 0.340451 | 0.036963 | |
| MSTRG.9695;A5:chr07:16439920-16440048:16439920-16440056:- | Vitvi07g01181 | P19672|Putative rRNA methyltransferase YqxC | -0.214630 | 0.032967 | |
| A3 vs. C3 | MSTRG.3140;A3:chr02:6450274-6450465:6450274-6450502:+ | Vitvi02g01481 | Q8GT41|PLA1_PLAAC(Putative invertase inhibitor | -0.298220 | 0.047952 |
| MSTRG.5600;RI:chr04:19046492:19046896-19046989:19047215:- | Vitvi04g01339 | Q9FYZ9|Benzoate carboxyl methyltransferase | -0.425830 | 0.018482 | |
| A4 vs. C4 | MSTRG.4755;A5:chr04:1209673-1211033:1209641-1211033:+ | Vitvi04g00134 | Q96P11|Probable 28S rRNA(cytosine-C(5))- methyltransferase | 0.922624 | 0.044955 |
| A5 vs. C5 | MSTRG.17335;A3:chr13:1446344-1446866:1446344-1446940:+ | Vitvi13g00159 | B6YUU9|tRNA(guanine(26)- N(2))-dimethyltransferase | 0.482021 | 0.040460 |
| MSTRG.17774;A3:chr13:6735007-6735041:6734974-6735041:- | Vitvi13g00674 | F4JW79|Protein RNA-directed DNA methylation 3 | 0.638740 | 0.016983 |
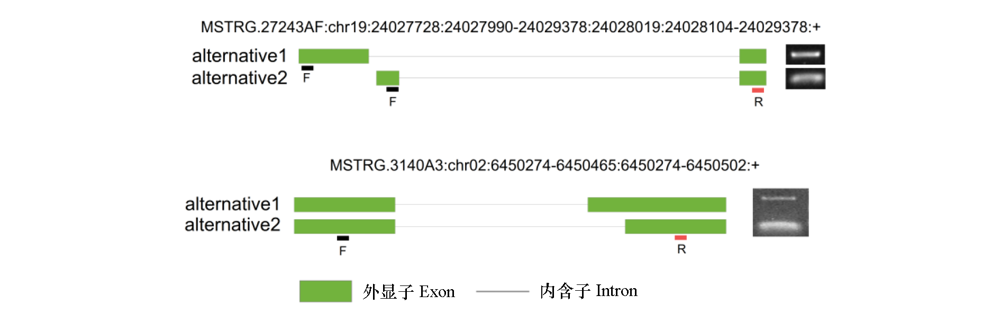
Fig. 6 Alternative splicing events validation The light green rectangle represents the exon region,the black line represents the intron region,“F”represents the forward primer position,“R”represents the reverse primer position,and“alternative1”and“alternative2”represent two different splicing forms of the alternative splicing events.
| [1] | Bian Lu, Guo Dalong, Yu Keke, Wei Tonglu, Pei Maosong, Liu Hainan, Yu Yihe. 2021. Cloning and expression analysis of the cytokinin response regulator VlRR5 in Kyoho grapevine. Acta Horticulturae Sinica, 48 (8):1437-1445. (in Chinese) |
|
边璐, 郭大龙, 于可可, 韦同路, 裴茂松, 刘海楠, 余义和. 2021. ‘巨峰’葡萄细胞分裂素响应调节因子VlRR5的克隆与表达分析. 园艺学报, 48 (8):1437-1445.
doi: 10.16420/j.issn.0513-353x.2020-0668 |
|
| [2] | Canaguier A, Grimplet J, Di Gaspero G, Scalabrin S, Duchene E, Choisne N, Mohellibi N, Guichard C, Rombauts S, Le Clainche I, Berard A, Chauveau A, Bounon R, Rustenholz C, Morgante M, Le Paslier M C, Brunel D, Adam-Blondon A F. 2017. A new version of the grapevine reference genome assembly(12X.v2) and of its annotation(VCost.v3). Genom Data, 14:56-62. |
| [3] |
Liu Chenghui, Shi Lijia, Jiang Aili, Hu Wwnzhong. 2021. An analysis of alternative splicing events during browning inhibition of fresh-cut apples by hydrogen sulfide treatment. Acta Hortic Sin, 48 (11):2121-2132. (in Chinese)
doi: 10.16420/j.issn.0513-353x.2020-0847 URL |
|
陈晨, 刘程惠, 石立佳, 姜爱丽, 胡文忠. 2021. 硫化氢控制鲜切苹果褐变的可变剪切基因分析. 园艺学报, 48 (11):2121-2132.
doi: 10.16420/j.issn.0513-353x.2020-0847 URL |
|
| [4] |
Dobin A, Davis C A, Schlesinger F, Drenkow J, Zaleski C, Jha S, Batut P, Chaisson M, Gingeras T R. 2013. STAR:ultrafast universal RNA-seq aligner. Bioinformatics, 29 (1):15-21.
doi: 10.1093/bioinformatics/bts635 URL |
| [5] |
Du J, Kirui A, Huang S X, Wang L L, Barnes W J, Kiemle S, Zheng Y Z, Rui Y, Ruan M, Qi S Q, Kim S H, Wang T, Cosgrove D J, Anderson C T, Xiao C W. 2020. Mutations in the pectin methyltransferase QUASIMODO 2 influence cellulose biosynthesis and wall integrity in Arabidopsis thaliana. The Plant Cell, 32 (11):3576-3597.
doi: 10.1105/tpc.20.00252 URL |
| [6] |
Gao P, Quilichini T D, Zhai C, Qin L, Nilsen K T, Li Q, Sharpe A G, Kochian L V, Zou J, Reddy A, Wei Y, Pozniak C, Patterson N, Gillmor C S, Datla R, Xiang D. 2021. Alternative splicing dynamics and evolutionary divergence during embryogenesis in wheat species. Plant Biotechnol J, 19 (8):1624-1643.
doi: 10.1111/pbi.13579 pmid: 33706417 |
| [7] |
Guo D L, Li Q, Ji X R, Wang Z G, Yu Y H. 2019a. Transcriptome profiling of‘Kyoho’grape at different stages of berry development following 5-azaC treatment. BMC Genomics, 20:825.
doi: 10.1186/s12864-019-6204-1 |
| [8] |
Guo D, Li Q, Zhao H, Wang Z, Zhang G, Yu Y. 2019b. The variation of berry development and DNA methylation after treatment with 5-azaC on ‘Kyoho’grape. Scientia Horticulturae, 246:265-271
doi: 10.1016/j.scienta.2018.11.006 URL |
| [9] |
Hu Y, Mesihovic A, Jimenez-Gomez J M, Roth S, Gebhardt P, Bublak D, Bovy A, Scharf K D, Schleiff E, Fragkostefanakis S. 2020. Natural variation in HsfA 2 pre-mRNA splicing is associated with changes in thermotolerance during tomato domestication. New Phytol, 225 (3):1297-1310.
doi: 10.1111/nph.v225.3 URL |
| [10] |
Huang H, Liu R, Niu Q, Tang K, Zhang B, Zhang H, Chen K, Zhu J K, Lang Z. 2019. Global increase in DNA methylation during orange fruit development and ripening. Proc Natl Acad Sci USA, 116 (4):1430-1436.
doi: 10.1073/pnas.1815441116 pmid: 30635417 |
| [11] | Huang Ling, Li Q, Wei S, Lai J, Dai S, Zhang Q, Ceng H, Liu J, Ye P. 2019. Identification and difference analysis of the alternative splicing event in the hermaphroditic flowers and male flowers of Asparagus officinalis. Acta Hortic Sin, 46 (8):1503-1518. (in Chinese) |
|
黄玲, 李琼英, 韦树谷, 赖佳, 代顺冬, 张骞方, 曾华兰, 刘佳, 叶鹏盛. 2019. 芦笋两性花与雄花中发生可变剪接基因的差异分析. 园艺学报, 46 (8):1503-1518.
doi: 10.16420/j.issn.0513-353x.2018-1025 URL |
|
| [12] |
Jiang G, Zhang D, Li Z, Liang H, Deng R, Su X, Jiang Y, Duan X. 2021. Alternative splicing of MaMYB16L regulates starch degradation in banana fruit during ripening. J Integr Plant Biol, 63 (7):1341-1352.
doi: 10.1111/jipb.v63.7 URL |
| [13] |
Jin Z, Lv X, Sun Y, Fan Z, Xiang G, Yao Y. 2021. Comprehensive discovery of salt-responsive alternative splicing events based on Iso-Seq and RNA-seq in grapevine roots. Environmental and Experimental Botany, 192:104645.
doi: 10.1016/j.envexpbot.2021.104645 URL |
| [14] |
Katoh A, Uenohara K, Akita M, Hashimoto T. 2006. Early steps in the biosynthesis of NAD in Arabidopsis start with aspartate and occur in the plastid. Plant Physiol, 141 (3):851-857.
doi: 10.1104/pp.106.081091 URL |
| [15] | Lang Z, Wang Y, Tang K, Tang D, Datsenka T, Cheng J, Zhang Y, Handa A K, Zhu J K. 2017. Critical roles of DNA demethylation in the activation of ripening-induced genes and inhibition of ripening-repressed genes in tomato fruit. Proc Natl Acad Sci U S A, 114 (22):E4511-E4519. |
| [16] |
Li Y, Dai C, Hu C, Liu Z, Kang C. 2017. Global identification of alternative splicing via comparative analysis of SMRT-and Illumina-based RNA-seq in strawberry. Plant J, 90 (1):164-176.
doi: 10.1111/tpj.2017.90.issue-1 URL |
| [17] |
Maillot P, Velt A, Rustenholz C, Butterlin G, Merdinoglu D, Duchene E. 2021. Alternative splicing regulation appears to play a crucial role in grape berry development and is also potentially involved in adaptation responses to the environment. BMC Plant Biol, 21 (1):487.
doi: 10.1186/s12870-021-03266-1 pmid: 34696712 |
| [18] |
Patro R, Duggal G, Love M I, Irizarry R A, Kingsford C. 2017. Salmon provides fast and bias-aware quantification of transcript expression. Nat Methods, 14 (4):417-419.
doi: 10.1038/nmeth.4197 pmid: 28263959 |
| [19] |
Pertea M, Kim D, Pertea G M, Leek J T, Salzberg S L. 2016. Transcript-level expression analysis of RNA-seq experiments with HISAT,StringTie and Ballgown. Nat Protoc, 11 (9):1650-1667.
doi: 10.1038/nprot.2016.095 |
| [20] |
Reddy A S N, Marquez Y, Kalyna M, Barta A. 2013. Complexity of the alternative splicing landscape in plants. The Plant Cell, 25 (10):3657-3683.
doi: 10.1105/tpc.113.117523 pmid: 24179125 |
| [21] |
Robinson J T, Thorvaldsdottir H, Wenger A M, Zehir A, Mesirov J P. 2017. Variant review with the integrative genomics viewer. Cancer Res, 77 (21):e31-e34.
doi: 10.1158/0008-5472.CAN-17-0337 URL |
| [22] |
Shangguan L, Fang X, Jia H, Chen M, Zhang K, Fang J. 2020. Characterization of DNA methylation variations during fruit development and ripening of Vitis vinifera(cv. ‘Fujiminori’). Physiol Mol Biol Plants, 26 (4):617-637.
doi: 10.1007/s12298-020-00759-5 |
| [23] |
Shao Fengqing, Luo Xiurong, Wang Qi, Zhang Xianzhi, Wang Wencai. 2023. Advances in research of DNA methylation regulation during fruit ripening. Acta Horticulturae Sinica, 50 (1):197-208. (in Chinese)
doi: 10.16420/j.issn.0513-353x.2021-1130 URL |
|
邵凤清, 罗秀荣, 王奇, 张宪智, 王文彩. 2023. 果实成熟过程中的DNA 甲基化调控研究进展. 园艺学报, 50 (1):197-208.
doi: 10.16420/j.issn.0513-353x.2021-1130 |
|
| [24] |
Takeshi H, Keiko K, E G M, I W C, Mika N, Yasuomi T, Sachihiro M. 2019. LSD1-LIKE1-Mediated H3K4me 2 demethylation is required for homologous recombination repair. Plant Physiology, 181 (2):499-509.
doi: 10.1104/pp.19.00530 URL |
| [25] |
Trincado J L, Entizne J C, Hysenaj G, Singh B, Skalic M, Elliott D J, Eyras E. 2018. SUPPA2:fast,accurate,and uncertainty-aware differential splicing analysis across multiple conditions. Genome Biol, 19 (1):40.
doi: 10.1186/s13059-018-1417-1 pmid: 29571299 |
| [26] |
Wang G, Pichersky E. 2007. Nicotinamidase participates in the salvage pathway of NAD biosynthesis in Arabidopsis. Plant J, 49 (6):1020-1029.
doi: 10.1111/tpj.2007.49.issue-6 URL |
| [27] |
Yan X, Bai D, Song H, Lin K, Pang E. 2021. Alternative splicing during fruit development among fleshy fruits. BMC Genomics, 22 (1):762.
doi: 10.1186/s12864-021-08111-1 pmid: 34702184 |
| [28] |
Zhang Y, Si F, Wang Y, Liu C, Zhang T, Yuan Y, Gai S. 2020. Application of 5-azacytidine induces DNA hypomethylation and accelerates dormancy release in buds of tree peony. Plant Physiol Biochem, 147:91-100.
doi: 10.1016/j.plaphy.2019.12.010 URL |
| [29] |
Zhao D, Luan Y, Shi W, Zhang X, Meng J, Tao J. 2021. A Paeonia ostii caffeoyl-CoA O-methyltransferase confers drought stress tolerance by promoting lignin synthesis and ROS scavenging. Plant Science, 303:110765.
doi: 10.1016/j.plantsci.2020.110765 URL |
| [30] |
Zhao Q, Du Y, Wang H, Rogers H J, Yu C, Liu W, Zhao M, Xie F. 2019. 5-Azacytidine promotes shoot regeneration during Agrobacterium-mediated soybean transformation. Plant Physiology and Biochemistry, 141:40-50.
doi: 10.1016/j.plaphy.2019.05.014 URL |
| [1] | HUANG Rong, DONG Chao, JIANG Jiao, QIN Yi, LIU Yanlin, SONG Yuyang. Investigation of the Effect of Rain-shelter Cultivation on the Microbial Diversity of Berry Surface for‘Cabernet Sauvignon’Grapes [J]. Acta Horticulturae Sinica, 2023, 50(3): 635-646. |
| [2] | SUN Lei, YAN Ailing, ZHANG Guojun, WANG Huiling, WANG Xiaoyue, REN Jiancheng, XU Haiying. A New Table Grape Cultivar‘Ruidu Mozhi’ [J]. Acta Horticulturae Sinica, 2023, 50(3): 685-686. |
| [3] | WANG Xiaochen, NIE Ziye, LIU Xianju, DUAN Wei, FAN Peige, LIANG Zhenchang. Effects of Abscisic Acid on Monoterpene Synthesis in‘Jingxiangyu’Grape Berries [J]. Acta Horticulturae Sinica, 2023, 50(2): 237-249. |
| [4] | SHAO Fengqing, LUO Xiurong, WANG Qi, ZHANG Xianzhi, WANG Wencai. Advances in Research of DNA Methylation Regulation During Fruit Ripening [J]. Acta Horticulturae Sinica, 2023, 50(1): 197-208. |
| [5] | WANG Baoliang, LIU Fengzhi, JI Xiaohao, WANG Xiaodi, SHI Xiangbin, ZHANG Yican, LI Peng, and WANG Haibo. A New Early Ripening Grape Cultivar‘Huapu Zaoyu’for Table [J]. Acta Horticulturae Sinica, 2022, 49(S2): 33-34. |
| [6] | WANG Baoliang, WANG Haibo, JI Xiaohao, WANG Xiaodi, SHI Xiangbin, WANG Zhiqiang, WANG Xiaolong, and LIU Fengzhi. A New Middle Ripening Grape Cultivar‘Huapu Huangyu’for Table [J]. Acta Horticulturae Sinica, 2022, 49(S2): 35-36. |
| [7] | A Late-maturing Seedless Grape Cultivar‘Zilongzhu’. A Late-maturing Seedless Grape Cultivar‘Zilongzhu’ [J]. Acta Horticulturae Sinica, 2022, 49(S2): 37-38. |
| [8] | SHI Xiaoxin, DU Guoqiang, YANG Lili, QIAO Yuelian, HUANG Chengli, WANG Suyue, ZHAO Yuexin, WEI Xiaohui, WANG Li, and QI Xiangli. A Late-ripening Seedless Grape Cultivar‘Hongfeng Wuhe’ [J]. Acta Horticulturae Sinica, 2022, 49(S2): 39-40. |
| [9] | WU Yueyan, CHEN Tianchi, WANG Liru, HAN Shanqi, and FU Tao. A New Table Grape Cultivar‘Yongzaohong’ [J]. Acta Horticulturae Sinica, 2022, 49(S2): 41-42. |
| [10] | WANG Xiaoyue, YAN Ailing, ZHANG Guojun, WANG Huiling, REN Jiancheng, LIU Zhenhua, SUN Lei, and XU Haiying, . A New Grape Cultivar‘Ruidu Wanhong’ [J]. Acta Horticulturae Sinica, 2022, 49(S1): 29-30. |
| [11] | ZHANG Wanqing, ZHANG Hongxiao, LIAN Xiaofang, LI Yuying, GUO Lili, HOU Xiaogai. Analysis of DNA Methylation Related to Callus Differentiation and Rooting Induction of Paeonia ostii‘Fengdan’ [J]. Acta Horticulturae Sinica, 2022, 49(8): 1735-1746. |
| [12] | WANG Yongjian, KONG Junhua, FAN Peige, LIANG Zhenchang, JIN Xiuliang, LIU Buchun, DAI Zhanwu. Grape Phenome High-throughput Acquisition and Analysis Methods:A Review [J]. Acta Horticulturae Sinica, 2022, 49(8): 1815-1832. |
| [13] | WEI Xiaoyu, WANG Yuejin. Correlation Between Anatomical Structure and Resistance to Powdery Mildew in Chinese Wild Vitis Species [J]. Acta Horticulturae Sinica, 2022, 49(6): 1200-1212. |
| [14] | LIU Zhongjie, ZHENG Ting, ZHAO Fanggui, FU Weihong, ZHUGE Yaxian, ZHANG Zhichang, FANG Jinggui. Resistance Difference and Physiological Response Mechanism of Grape Rootstocks to Osmotic Stress [J]. Acta Horticulturae Sinica, 2022, 49(5): 984-994. |
| [15] | LIANG Chen, SUN Ruyi, XIANG Rui, SUN Yimeng, SHI Xiaoxin, DU Guoqiang, WANG Li. Genome-wide Identification of Grape GRF Family and Expression Analysis [J]. Acta Horticulturae Sinica, 2022, 49(5): 995-1007. |
| Viewed | ||||||
|
Full text |
|
|||||
|
Abstract |
|
|||||
Copyright © 2012 Acta Horticulturae Sinica 京ICP备10030308号-2 国际联网备案号 11010802023439
Tel: 010-82109523 E-Mail: yuanyixuebao@126.com
Support by: Beijing Magtech Co.Ltd