
Acta Horticulturae Sinica ›› 2022, Vol. 49 ›› Issue (3): 519-532.doi: 10.16420/j.issn.0513-353x.2020-1046
• Research Papers • Previous Articles Next Articles
ZHOU Chenping1, YANG Min1, GUO Jinju2, KUANG Ruibin1, YANG Hu1, HUANG Bingxiong1, WEI Yuerong1,*( )
)
Received:2021-07-19
Revised:2021-08-20
Online:2022-03-25
Published:2022-03-25
Contact:
WEI Yuerong
E-mail:weid18@163.com
CLC Number:
ZHOU Chenping, YANG Min, GUO Jinju, KUANG Ruibin, YANG Hu, HUANG Bingxiong, WEI Yuerong. Dynamic Changes in DNA Methylome and Transcriptome Patterns During Papaya Fruit Ripening[J]. Acta Horticulturae Sinica, 2022, 49(3): 519-532.
Add to citation manager EndNote|Ris|BibTeX
URL: https://www.ahs.ac.cn/EN/10.16420/j.issn.0513-353x.2020-1046
| 用途 Application | 基因名称 Gene name | 上游引物(5′-3′) Forward primer | 下游引物(5′-3′) Reverse primer |
|---|---|---|---|
| McrBC-PCR | CpXTH30 | GGTTGGCTAATGATTGT | AGAAGGAAGGAGGGTGC |
| CpPDS | CTAGTTTCTAACTGCGTAT | AACCATTATCATGTGGG | |
| CpACO4 | GAAGACCATTAGCCACA | TTCGCTTAACTTAAACAC | |
| CpXTH31 | ATGAAGTAGGGTGAGAA | ATGGTGGAATAAGTGTT | |
| qRT-PCR | CpXTH30 | GTTCCATCGGTATAGCAT | CTTTGTATCTTCCACCTGA |
| CpPDS | AGTTGGCGTCCCTGTTA | GGATTGATTTGGGTTGTA | |
| CpACO4 | CACGAGCAGCCAAACGA | TCACCCACCCATCCCAC | |
| CpXTH31 | GGGAACCCTACACTTTG | CTTGGATACCTCCGAAT |
Table 1 Primers used for McrBC-PCR and qRT-PCR
| 用途 Application | 基因名称 Gene name | 上游引物(5′-3′) Forward primer | 下游引物(5′-3′) Reverse primer |
|---|---|---|---|
| McrBC-PCR | CpXTH30 | GGTTGGCTAATGATTGT | AGAAGGAAGGAGGGTGC |
| CpPDS | CTAGTTTCTAACTGCGTAT | AACCATTATCATGTGGG | |
| CpACO4 | GAAGACCATTAGCCACA | TTCGCTTAACTTAAACAC | |
| CpXTH31 | ATGAAGTAGGGTGAGAA | ATGGTGGAATAAGTGTT | |
| qRT-PCR | CpXTH30 | GTTCCATCGGTATAGCAT | CTTTGTATCTTCCACCTGA |
| CpPDS | AGTTGGCGTCCCTGTTA | GGATTGATTTGGGTTGTA | |
| CpACO4 | CACGAGCAGCCAAACGA | TCACCCACCCATCCCAC | |
| CpXTH31 | GGGAACCCTACACTTTG | CTTGGATACCTCCGAAT |
| 成熟期 Stage | 序列总数量 Total reads | 对比参考基 因组获得的 序列数量 Mapped reads | 匹配率/% Mapping rate | 重复序列 占比/% Duplication rate | ≥ Q30/% | 测序深度 Sequencing depth | 甲基化C位点 数量Number of Methylated C site | 平均甲基化水平/% Mean methylation level | |||
|---|---|---|---|---|---|---|---|---|---|---|---|
| C | CG | CHG | CHH | ||||||||
| 绿熟期 Green | 49 375 154 | 35 135 889 | 70.97 | 14.37 | 92.78 | 21.87 | 251 784 698 | 17.79 | 51.22 | 35.59 | 8.62 |
| 破色期 Color break | 49 361 724 | 34 881 444 | 70.67 | 12.75 | 91.56 | 22.08 | 258 718 931 | 17.51 | 50.64 | 35.09 | 8.44 |
| 熟化期 Half yellow | 49 461 069 | 34 774 513 | 70.36 | 12.62 | 92.57 | 21.95 | 232 054 629 | 17.04 | 48.60 | 33.76 | 8.26 |
| 腐熟期 Full yellow | 48 863 321 | 34 071 375 | 70.61 | 10.79 | 92.19 | 22.54 | 233 578 504 | 16.26 | 46.88 | 32.77 | 7.81 |
Table 2 Summary of WGBS data of papaya fruits at different ripening stages
| 成熟期 Stage | 序列总数量 Total reads | 对比参考基 因组获得的 序列数量 Mapped reads | 匹配率/% Mapping rate | 重复序列 占比/% Duplication rate | ≥ Q30/% | 测序深度 Sequencing depth | 甲基化C位点 数量Number of Methylated C site | 平均甲基化水平/% Mean methylation level | |||
|---|---|---|---|---|---|---|---|---|---|---|---|
| C | CG | CHG | CHH | ||||||||
| 绿熟期 Green | 49 375 154 | 35 135 889 | 70.97 | 14.37 | 92.78 | 21.87 | 251 784 698 | 17.79 | 51.22 | 35.59 | 8.62 |
| 破色期 Color break | 49 361 724 | 34 881 444 | 70.67 | 12.75 | 91.56 | 22.08 | 258 718 931 | 17.51 | 50.64 | 35.09 | 8.44 |
| 熟化期 Half yellow | 49 461 069 | 34 774 513 | 70.36 | 12.62 | 92.57 | 21.95 | 232 054 629 | 17.04 | 48.60 | 33.76 | 8.26 |
| 腐熟期 Full yellow | 48 863 321 | 34 071 375 | 70.61 | 10.79 | 92.19 | 22.54 | 233 578 504 | 16.26 | 46.88 | 32.77 | 7.81 |
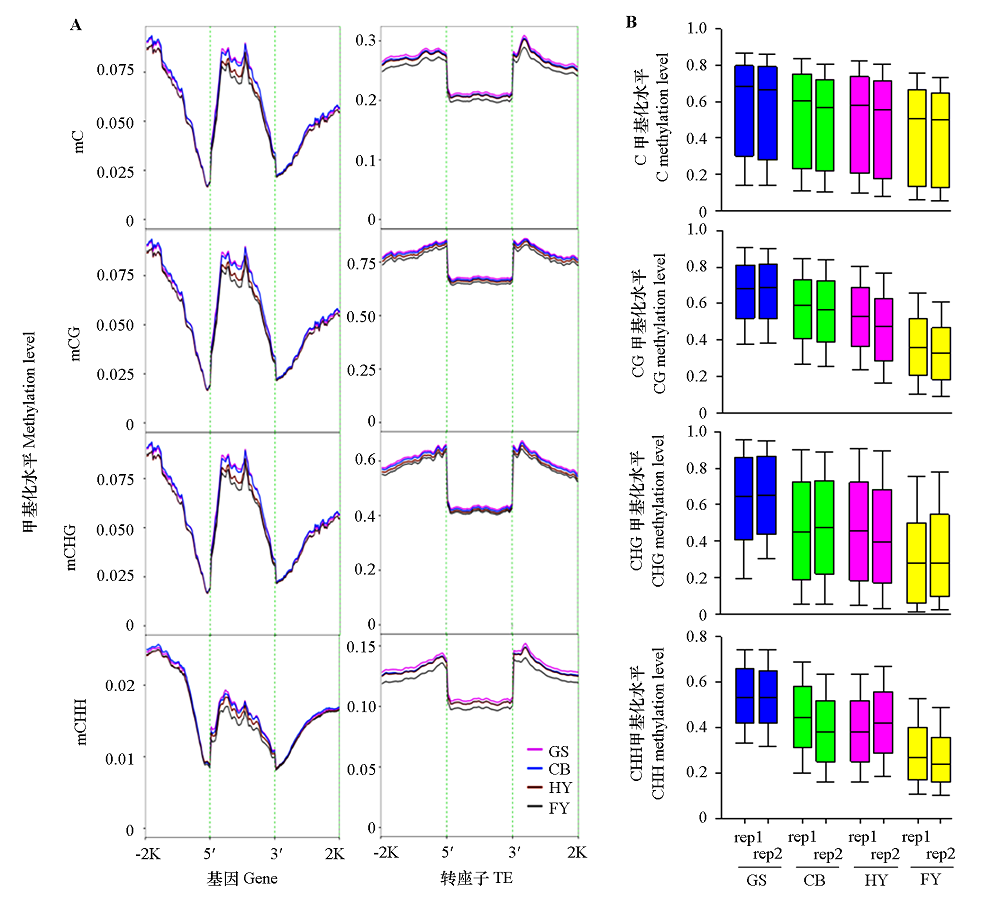
Fig. 2 DNA methylation levels of genes and TEs(A)and C,CG,CHG,and CHH contexts(B)at different ripening stages of papaya fruits GS:Green stage;CB:Color break stage;HY:Half yellow stage;FY:Full yellow stage;-2K and 2K:2 kb upstream and downstream regions of genes;5′ and 3′:5′ and 3′ flanking regions of genes;rep1 and rep2:Two biological replicates. The same below.
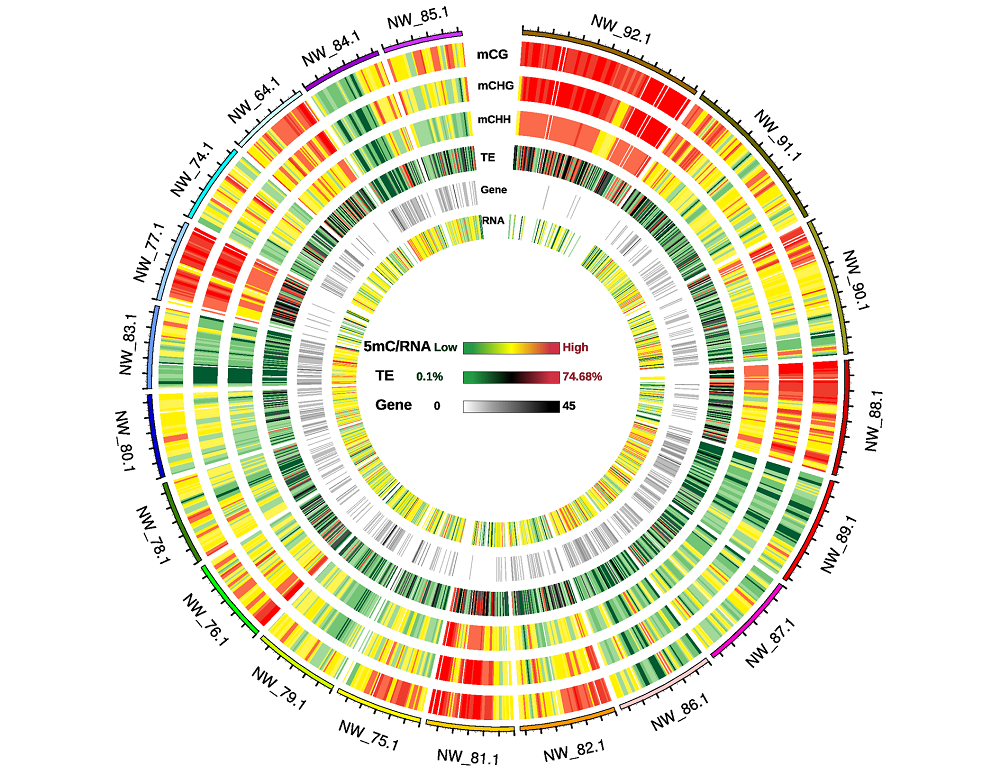
Fig. 4 Circos plot of twenty genome scaffold methylation density Number on outer rim:Number of the twenty genome scaffolds;Scales of 5mC/RNA,TE and Gene:The methylation levels of CG,CHG,CHH sequence context,RNA abundance,the ratio of transposon repeats and gene number were from low to high,respectively.
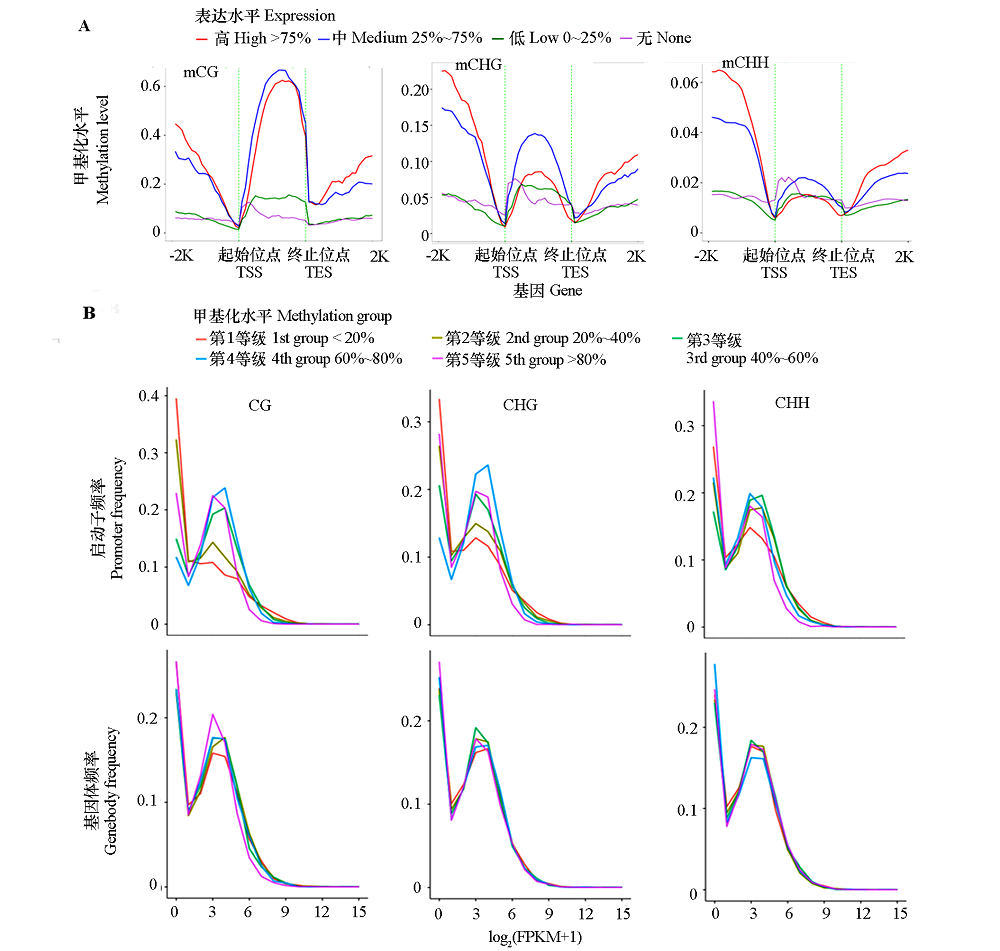
Fig. 5 Relationship between distribution of methylation levels in three contexts within the genebody,upstream,and downstream 2 kb rigions(A)and methylation levels of genebody and promoterwith the gene expression levels(B) -2K and 2K:2 kb upstream and downstream regions of genes;Expression:genes with FPKM value < 1 were considered as none(non-expressed),expressed genes(FPKM value ≥ 1)were ranked from small to large and divided into three groups based on the upper and lower quartile;Methylation group:Methylation levels of genebodies or promoters were ranked from small to large and divided into quinitiles.
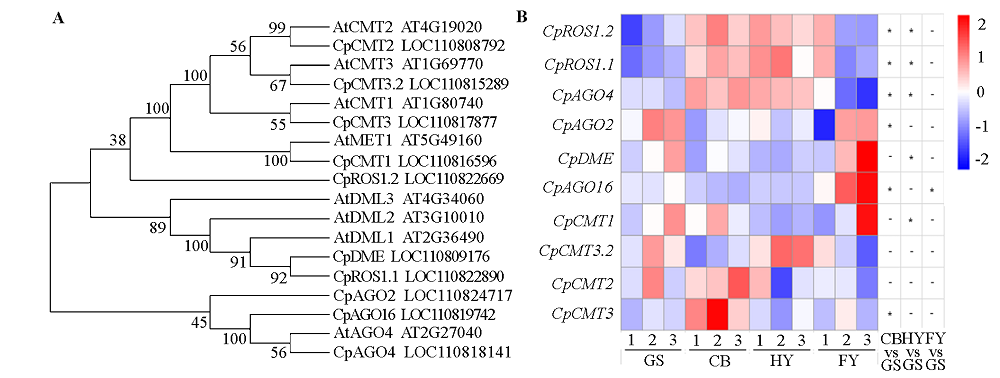
Fig. 7 Homologous alignment of DNA methylation genes(A)and their expression changes at different ripening stages in papaya fruits(B) Scale:From blue to red represent the gene expression levels was from low to high. 1-3:Three biological replicates.*:P < 0.05;-:No significant difference.
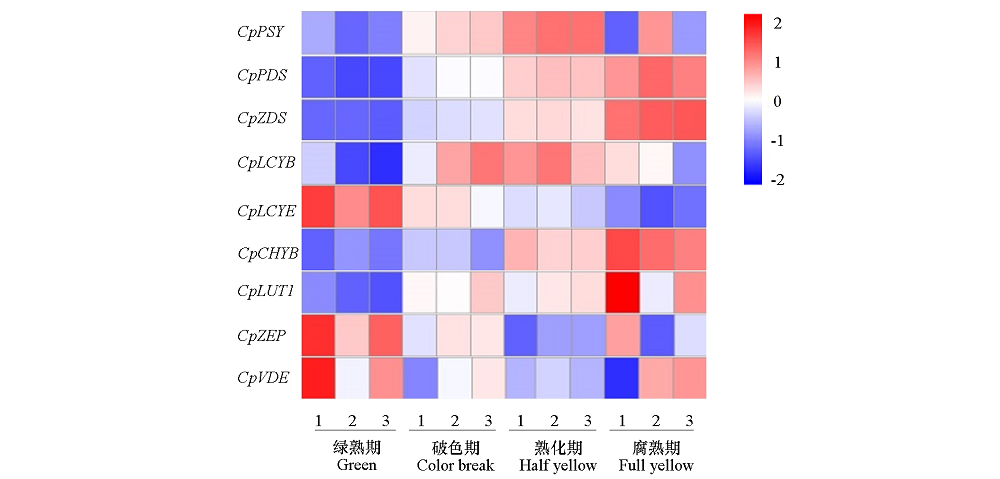
Fig. 8 Expression changes of carotenoid biosynthetic-related genes at different ripening stages of papaya fruits Scale:From blue to red represent the gene expression levels was from low to high;1-3:Three biological replicates.
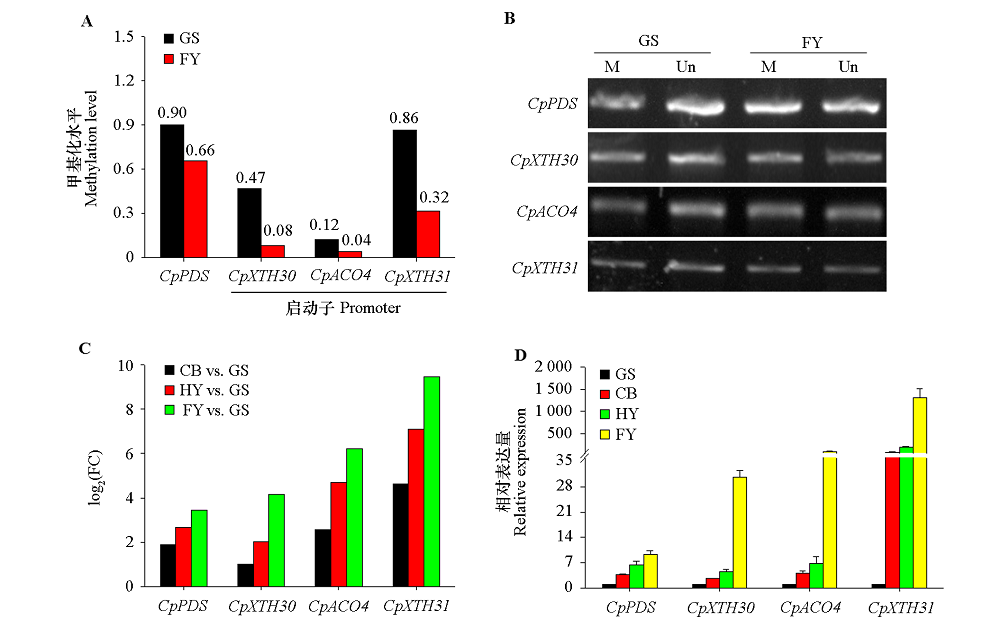
Fig. 9 The DMRs methylation and expression levels of four candidate genes at different maturation stages of papaya fruits A:DMRs methylation levels of CpACO4,CpXTH30 and CpXTH31 promoters and CpPDS genebody;B:The McrBC-PCR validation(M:PCR products been digested by McrBC,Un:PCR products not been digested);C:Fold change(FC);D:qRT-PCR validation.
| [1] |
Ahmad A, Zhang Y, Cao X F. 2010. Decoding the epigenetic language of plant development. Molecular Plant, 3(4):719-728.
doi: 10.1093/mp/ssq026 pmid: 20663898 |
| [2] |
Bolger A M, Lohse M, Usadel B. 2014. Trimmomatic:a flexible trimmer for Illumina sequence data. Bioinformatics, 30(15):2114-2120.
doi: 10.1093/bioinformatics/btu170 URL |
| [3] |
Bonasio R, Tu S, Reinberg D. 2010. Molecular signals of epigenetic states. Science, 330(6004):612-616.
doi: 10.1126/science.1191078 URL |
| [4] | Chen Duanfen, Peng Zhenhua, Gao Zhimin. 2008. Cloning and expression analysis of PDS gene in Narcissus tazetta var. chinensis. Molecular Plant Breeding, 6(3):574-578. (in Chinese) |
| 陈段芬, 彭镇华, 高志民. 2008. 中国水仙八氢番茄红素脱氢酶基因(PDS)的克隆及表达分析. 分子植物育种, 6(3):574-578. | |
| [5] | Chen Yong-ping, Gao Feng, Shen Yan-hong, Zhao Wan-wan, Chen Gui-xin, Wang Ping. 2019. The analysis of ERFs related to fruit ripening in papaya. Acta Horticulturae Sinica, 46(2):252-264. (in Chinese) |
| 陈永萍, 高峰, 申艳红, 赵弯弯, 陈桂信, 王平. 2019. 番木瓜ERF家族与果实成熟相关成员的分析. 园艺学报, 46(2):252-264. | |
| [6] |
Cheng J, Niu Q, Zhang B, Chen K, Yang R, Zhu J K, Zhang Y, Lang Z. 2018. Downregulation of RdDM during strawberry fruit ripening. Genome Biology, 19(1):212.
doi: 10.1186/s13059-018-1587-x URL |
| [7] |
Ding X, Zhu X, Ye L, Xiao S, Wu Z, Chen W, Li X. 2019. The interaction of CpEBF1 with CpMADSs is involved in cell wall degradation during papaya fruit ripening. Horticulture Research, 6(1):13.
doi: 10.1038/s41438-018-0095-1 URL |
| [8] | Du Xiaoyun, Song Laiqing, Zhao Lingling, Liu Meiying, Tang Yan, Sun Yanxia, Jiang Zhongwu, Shu Huairui. 2019. DNA methylation in Red Fuji apple bud sports lines. Acta Horticulturae Sinica, 46(1):107-120. (in Chinese) |
| 杜晓云, 宋来庆, 赵玲玲, 刘美英, 唐岩, 孙燕霞, 姜中武, 束怀瑞. 2019. 红富士苹果芽变系DNA甲基化研究. 园艺学报, 46(1):107-120. | |
| [9] |
Fabi J P, Do Prado S B R. 2019. Fast and furious:ethylene-triggered changes in the metabolism of papaya fruit during ripening. Frontiers in Plant Science, 10:535.
doi: 10.3389/fpls.2019.00535 URL |
| [10] | Fan Zhong-qi, Kuang Jian-fei, Lu Wang-jin, Chen Jian-ye. 2015. Advances in research of the mechanism of transcription factors involving in regulating fruit ripening and senescence. Acta Horticulturae Sinica, 42(9):1649-1663. (in Chinese) |
| 范中奇, 邝健飞, 陆旺金, 陈建业. 2015. 转录因子调控果实成熟和衰老机制研究进展. 园艺学报, 42(9):1649-1663. | |
| [11] |
Finnegan E J, Kovac K A. 2000. Plant DNA methyltransferases. Plant Molecular Biology, 43:189-201.
pmid: 10999404 |
| [12] | Grafi G. 2011. Epigenetics in plant development and response to stress. Biochimica et Biophysica Acta, 1809:351-352. |
| [13] |
He X J, Chen T, Zhu J K. 2011. Regulation and function of DNA methylation in plants and animals. Cell Research, 21:442-465.
doi: 10.1038/cr.2011.23 URL |
| [14] | Huang H, Liu R, Niu Q, Tang K, Zhang B, Zhang H, Chen K, Zhu J K, Lang Z. 2019. Global increase in DNA methylation during orange fruit development and ripening. Proceedings of the National Academy of Sciences of the United States of America, 116(4):1430-1436. |
| [15] | Huang Wen-jun, Zhong Cai-hong. 2017. Research advances in the postharvest physiology of kiwifruit. Plant Science Journal, 35(4):622-630. (in Chinese) |
| 黄文俊, 钟彩虹. 2017. 猕猴桃果实采后生理研究进展. 植物科学学报, 35(4):622-630. | |
| [16] |
Krueger F, Andrews S R. 2011. Bismark:a flexible aligner and methylation caller for Bisulfite-Seq applications. Bioinformatics, 27(11):1571-1572.
doi: 10.1093/bioinformatics/btr167 pmid: 21493656 |
| [17] |
Krzywinski M, Schein J, Birol I, Connors J, Gascoyne R, Horsman D, Jones S J, Marra M A. 2009. Circos:an information aesthetic for comparative genomics. Genome Research, 19:1639-1645.
doi: 10.1101/gr.092759.109 pmid: 19541911 |
| [18] | Lang Z, Wang Y, Tang K, Tang D, Datsenka T, Cheng J, Zhang Y, Handa A K, Zhu J K. 2017. Critical roles of DNA demethylation in the activation of ripeninginduced genes and inhibition of ripening-repressed genes in tomato fruit. Proceedings of the National Academy of Sciences of the United States of America, 114:E4511-E4519. |
| [19] |
Law J A, Jacobsen S E. 2010. Establishing,maintaining and modifying DNA methylation patterns in plants and animals. Nature Reviews Genetics, 11:204-220.
doi: 10.1038/nrg2719 URL |
| [20] |
Li X, Zhu J, Hu F, Ge S, Ye M, Xiang H, Zhang G, Zheng X, Zhang H, Zhang S, Li Q, Luo R, Yu C, Yu J, Sun J, Zou X, Cao X, Xie X, Wang J, Wang W. 2012. Single-base resolution maps of cultivated and wild rice methylomes and regulatory roles of DNA methylation in plant gene expression. BMC Genomics, 13:300.
doi: 10.1186/1471-2164-13-300 URL |
| [21] |
Lister R, O’Malley R C, Tonti-Filippini J, Gregory B D, Berry C C, Millar A H, Ecker J R. 2008. Highly integrated single-base resolution maps of the epigenome in Arabidopsis. Cell, 133:523-536.
doi: 10.1016/j.cell.2008.03.029 pmid: 18423832 |
| [22] | Liu R, How-Kit A, Stammitti L, Teyssier E, Rolin D, Mortain-Bertrand A, Halle S, Liu M, Kong J, Wu C, Degraeve-Guibault C, Chapman N H, Maucourt M, Hodgman T C, Tost J, Bouzayen M, Hong Y, Seymour G B, Giovannoni J J, Gallusci P. 2015. A DEMETER-like DNA demethylase governs tomato fruit ripening. Proceedings of the National Academy of Sciences of the United States of America, 112:10804-10809. |
| [23] | Liu Xin, Chen Yunzhu, Kim Pyol, Kim Min-Jun, Song Hyondok, Li Yuhua, Wang Yu. 2020. Progress on molecular mechanism and regulation of tomato fruit color formation. Acta Horticulturae Sinica, 47(9):1689-1704. (in Chinese) |
| 刘昕, 陈韵竹, Kim Pyol, Kim Min-Jun, Song Hyondok, 李玉花, 王宇. 2020. 番茄果实颜色形成的分子机制及调控研究进展. 园艺学报, 47(9):1689-1704. | |
| [24] |
Lu X, Wang X, Chen X, Shu N, Wang J, Wang D, Wang S, Fan W, Guo L, Guo X, Ye W. 2017. Single-base resolution methylomes of upland cotton(Cossypium hirsutum L.)reveal epigenome modification in response to drought stress. BMC Genomics, 18(1):297.
doi: 10.1186/s12864-017-3681-y URL |
| [25] |
Manning K, Tör M, Poole M, Hong Y, Thompson A J, King G J, Giovannoni J J, Seymour G B. 2006. A naturally occurring epigenetic mutation in a gene encoding an SBP-box transcription factor inhibits tomato fruit ripening. Nature Genetics, 38(8):948.
doi: 10.1038/ng1841 URL |
| [26] |
Marty I, Bureau S, Sarkissian G, Gouble B, Audergon J M, Albagnac G. 2005. Ethylene regulation of carotenoid accumulation and carotenogenic gene expression in colour-contrasted apricot varieties(Prunus armeniaca). Journal of Experimental Botany, 56(417):1877-1886.
pmid: 15911563 |
| [27] |
Muñoz-Bertomeu J, Miedes E, Lorences E P. 2013. Expression of xyloglucan endotransglucosylase/hydrolase(XTH)genes and XET activity in ethylene treated apple and tomato. Journal of Plant Physiology, 170(13):1194-1201.
doi: 10.1016/j.jplph.2013.03.015 pmid: 23628624 |
| [28] |
Narsaiah K, Wilson R A, Gokul K, Mandge H M, Jha S N, Bhadwal S, Anurag R K, Malik RK, Vij S. 2015. Effect of bacteriocin-incorporated alginate coating on shelf-life of minimally processed papaya(Carica papaya L.). Postharvest Biology and Technology, 100:212-218.
doi: 10.1016/j.postharvbio.2014.10.003 URL |
| [29] |
Niederhuth C E, Bewick A J, Ji L, Alabady M S, Kim K D, Li Q, Rohr N A, Rambani A, Burke J M, Udall J A, Egesi C, Schmutz J, Grimwood J, Jackson S A, Springer N M, Schmitz R J. 2016. Widespread natural variation of DNA methylation within angiosperms. Genome Biology, 17:194.
pmid: 27671052 |
| [30] | Pesaresi P, Mizzotti C, Colombo M, Masiero S. 2014. Genetic regulation and structural changes during tomato fruit development and ripening. Frontiers in Plant Science, 23(5):124. |
| [31] |
Pfaffl M W. 2001. A new mathematical model for relative quantification in real-time RT-PCR. Nucleic Acids Research, 29(9):e45.
doi: 10.1093/nar/29.9.e45 pmid: 11328886 |
| [32] |
Qi W, Wang H, Zhou Z, Yang P, Wu W, Li Z, Li X. 2020. Ethylene emission as a potential indicator of Fuji apple flavor quality evaluation under low temperature. Horticultural Plant Journal, 6(4):231-239.
doi: 10.1016/j.hpj.2020.03.007 URL |
| [33] | Seymour DK, Koenig D, Hagmann J, Becker C, Weigel D. 2014. Evolution of DNA methylation patterns in the Brassicaceae is driven by differences in genome organization. PLoS Genetics, 10:e1004785. |
| [34] |
Shen Y H, Lu B G, Feng L, Yang F Y, Geng J J, Ming R, Chen X J. 2017. Isolation of ripening-related genes from ethylene/1-MCP treated papaya through RNA-seq. BMC Genomics, 18(1):671.
doi: 10.1186/s12864-017-4072-0 URL |
| [35] |
Tang K, Lang Z, Zhang H, Zhu J K. 2016. The DNA demethylase ROS1 targets genomic regions with distinct chromatin modifications. Nature Plants, 2:16169.
doi: 10.1038/nplants.2016.169 pmid: 27797352 |
| [36] |
Tian Shi-ping. 2013. Molecular mechanisms of fruit ripening and senescence. Chinese Bulletin of Botany, 48(5):481-488. (in Chinese)
doi: 10.3724/SP.J.1259.2013.00481 URL |
| 田世平. 2013. 果实成熟和衰老的分子调控机制. 植物学报, 48(5):481-488. | |
| [37] | Wang Zhong-feng. 2009. Research advancement in relation of enzymes for cell wall metabolism with fruit softening. Chinese Agricultural Science Bulletin, 25(18):126-130. (in Chinese) |
| 王中凤. 2009. 细胞壁分解酶与果实软化的关系研究进展. 中国农学通报, 25(18):126-130. | |
| [38] |
Xu J, Wang X, Cao H, Xu H, Xu Q, Deng X. 2017. Dynamic changes in methylome and transcriptome patterns in response to methyltransferase inhibitor 5-azacytidine treatment in citrus. DNA Research, 24(5):509-522.
doi: 10.1093/dnares/dsx021 URL |
| [39] |
Xu J, Zhou S, Gong X, Song Y, van Nocker S, Ma F, Guan Q. 2018. Single-base methylome analysis reveals dynamic epigenomic differences associated with water deficit in apple. Plant Biotechnology Journal, 16:672-687.
doi: 10.1111/pbi.2018.16.issue-2 URL |
| [40] | Xu Sheng, Zhang Xin, Liu Decai, Gu Tingting. 2020. Identification and functional analysis of N6-adenine methylation(6mA)methyltransferase genes in Fragaria vesca. Acta Horticulturae Sinica, 47(11):2194-2206. (in Chinese) |
| 徐昇, 张鑫, 刘德才, 顾婷婷. 2020. 森林草莓6mA甲基转移酶基因鉴定及功能分析. 园艺学报, 47(11):2194-2206. | |
| [41] |
Zhang M, Zhang X, Guo L, Qi T, Liu G, Feng J, Shahzad K, Zhang B, Li X, Wang H, Tang H, Qiao X, Wu J, Xing C. 2020. Single-base resolution methylome of cotton cytoplasmic male sterility system reveals epigenomic changes in response to high-temperature stress during anther development. Journal of Experimental Botany, 71:951-969.
doi: 10.1093/jxb/erz470 pmid: 31639825 |
| [42] |
Zhang X. 2008. The epigenetic landscape of plants. Science, 320(5875):489-492.
doi: 10.1126/science.1153996 URL |
| [43] |
Zhong S, Fei Z, Chen Y R, Zheng Y, Huang M, Vrebalov J, McQuinn R, Gapper N, Liu B, Xiang J, Shao Y, Giovannoni J J. 2013. Single-base resolution methylomes of tomato fruit development reveal epigenome modifications associated with ripening. Nature Biotechnology, 31(2):154-159.
doi: 10.1038/nbt.2462 URL |
| [44] | Zhou C, Yang M, Luo X, Kuang R, Yang H, Yao J, Huang B, Wei Y. 2021. Transcriptomic analysis of papaya(Carica papaya L.)shoot explants obtained by leaf- and stem-inoculation methods for adventitious roots induction. Scientia Horticulturae, 276:109762. |
| [45] |
Zhu J K. 2009. Active DNA demethylation mediated by DNA glycosylases. Annual Review of Genetics, 43:143-66.
doi: 10.1146/genet.2009.43.issue-1 URL |
| [1] | WANG Xiaochen, NIE Ziye, LIU Xianju, DUAN Wei, FAN Peige, and LIANG Zhenchang, . Effects of Abscisic Acid on Monoterpene Synthesis in‘Jingxiangyu’Grape Berries [J]. Acta Horticulturae Sinica, 2023, 50(2): 237-249. |
| [2] | WANG Rui, HONG Wenjuan, LUO Hua, ZHAO Lina, CHEN Ying, and WANG Jun, . Construction of SSR Fingerprints of Pomegranate Cultivars and Male Parent Identification of Hybrids [J]. Acta Horticulturae Sinica, 2023, 50(2): 265-278. |
| [3] | ZHANG Xin, QI Yanxiang, ZENG Fanyun, WANG Yanwei, XIE Peilan, XIE Yixian, and PENG Jun. Functional Analysis of Dicer-like Genes in Fusarium oxysporum f. sp. cubense Race 4 [J]. Acta Horticulturae Sinica, 2023, 50(2): 279-294. |
| [4] | YU Tingting, LI Huan, NING Yuansheng, SONG Jianfei, PENG Lulin, JIA Junqi, ZHANG Weiwei, and YANG Hongqiang. Genome-wide Identification of GRAS Gene Family in Apple and Expression Analysis of Its Response to Auxin [J]. Acta Horticulturae Sinica, 2023, 50(2): 397-409. |
| [5] | WANG Mengmeng, SUN Deling, CHEN Rui, YANG Yingxia, ZHANG Guan, LÜ Mingjie, WANG Qian, XIE Tianyu, NIU Guobao, SHAN Xiaozheng, TAN Jin, and YAO Xingwei, . Construction and Evaluation of Cauliflower Core Collection [J]. Acta Horticulturae Sinica, 2023, 50(2): 421-431. |
| [6] | ZHAI Hanhan, ZHAI Yujie, TIAN Yi, ZHANG Ye, YANG Li, WEN Zhiliang, CHEN Haijiang. Genome-wide Identification of Peach SAUR Gene Family and Characterization of PpSAUR5 Gene [J]. Acta Horticulturae Sinica, 2023, 50(1): 1-14. |
| [7] | YANG Zhi, ZHANG Chuanjiang, YANG Xinfang, DONG Mengyi, WANG Zhenlei, YAN Fenfen, WU Cuiyun, WANG Jiurui, LIU Mengjun, LIN Minjuan. Analysis of Fruit Genetic Tendency and Mixed Inheritance in Hybrid Progeny of Jujube and Wild Jujube [J]. Acta Horticulturae Sinica, 2023, 50(1): 36-52. |
| [8] | YUAN Xin, XU Yunhe, ZHANG Yupei, SHAN Nan, CHEN Chuying, WAN Chunpeng, KAI Wenbin, ZHAI Xiawan, CHEN Jinyin, GAN Zengyu. Studies on AcAREB1 Regulating the Expression of AcGH3.1 During Postharvest Ripening of Kiwifruit [J]. Acta Horticulturae Sinica, 2023, 50(1): 53-64. |
| [9] | LUO Hailin, YUAN Lei, WENG Hua, YAN Jiahui, GUO Qingyun, WANG Wenqing, MA Xinming. Identification and Analysis of Complete Genomic Sequence of Broad Bean Wilt Virus 2 Pepper Isolate in Qinghai Province [J]. Acta Horticulturae Sinica, 2023, 50(1): 161-169. |
| [10] | HU Jingyu, QUE Kaijuan, MIAO Tianli, WU Shaozheng, WANG Tiantian, ZHANG Lei, DONG Xian, JI Pengzhang, DONG Jiahong. Identification of Tomato Spotted Wilt Orthotospovirus Infecting Iris tectorum [J]. Acta Horticulturae Sinica, 2023, 50(1): 170-176. |
| [11] | SHAO Fengqing, LUO Xiurong, WANG Qi, ZHANG Xianzhi, WANG Wencai. Advances in Research of DNA Methylation Regulation During Fruit Ripening [J]. Acta Horticulturae Sinica, 2023, 50(1): 197-208. |
| [12] | HAN Xiaolei, ZHANG Caixia, LIU Kai, YANG An, YAN Jiadi, LI Wuxing, KANG Liqun, and CONG Peihua. A New Mid-ripening Apple Cultivar‘Zhongping Youlei’ [J]. Acta Horticulturae Sinica, 2022, 49(S2): 1-2. |
| [13] | OU Chunqing, JIANG Shuling, WANG Fei, MA Li, ZHANG Yanjie, and LIU Zhenjie. A New Early-ripening Pear Cultivar‘Xingli Mishui’ [J]. Acta Horticulturae Sinica, 2022, 49(S2): 7-8. |
| [14] | YANG Xingwang, LIU Fengzhi, WANG Haibo, WANG Yingying, WANG Zhiqiang, SHI Xiangbin, JI Xiaohao, LIU Wanchun, and WANG Xiaodi. A New Mid-ripening Cold Resistant Peach Cultivar‘Zhongnong Hanshuimi’ [J]. Acta Horticulturae Sinica, 2022, 49(S2): 19-20. |
| [15] | YANG Xingwang, LIU Fengzhi, WANG Haibo, WANG Yingying, ZHANG Yican, LI Peng, WANG Xiaolong, LIU Wanchun, and WANG Xiaodi. A New Late-ripening Cold Resistant Peach Cultivar‘Zhongnong Qiuxiang’ [J]. Acta Horticulturae Sinica, 2022, 49(S2): 21-22. |
| Viewed | ||||||
|
Full text |
|
|||||
|
Abstract |
|
|||||
Copyright © 2012 Acta Horticulturae Sinica 京ICP备10030308号-2 国际联网备案号 11010802023439
Tel: 010-82109523 E-Mail: yuanyixuebao@126.com
Support by: Beijing Magtech Co.Ltd