
Acta Horticulturae Sinica ›› 2025, Vol. 52 ›› Issue (4): 933-946.doi: 10.16420/j.issn.0513-353x.2024-0376
• Genetic & Breeding·Germplasm Resources·Molecular Biology • Previous Articles Next Articles
SHAO Yifan1, ZHU Baoqing1, WANG Tongxin1, LIAO Jianhe2, WU Fanhua3, YANG Siyi1, FENG Jianhang1, YU Xudong1,*( )
)
Received:2025-01-08
Revised:2025-02-20
Online:2025-05-08
Published:2025-04-25
Contact:
YU Xudong
SHAO Yifan, ZHU Baoqing, WANG Tongxin, LIAO Jianhe, WU Fanhua, YANG Siyi, FENG Jianhang, YU Xudong. Research of Genetic Regulation of Bombax ceiba Prickle Based on Hormone,Transcriptome and Metabolome[J]. Acta Horticulturae Sinica, 2025, 52(4): 933-946.
Add to citation manager EndNote|Ris|BibTeX
URL: https://www.ahs.ac.cn/EN/10.16420/j.issn.0513-353x.2024-0376
| 引物 Primer | 正引物序列(5′-3′) Forward primer | 反引物序列(5′-3′) Reverse primer | 基因ID Gene ID |
|---|---|---|---|
| BRI1 | GGAGGTCATAAACAGCCC | TCCTTCAAGTCCCCAATC | evm.model.Scaffold196.5 |
| BZR1/2 | ACCTCTATCTTCCCCTAC | CATCAGACTCATCGCATT | evm.model.Scaffold21.213 |
| AUX1 | ACCCGAGAAGAGGGAAAA | AGAAGGAATAGGGCAGCG | evm.model.Scaffold500.291 |
| AUX/IAA | GCGAACTCACCAAAAACA | AATAGAGCGAGCAGCAAC | evm.model.Scaffold262.58 |
| ARF | TGTGGAATCAGACGAAGT | TCCCGATAGAGTAAATGG | evm.model.Scaffold181.29 |
| DELLA | ATGGAGGTAAAAGGAACG | CATCAGCAAAATAGTGGG | evm.model.Scaffold132.216 |
| JAR1 | GCTCTGGCACTACTCAA | CAGCAAACAAACCACCTT | evm.model.Scaffold221.79 |
| TGA | AGATTGGTATGTGGGGAG | TCGTCAGCATCACTTCCT | evm.model.Scaffold127.460 |
| PR-1 | CCCAATCTATTCCACCTC | ATCGGGCACATTTCTCAA | evm.model.Scaffold2351.33 |
| SIMKK | TCCCAGAAACGGAGAGAC | AGTAGAAGGAGTAGGCGG | evm.model.Scaffold66.339 |
| MYB62 | GGGAGAACGGAAAATGAG | TGGATGAACAAGAGGATG | evm.model.Scaffold144.195 |
| MYB123 | CGTTACAGCCACCACCAA | CATCAAGCACTGCGAATC | evm.model.Scaffold498.25 |
| WRKY28 | TCAGTTTCTTCCTCCTCT | TCCTCCACCTGTATCCAT | evm.model.Scaffold46.3 |
Table 1 Primer sequences of candidate genes
| 引物 Primer | 正引物序列(5′-3′) Forward primer | 反引物序列(5′-3′) Reverse primer | 基因ID Gene ID |
|---|---|---|---|
| BRI1 | GGAGGTCATAAACAGCCC | TCCTTCAAGTCCCCAATC | evm.model.Scaffold196.5 |
| BZR1/2 | ACCTCTATCTTCCCCTAC | CATCAGACTCATCGCATT | evm.model.Scaffold21.213 |
| AUX1 | ACCCGAGAAGAGGGAAAA | AGAAGGAATAGGGCAGCG | evm.model.Scaffold500.291 |
| AUX/IAA | GCGAACTCACCAAAAACA | AATAGAGCGAGCAGCAAC | evm.model.Scaffold262.58 |
| ARF | TGTGGAATCAGACGAAGT | TCCCGATAGAGTAAATGG | evm.model.Scaffold181.29 |
| DELLA | ATGGAGGTAAAAGGAACG | CATCAGCAAAATAGTGGG | evm.model.Scaffold132.216 |
| JAR1 | GCTCTGGCACTACTCAA | CAGCAAACAAACCACCTT | evm.model.Scaffold221.79 |
| TGA | AGATTGGTATGTGGGGAG | TCGTCAGCATCACTTCCT | evm.model.Scaffold127.460 |
| PR-1 | CCCAATCTATTCCACCTC | ATCGGGCACATTTCTCAA | evm.model.Scaffold2351.33 |
| SIMKK | TCCCAGAAACGGAGAGAC | AGTAGAAGGAGTAGGCGG | evm.model.Scaffold66.339 |
| MYB62 | GGGAGAACGGAAAATGAG | TGGATGAACAAGAGGATG | evm.model.Scaffold144.195 |
| MYB123 | CGTTACAGCCACCACCAA | CATCAAGCACTGCGAATC | evm.model.Scaffold498.25 |
| WRKY28 | TCAGTTTCTTCCTCCTCT | TCCTCCACCTGTATCCAT | evm.model.Scaffold46.3 |
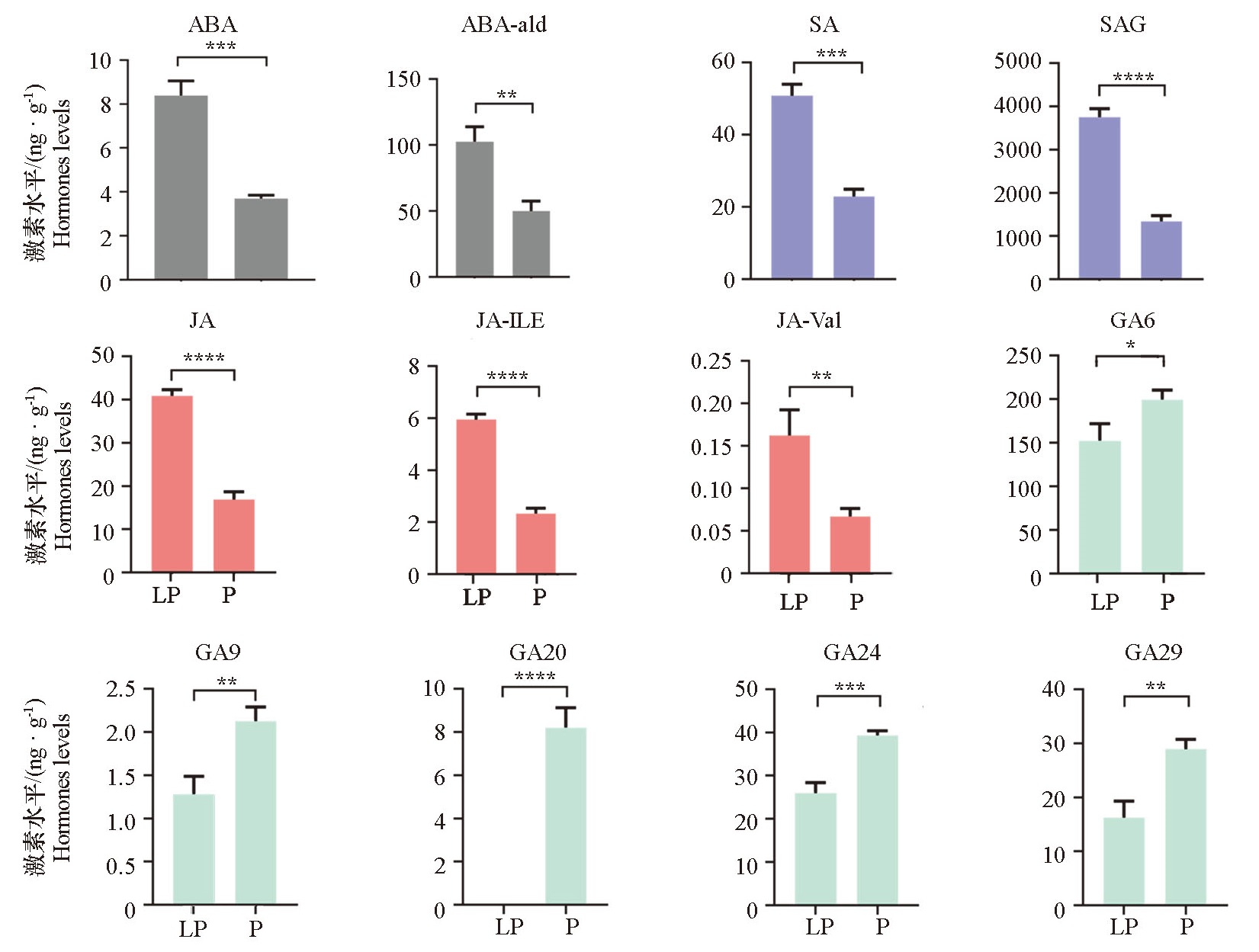
Fig. 1 Analysis diagram of hormone determination in less prickly area and more prickly area of Bombax ceiba LP:Upper less prickly area;P:Basal prickly area. ABA:Abscisic acid;ABA-ald:Abscisic aldehyde;SA:Salicylic acid;SAG:Salicylic acid 2-O-β-glucoside;JA:Jasmonic acid;JA-ILE:Jasmonoyl-L-isoleucine;JA-Val:N-[(-)-Jasmonoyl]-(L)-valine;GA6:Gibberellin A6;GA9:Gibberellin A9;GA20:Gibberellin A20; GA24:Gibberellin A24;GA29:Gibberellin A29. *:P < 0.05,**:P < 0.01,***:P < 0.001,****:P < 0.0001
| 样品 Sample | 总读取数Total Reads | 比对数 Reads mapped | 唯一比对数 Unique mapped | 多重比对数 Multi mapped | Read1比对数 Read1 mapped | Read2比对数 Read2 mapped | 正链比对数 ‘+’mapped | 负链比对数 ‘-’mapped |
|---|---|---|---|---|---|---|---|---|
| LP-1 | 43 929 340 | 29 341 819(66.79%) | 28 260 900(64.33%) | 10 809 19(2.46%) | 14 203 787(32.33%) | 14 057 113(32.00%) | 14 115 917(32.13%) | 14 144 983(32.20%) |
| LP-2 | 47 777 116 | 32 339 797(67.69%) | 31 122 901(65.14%) | 12 168 96(2.55%) | 15 637 980(32.73%) | 15 484 921(32.41%) | 15 545 737(32.54%) | 15 577 164(32.60%) |
| LP-3 | 46 733 462 | 30 526 628(65.32%) | 29 400 369(62.91%) | 11 262 59(2.41%) | 14 773 962(31.61%) | 14 626 407(31.30%) | 14 684 755(31.42%) | 14 715 614(31.49%) |
| P-1 | 45 162 140 | 28 053 596(62.12%) | 26 901 920(59.57%) | 11 516 76(2.55%) | 13 517 914(29.93%) | 13 384 006(29.64%) | 13 440 797(29.76%) | 13 461 123(29.81%) |
| P-2 | 44 355 554 | 28 766 857(64.86%) | 27 560 164(62.13%) | 12 066 93(2.72%) | 13 849 789(31.22%) | 13 710 375(30.91%) | 13 770 449(31.05%) | 13 789 715(31.09%) |
| P-3 | 43 934 068 | 27 565 859(62.74%) | 26 383 331(60.05%) | 11 825 28(2.69%) | 13 135 656(29.90%) | 13 247 675(30.15%) | 13 184 883(30.01%) | 13 198 448(30.04%) |
Table 2 Comparison statistics table of transcriptome data and reference genome
| 样品 Sample | 总读取数Total Reads | 比对数 Reads mapped | 唯一比对数 Unique mapped | 多重比对数 Multi mapped | Read1比对数 Read1 mapped | Read2比对数 Read2 mapped | 正链比对数 ‘+’mapped | 负链比对数 ‘-’mapped |
|---|---|---|---|---|---|---|---|---|
| LP-1 | 43 929 340 | 29 341 819(66.79%) | 28 260 900(64.33%) | 10 809 19(2.46%) | 14 203 787(32.33%) | 14 057 113(32.00%) | 14 115 917(32.13%) | 14 144 983(32.20%) |
| LP-2 | 47 777 116 | 32 339 797(67.69%) | 31 122 901(65.14%) | 12 168 96(2.55%) | 15 637 980(32.73%) | 15 484 921(32.41%) | 15 545 737(32.54%) | 15 577 164(32.60%) |
| LP-3 | 46 733 462 | 30 526 628(65.32%) | 29 400 369(62.91%) | 11 262 59(2.41%) | 14 773 962(31.61%) | 14 626 407(31.30%) | 14 684 755(31.42%) | 14 715 614(31.49%) |
| P-1 | 45 162 140 | 28 053 596(62.12%) | 26 901 920(59.57%) | 11 516 76(2.55%) | 13 517 914(29.93%) | 13 384 006(29.64%) | 13 440 797(29.76%) | 13 461 123(29.81%) |
| P-2 | 44 355 554 | 28 766 857(64.86%) | 27 560 164(62.13%) | 12 066 93(2.72%) | 13 849 789(31.22%) | 13 710 375(30.91%) | 13 770 449(31.05%) | 13 789 715(31.09%) |
| P-3 | 43 934 068 | 27 565 859(62.74%) | 26 383 331(60.05%) | 11 825 28(2.69%) | 13 135 656(29.90%) | 13 247 675(30.15%) | 13 184 883(30.01%) | 13 198 448(30.04%) |

Fig. 5 Hormonal biosynthesis pathway map in the less prickly and more prickly regions of Bombax ceiba LP:Upper less prickly area;P:Basal prickly area。The same below
| [1] |
|
| [2] |
|
| [3] |
|
|
冯发玉, 王毅, 陆斌, 何家妮, 原晓龙. 2022. 基于转录组数据竹叶花椒WRKY基因家族的鉴定及表达分析. 分子植物育种, 20 (18):5984-5994.
|
|
| [4] |
|
|
冯发玉, 王毅, 王丽娟, 罗玛妮娅, 徐令文, 杨彦萍, 陆斌. 2021. 竹叶花椒MYB基因家族的鉴定及其表达特性的分析. 经济林研究, 39 (4):148-157.
|
|
| [5] |
|
| [6] |
|
|
关淑文. 2022. 竹叶花椒的皮刺发育分子机理研究[硕士论文]. 昆明: 西南林业大学.
|
|
| [7] |
|
| [8] |
|
| [9] |
|
| [10] |
|
|
李旭辉, 陈卫卫, 齐永文, 陆思奇. 2023. 玉米ZmWRKY28基因在调控根毛长度中的应用. CN116640773A,1-11.
|
|
| [11] |
|
|
刘金秋, 陈凯, 张珍珠, 陈秀玲, 王傲雪. 2016. 外施GA、MeJA、IAA、SA和KT对番茄表皮毛发生的作用. 园艺学报, 43 (11):2151-2160.
|
|
| [12] |
|
| [13] |
|
| [14] |
|
| [15] |
|
| [16] |
|
| [17] |
|
| [18] |
|
| [19] |
|
| [20] |
|
|
吴金山, 于旭东, 许泽康, 王亚沉, 高豪杰, 赵莹. 2017. 海南木棉不同月份光合特性及碳汇能力研究. 中国热带农业,(3):67-71.
|
|
| [21] |
|
|
武延生, 严文培, 张晓丽, 牛伟涛, 张鹏飞, 武丽娜, 李敏, 邢翠娟, 王僧虎. 2021. 植物刺的类型与功能辨析. 生物学通报, 56 (4):13-15.
|
|
| [22] |
|
| [23] |
|
|
夏小丛. 2017. 棉花(Gossypium hirsutum)GhJAZ3和GhGAI1在JA和GA介导的转基因拟南芥表皮毛分化和伸长中的功能研究[硕士论文]. 武汉: 华中师范大学.
|
|
| [24] |
|
|
姚珂心. 2023. 四季秋海棠BsMYB62转录因子的功能研究[硕士论文]. 郑州: 河南农业大学.
|
|
| [25] |
|
| [26] |
|
|
庄园. 2015. 红松查尔酮合酶基因和肉桂酰辅酶A还原酶基因的克隆及功能分析[硕士论文]. 长春: 吉林大学.
|
| [1] | YUAN Juanwei, JIA Li, WANG Han, YAN Congsheng, ZHANG Qi’an, YU Feifei, GAN Defang, JIANG Haikun. Screening of Resistant Germplasm and Identifying Candidate Gene for Phytophthora capsici Resistance in Pepper Germplasm Resources [J]. Acta Horticulturae Sinica, 2025, 52(4): 857-871. |
| [2] | TANG Lingli, HE Yuhua, WANG Qingtao, AN Lulu, XU Yongyang, ZHANG Jian, KONG Weihu, HU Keyun, ZHAO Guangwei. Hormonal and Transcriptome Analysis of Ripe Fruits of Non-Climacteric and Climacteric Melon [J]. Acta Horticulturae Sinica, 2025, 52(4): 883-896. |
| [3] | LIN Ruxue, WANG Fei, ZHANG Yanjie, MA Li, LIU Xiaofeng, LI Shuran, LIU Yalong, WANG Chen, JIANG Shuling, OU Chunqing. Transcriptome Screening and Functional Analysis of Dwarf Related Genes in Pear [J]. Acta Horticulturae Sinica, 2025, 52(3): 545-560. |
| [4] | KUANG Meimei, LI Li, MA Jianwei, LIU Yuan, JIANG Hongfei, LEI Rui, MAN Yuping, WANG Yifan, HUANG Bo, WANG Yanchang, LIU Shibiao. Screening of Kiwifruit Canker Resistance-Related Genes Based on the Transcriptome Sequencing Analysis of Hybrids from Actinidia chinensis [J]. Acta Horticulturae Sinica, 2024, 51(8): 1743-1757. |
| [5] | YAO Fengping, WANG Yanbin, QIN Yuchuan, WANG Liling. Analysis of the Difference of Flavonoids in Dendropanax dentiger Leaves with Different Drying Processes Based on Untargeted Metabolomics [J]. Acta Horticulturae Sinica, 2024, 51(5): 1069-1082. |
| [6] | ZHANG Wenhao, ZHANG Hui, LIU Yuting, WANG Yan, ZHANG Yingying, WANG Xinman, WANG Quanhua, ZHU Weimin, YANG Xuedong. Preliminary Transcriptome Analysis in Two Tomato Fruits Materials with Different Sugar Content [J]. Acta Horticulturae Sinica, 2024, 51(2): 281-294. |
| [7] | WANG Hong, MIAO Chen, DING Xiaotao, SHEN Haibin, ZHU Weimin. Growth,Development and Transcriptome Analysis of Tomato Cultivars with Different Low Light Tolerances Under Low Light Condition [J]. Acta Horticulturae Sinica, 2024, 51(2): 335-345. |
| [8] | BU Wenxuan, YAO Yiping, HUANG Yu, YANG Xingyu, LUO Xiaoning, ZHANG Minhuan, LEI Weiqun, WANG Zheng, TIAN Jianing, CHEN Lujie, QIN Liping. Transcriptome Analysis of the Response of Tree Peony‘Hu Hong’Under High Temperature Stress [J]. Acta Horticulturae Sinica, 2024, 51(12): 2800-2816. |
| [9] | LU Yanqing, LIN Yanjin, LU Xinkun. Cell Wall Material Metabolism and the Response to High Temperature and Water Deficiency Stresses in Pericarp are Associated with Fruit Cracking of‘Duwei’Pummelo [J]. Acta Horticulturae Sinica, 2023, 50(8): 1747-1768. |
| [10] | RUAN Ruoxin, LUO Huifeng, ZHANG Chen, HUANG Kangkang, XI Dujun, PEI Jiabo, XING Mengyun, LIU Hui. Transcriptome Analysis of Flower Buds of Sweet Cherry Cultivars with Different Chilling Requirements During Dormancy Stages in Hangzhou [J]. Acta Horticulturae Sinica, 2023, 50(6): 1187-1202. |
| [11] | XU Yue, LI Zixiong, CHEN Jie, SUN Liang. Transcriptional Bases of the Regulation of 2,4-D on Tomato Fruit Shape [J]. Acta Horticulturae Sinica, 2023, 50(4): 802-814. |
| [12] | WANG Zehan, YU Wentao, WANG Pengjie, LIU Caiguo, FAN Xiaojing, GU Mengya, CAI Chunping, WANG Pan, YE Naixing. WGCNA Analysis of Differentially Expressed Genes in Floral Organs of Tea Germplasms with Ovary-glabrous and Ovary-trichome [J]. Acta Horticulturae Sinica, 2023, 50(3): 620-634. |
| [13] | JIANG Yu, TU Xunliang, HE Junrong. Analysis of Differential Expression Genes in Leaves of Leaf Color Mutant of Chinese Orchid [J]. Acta Horticulturae Sinica, 2023, 50(2): 371-381. |
| [14] | XIE Qian, ZHANG Shiyan, JIANG Lai, DING Mingyue, LIU Lingling, WU Rujian, CHEN Qingxi. Analysis of Canarium album Transcriptome SSR Information and Molecular Marker Development and Application [J]. Acta Horticulturae Sinica, 2023, 50(11): 2350-2364. |
| [15] | ZHAO Xueyan, WANG Qi, WANG Li, WANG Fangyuan, WANG Qing, LI Yan. Comparative Transcriptome Analysis of Differential Expression in Different Tissues of Corydalis yanhusuo [J]. Acta Horticulturae Sinica, 2023, 50(1): 177-187. |
| Viewed | ||||||
|
Full text |
|
|||||
|
Abstract |
|
|||||
Copyright © 2012 Acta Horticulturae Sinica 京ICP备10030308号-2 国际联网备案号 11010802023439
Tel: 010-82109523 E-Mail: yuanyixuebao@126.com
Support by: Beijing Magtech Co.Ltd