
Acta Horticulturae Sinica ›› 2024, Vol. 51 ›› Issue (8): 1743-1757.doi: 10.16420/j.issn.0513-353x.2023-0593
• Genetic & Breeding·Germplasm Resources·Molecular Biology • Previous Articles Next Articles
KUANG Meimei1,2, LI Li2, MA Jianwei3, LIU Yuan3, JIANG Hongfei3, LEI Rui2, MAN Yuping2, WANG Yifan4, HUANG Bo3, WANG Yanchang2,*( ), LIU Shibiao1,*(
), LIU Shibiao1,*( )
)
Received:2024-01-12
Revised:2024-05-20
Online:2024-08-25
Published:2024-08-21
Contact:
WANG Yanchang, LIU Shibiao
KUANG Meimei, LI Li, MA Jianwei, LIU Yuan, JIANG Hongfei, LEI Rui, MAN Yuping, WANG Yifan, HUANG Bo, WANG Yanchang, LIU Shibiao. Screening of Kiwifruit Canker Resistance-Related Genes Based on the Transcriptome Sequencing Analysis of Hybrids from Actinidia chinensis[J]. Acta Horticulturae Sinica, 2024, 51(8): 1743-1757.
Add to citation manager EndNote|Ris|BibTeX
URL: https://www.ahs.ac.cn/EN/10.16420/j.issn.0513-353x.2023-0593
| 基因编号 Gene ID | 引物序列 Primer sequence | 产物大小/bp Product length | 退火温度/℃ Tm |
|---|---|---|---|
| Actin | F:5′-TGAGAGATTCCGTTGCCCAGAAGT-3′/ R:5′-TTCCTTACTCATGCGGTCTGCGAT-3′ | 193 | 57.4/57.4 |
| Acc23292 | F:5′-GGGAGCTGTCATTCGAGGAA-3′/ R:5′-GATAATCGCCCAGCCTTTCG-3′ | 140 | 53.8/55.0 |
| Acc14142 | F:5′-TCAATCCACCATGCCAGAGT-3′/ R:5′-CAGCTGCCTTCGTGTTCTTT-3′ | 181 | 51.8/51.8 |
| Acc28207 | F:5′-ATGGACCATCTACTTTGCTT-3′/ R:5′-GTGCTGAGTGCAATCTGCGG-3′ | 128 | 51.4/55.3 |
| Acc04756 | F:5′-ATGTCGACGAGGAAGGATGT-3′/ R:5′-CGGATCGATCTTGCTAATGC-3′ | 126 | 56.5/58.3 |
| Acc26787 | F:5′-CGTATGCTCTCCGAGTCACC-3′/ R:5′-ACTTGGCCGGAGAATTGGTT-3′ | 221 | 57.8/60.9 |
| Acc04136 | F:5′-GACGAGTCGTGGGTTGTAGG-3′/ R:5′-TTTGGTCACTCCAGCTTCCG-3′ | 159 | 57.6/60.9 |
| Acc06864 | F:5′-CTAGCCACGGTTCACTCCTC-3′/ R:5′-ATGTTCTCGCCGTACTTCCC-3′ | 203 | 55.9/53.8 |
| Acc06865 | F:5′-AACACCGTTCGTGACCAAGT-3′/ R:5′-TGGTTAGCGTAGTTCTCGGC-3′ | 83 | 51.8/53.8 |
| Acc06866 | F:5′-AACCAGAGGTCTGGCGATTG-3′/ R:5′-AACCTTACCTGAGGCACACG-3′ | 171 | 53.8/53.8 |
| Acc06484 | F:5′-ATCACTCAGCCTGCCTCAAC-3′/ R:5′-CTTGGTCACGCAATACGCAG-3′ | 251 | 53.8/53.8 |
Table 1 Primers for qRT-PCR
| 基因编号 Gene ID | 引物序列 Primer sequence | 产物大小/bp Product length | 退火温度/℃ Tm |
|---|---|---|---|
| Actin | F:5′-TGAGAGATTCCGTTGCCCAGAAGT-3′/ R:5′-TTCCTTACTCATGCGGTCTGCGAT-3′ | 193 | 57.4/57.4 |
| Acc23292 | F:5′-GGGAGCTGTCATTCGAGGAA-3′/ R:5′-GATAATCGCCCAGCCTTTCG-3′ | 140 | 53.8/55.0 |
| Acc14142 | F:5′-TCAATCCACCATGCCAGAGT-3′/ R:5′-CAGCTGCCTTCGTGTTCTTT-3′ | 181 | 51.8/51.8 |
| Acc28207 | F:5′-ATGGACCATCTACTTTGCTT-3′/ R:5′-GTGCTGAGTGCAATCTGCGG-3′ | 128 | 51.4/55.3 |
| Acc04756 | F:5′-ATGTCGACGAGGAAGGATGT-3′/ R:5′-CGGATCGATCTTGCTAATGC-3′ | 126 | 56.5/58.3 |
| Acc26787 | F:5′-CGTATGCTCTCCGAGTCACC-3′/ R:5′-ACTTGGCCGGAGAATTGGTT-3′ | 221 | 57.8/60.9 |
| Acc04136 | F:5′-GACGAGTCGTGGGTTGTAGG-3′/ R:5′-TTTGGTCACTCCAGCTTCCG-3′ | 159 | 57.6/60.9 |
| Acc06864 | F:5′-CTAGCCACGGTTCACTCCTC-3′/ R:5′-ATGTTCTCGCCGTACTTCCC-3′ | 203 | 55.9/53.8 |
| Acc06865 | F:5′-AACACCGTTCGTGACCAAGT-3′/ R:5′-TGGTTAGCGTAGTTCTCGGC-3′ | 83 | 51.8/53.8 |
| Acc06866 | F:5′-AACCAGAGGTCTGGCGATTG-3′/ R:5′-AACCTTACCTGAGGCACACG-3′ | 171 | 53.8/53.8 |
| Acc06484 | F:5′-ATCACTCAGCCTGCCTCAAC-3′/ R:5′-CTTGGTCACGCAATACGCAG-3′ | 251 | 53.8/53.8 |

Fig. 1 Comparison of the phenotypes of disease-resistant individual‘21-2-4’and susceptible individual‘19-3-11’ isolated branches of kiwifruit hybrid progeny 35 d after inoculation with Psa

Fig. 2 GO and KEGG enrichment analysis of DEGs in kiwifruit disease resistant monocot‘21-2-4’at 6 h and 24 h after inoculation with Psa versus before inoculation
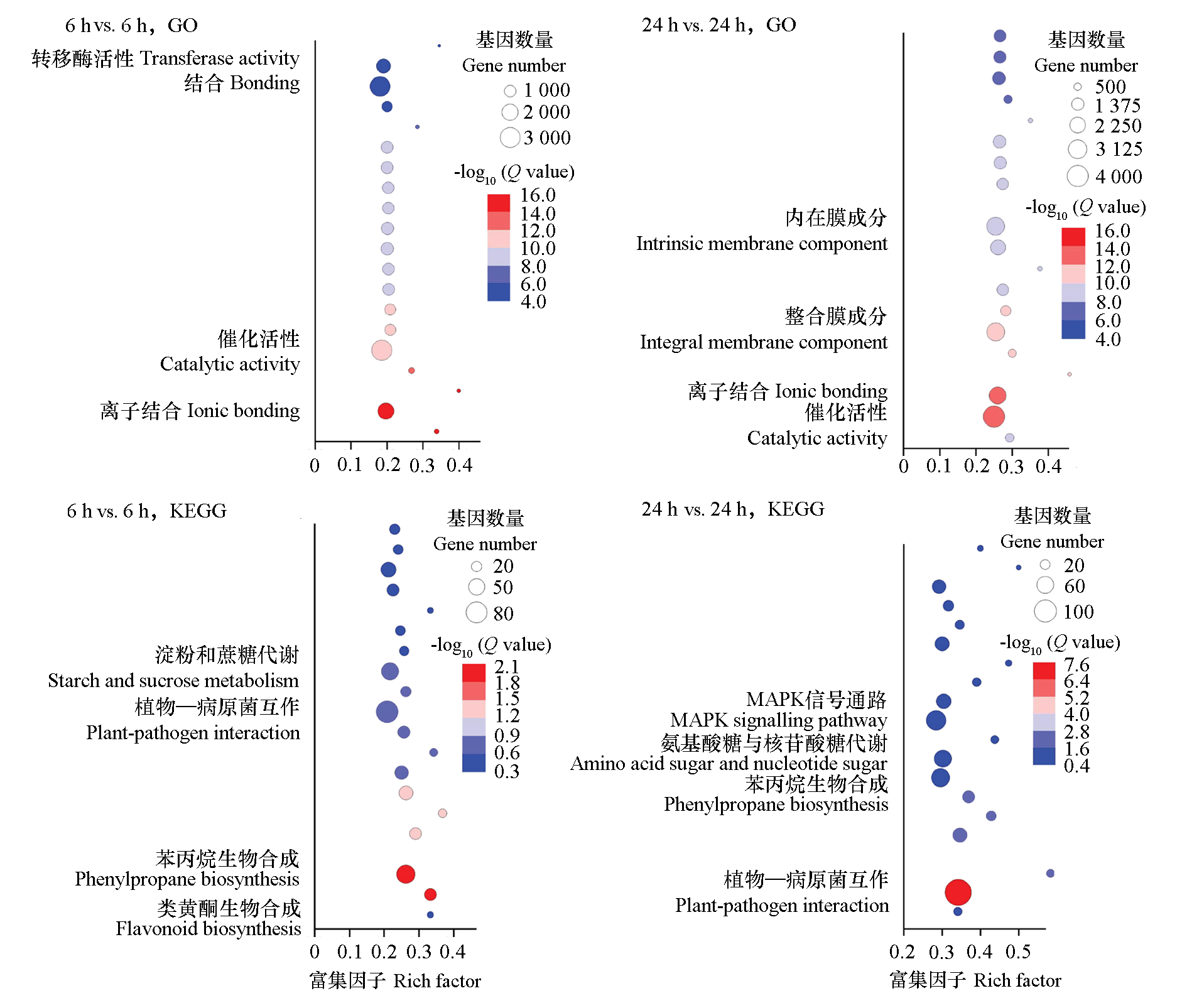
Fig. 3 GO and KEGG Enrichment analysis of DEGs between kiwifruit disease-resistant monocot‘21-2-4’ and susceptible monocot‘19-3-11’at 6 h and 24 h after inoculation with Psa
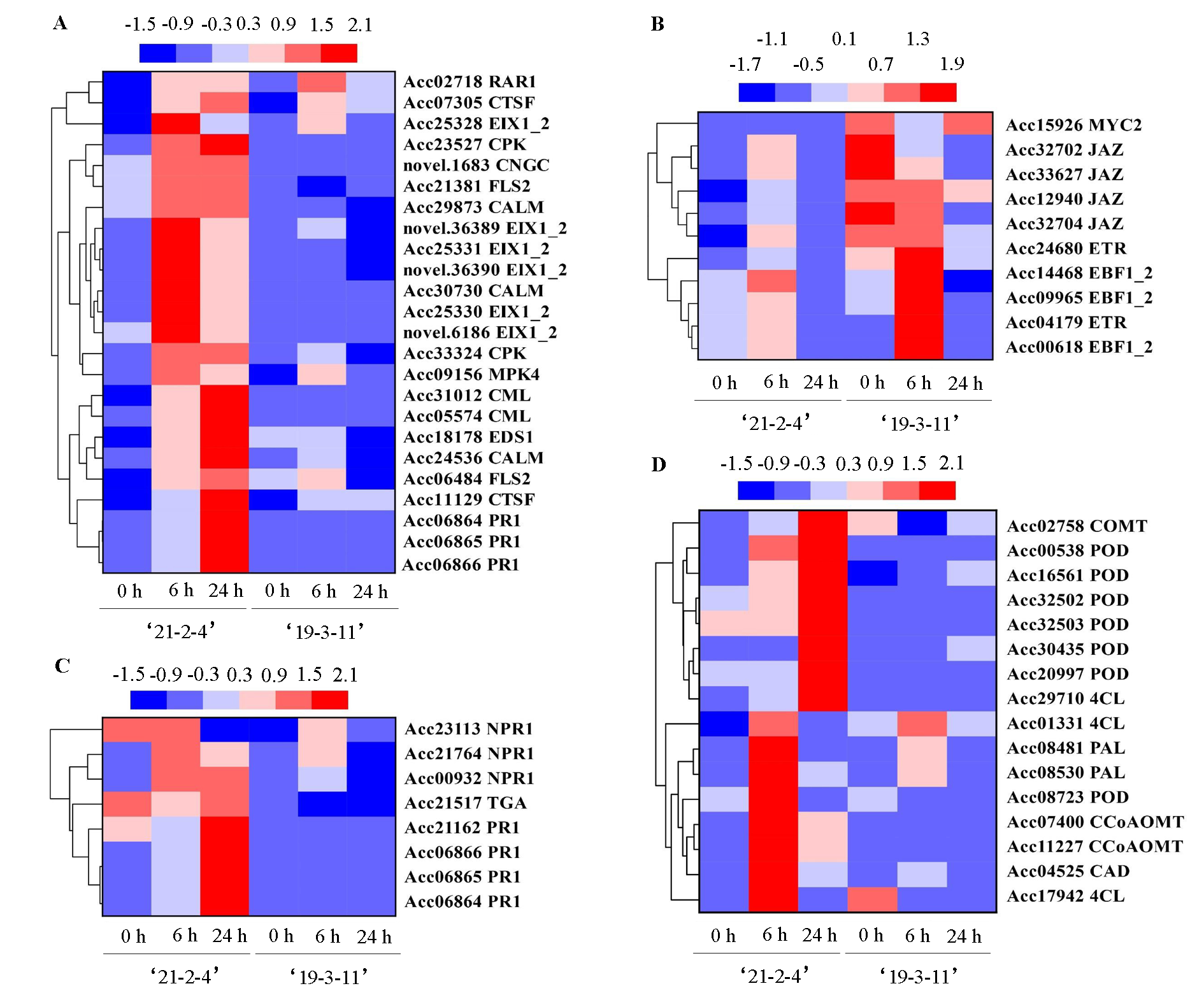
Fig. 4 Expression patterns of DEGs involved in plant-pathogen interaction pathway in‘21-2-4’and‘19-3-11’ A:Involved in the plant-pathogen interaction pathway;B:Involved in the ethylene/jasmonate signalling pathway;C:Involved in the salicylic acid signalling pathway;D:Involved in the phenylpropane biosynthesis pathway.
| 基因编号 Gene ID | 表达量 FPKM(‘21-2-4’) | KO | 途径 Pathway | NR注释 NR_annotation | ||
|---|---|---|---|---|---|---|
| 0 h | 6 h | 24 h | ||||
| Acc06866 | 33.27 b | 305.75 b | 2 308.34 a | PR1 | PTI、SA | 病程相关蛋白Pathogenesis-related protein |
| Acc06865 | 64.83 b | 537.47 b | 2 678.93 a | PR1 | PTI、SA | 病程相关蛋白Pathogenesis-related protein |
| Acc06864 | 367.59 b | 2 171.17 b | 10 865.11 a | PR1 | PTI、SA | 病程相关蛋白Pathogenesis-related protein |
| Acc06484 | 14.20 b | 111.05 a | 25.56 b | FLS2 | PTI | LRR受体样激酶LRR receptor-like serine/threonine-protein kinase |
| Acc25328 | 2.92 b | 15.19 a | 7.70 ab | EIX1_2 | PTI | 受体样蛋白Receptor-like protein |
| novel.6186 | 2.69 b | 12.26 a | 6.08 ab | EIX1_2 | PTI | 受体样蛋白Receptor-like protein |
| novel.36390 | 2.59 b | 11.14 a | 6.99 ab | EIX1_2 | PTI | 受体样蛋白Receptor-like protein |
| Acc25330 | 3.16 b | 12.70 a | 7.51 ab | EIX1_2 | PTI | 受体样蛋白Receptor-like protein |
| Acc23527 | 2.24 b | 7.74 a | 10.43 ab | CPK | PTI | 钙依赖蛋白激酶Calcium-dependent protein kinase |
| Acc18178 | 11.18 b | 30.83 a | 39.39 a | EDS1 | ETI | EDS1样蛋白 Protein EDS1B like |
| Acc07305 | 28.87 b | 86.32 a | 89.80 a | CTSF | ETI | 半胱氨酸蛋白酶Cysteine protease |
| Acc11129 | 78.04 b | 159.43 a | 305.97 a | CTSF | ETI | 半胱氨酸蛋白酶Cysteine protease |
| Acc02718 | 10.53 b | 32.00 a | 31.63 a | RAR1 | ETI | 富含半胱氨酸和组氨酸结构域蛋白 Cysteine and histidine-rich domain-containing protein |
| Acc21162 | 321.06 ab | 193.38 b | 692.83 a | PR1 | SA | 病程相关蛋白基本形式Basic form of pathogenesis-related protein like |
| Acc29710 | 2.74 b | 5.29 ab | 11.16 a | 4CL | PAL | 类4香豆酸辅酶 A 4-coumarate-CoA ligase-like |
| Acc01331 | 7.41 b | 23.73 a | 12.13 ab | 4CL | PAL | 4香豆酸辅酶 A 4-coumarate-CoA ligase |
| Acc16561 | 160.02 b | 284.77 ab | 449.28 a | POD | PAL | 过氧化物酶Peroxidase |
| Acc20997 | 1.91 b | 1.70 b | 10.94 a | POD | PAL | 过氧化物酶Peroxidase |
| Acc00538 | 5.41 b | 28.43 a | 37.94 a | POD | PAL | 过氧化物酶Peroxidase |
| Acc32503 | 362.30 b | 390.84 b | 777.05 a | POD | PAL | 过氧化物酶超家族蛋白Peroxidase superfamily protein |
| Acc32502 | 219.54 b | 247.40 ab | 544.78 a | POD | PAL | 过氧化物酶超家族阴性蛋白Peroxidase superfamily negative protein |
| Acc30435 | 47.41 b | 60.30 b | 623.97 a | POD | PAL | 木质素阴性过氧化物酶Lignin-forming anionic peroxidase |
Table 2 DEGs in disease resistance-related pathways
| 基因编号 Gene ID | 表达量 FPKM(‘21-2-4’) | KO | 途径 Pathway | NR注释 NR_annotation | ||
|---|---|---|---|---|---|---|
| 0 h | 6 h | 24 h | ||||
| Acc06866 | 33.27 b | 305.75 b | 2 308.34 a | PR1 | PTI、SA | 病程相关蛋白Pathogenesis-related protein |
| Acc06865 | 64.83 b | 537.47 b | 2 678.93 a | PR1 | PTI、SA | 病程相关蛋白Pathogenesis-related protein |
| Acc06864 | 367.59 b | 2 171.17 b | 10 865.11 a | PR1 | PTI、SA | 病程相关蛋白Pathogenesis-related protein |
| Acc06484 | 14.20 b | 111.05 a | 25.56 b | FLS2 | PTI | LRR受体样激酶LRR receptor-like serine/threonine-protein kinase |
| Acc25328 | 2.92 b | 15.19 a | 7.70 ab | EIX1_2 | PTI | 受体样蛋白Receptor-like protein |
| novel.6186 | 2.69 b | 12.26 a | 6.08 ab | EIX1_2 | PTI | 受体样蛋白Receptor-like protein |
| novel.36390 | 2.59 b | 11.14 a | 6.99 ab | EIX1_2 | PTI | 受体样蛋白Receptor-like protein |
| Acc25330 | 3.16 b | 12.70 a | 7.51 ab | EIX1_2 | PTI | 受体样蛋白Receptor-like protein |
| Acc23527 | 2.24 b | 7.74 a | 10.43 ab | CPK | PTI | 钙依赖蛋白激酶Calcium-dependent protein kinase |
| Acc18178 | 11.18 b | 30.83 a | 39.39 a | EDS1 | ETI | EDS1样蛋白 Protein EDS1B like |
| Acc07305 | 28.87 b | 86.32 a | 89.80 a | CTSF | ETI | 半胱氨酸蛋白酶Cysteine protease |
| Acc11129 | 78.04 b | 159.43 a | 305.97 a | CTSF | ETI | 半胱氨酸蛋白酶Cysteine protease |
| Acc02718 | 10.53 b | 32.00 a | 31.63 a | RAR1 | ETI | 富含半胱氨酸和组氨酸结构域蛋白 Cysteine and histidine-rich domain-containing protein |
| Acc21162 | 321.06 ab | 193.38 b | 692.83 a | PR1 | SA | 病程相关蛋白基本形式Basic form of pathogenesis-related protein like |
| Acc29710 | 2.74 b | 5.29 ab | 11.16 a | 4CL | PAL | 类4香豆酸辅酶 A 4-coumarate-CoA ligase-like |
| Acc01331 | 7.41 b | 23.73 a | 12.13 ab | 4CL | PAL | 4香豆酸辅酶 A 4-coumarate-CoA ligase |
| Acc16561 | 160.02 b | 284.77 ab | 449.28 a | POD | PAL | 过氧化物酶Peroxidase |
| Acc20997 | 1.91 b | 1.70 b | 10.94 a | POD | PAL | 过氧化物酶Peroxidase |
| Acc00538 | 5.41 b | 28.43 a | 37.94 a | POD | PAL | 过氧化物酶Peroxidase |
| Acc32503 | 362.30 b | 390.84 b | 777.05 a | POD | PAL | 过氧化物酶超家族蛋白Peroxidase superfamily protein |
| Acc32502 | 219.54 b | 247.40 ab | 544.78 a | POD | PAL | 过氧化物酶超家族阴性蛋白Peroxidase superfamily negative protein |
| Acc30435 | 47.41 b | 60.30 b | 623.97 a | POD | PAL | 木质素阴性过氧化物酶Lignin-forming anionic peroxidase |
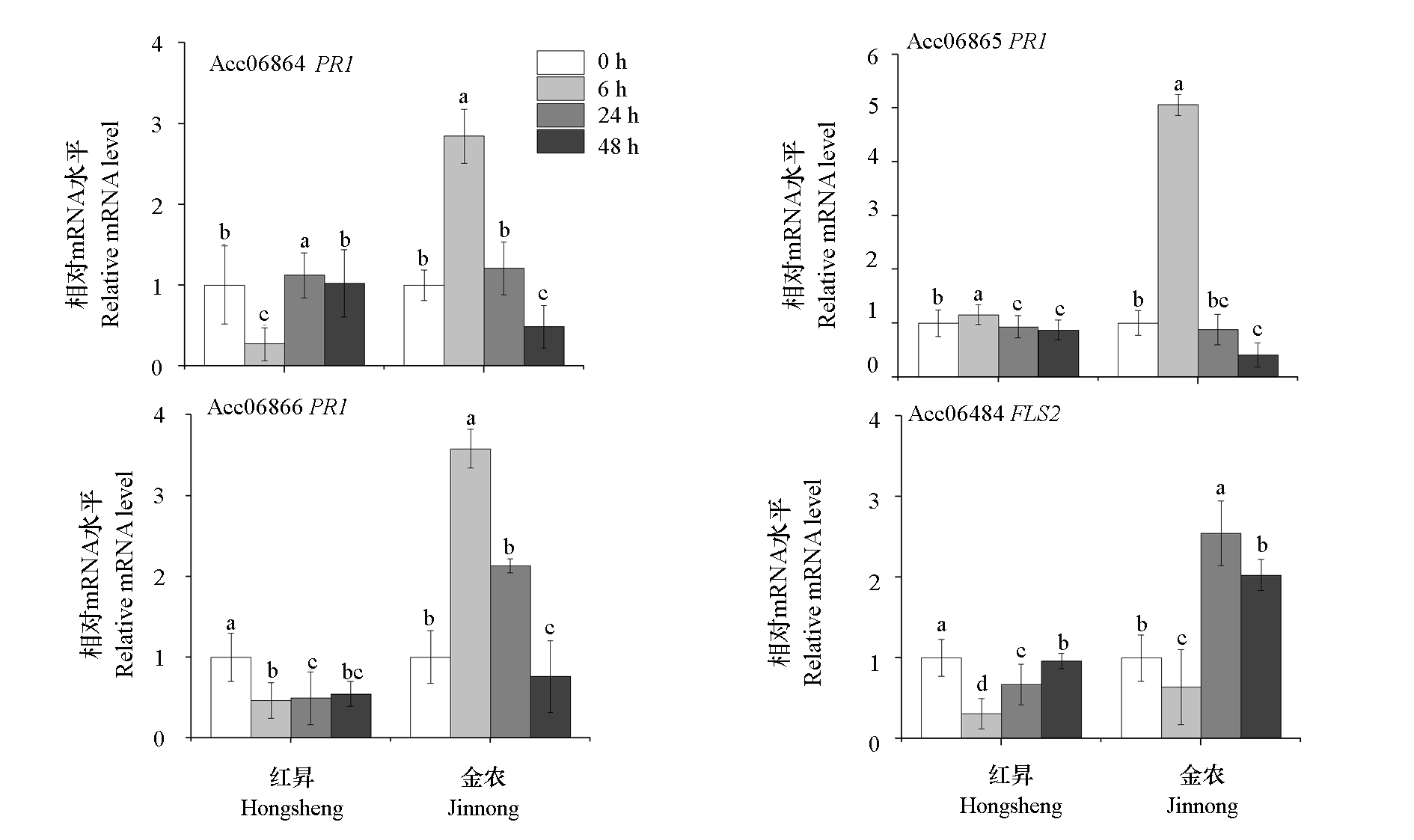
Fig. 5 qRT-PCR validation of the expression levels of disease resistance candidate genes in kiwifruit resistant variety‘Jinnong’ and susceptible variety‘Hongsheng’after inoculation with Psa Different lowercase letters indicate significant differences between periods(P < 0.05).
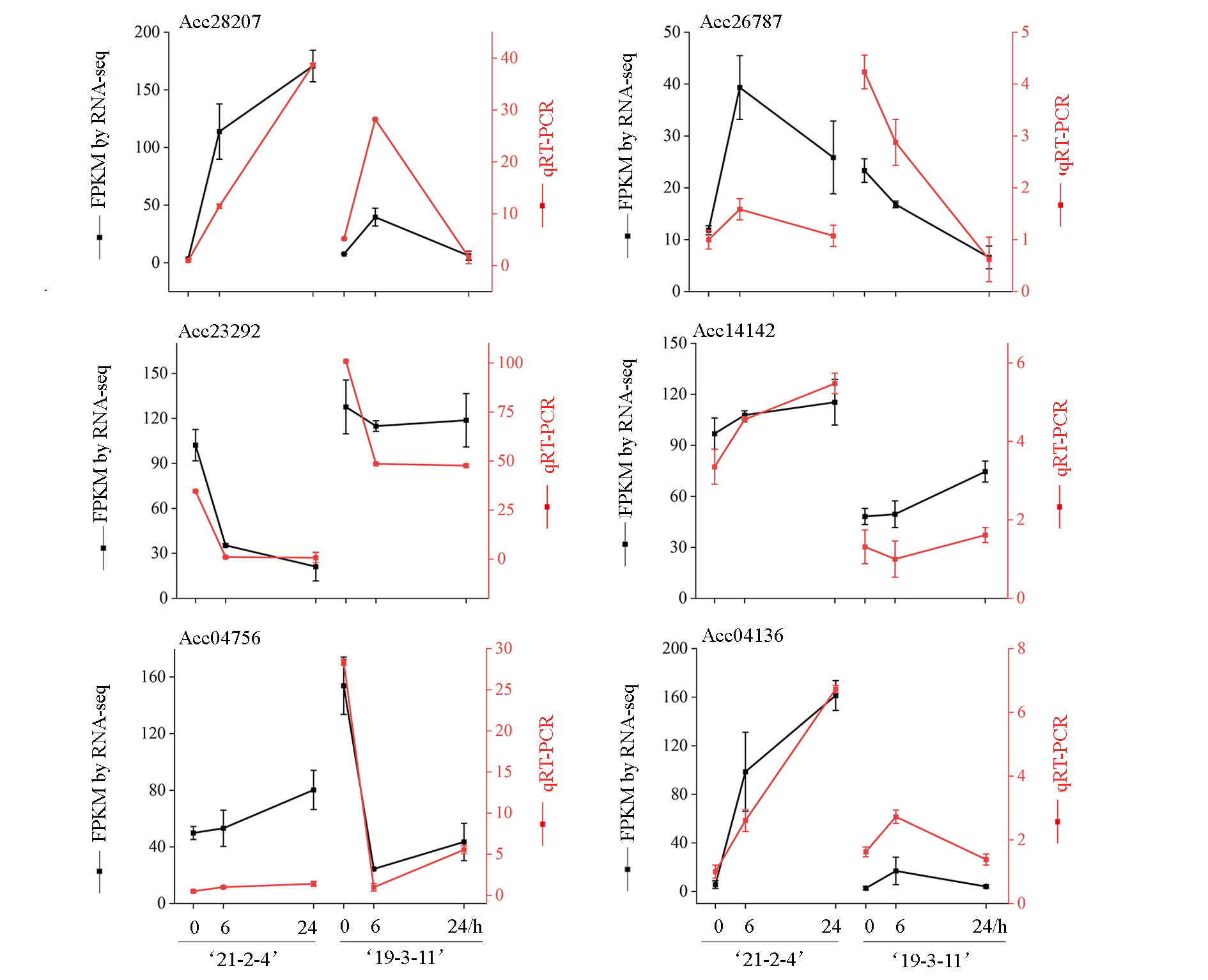
Fig. 6 RNA-Seq and qRT-PCR results of differentially expressed genes in kiwifruit resistant monocot‘21-2-4’ and susceptible monocot‘19-3-11’inoculated with pathogenic bacterium Psa
| [1] |
doi: 10.1021/acs.jafc.0c06283 pmid: 33594880 |
| [2] |
|
|
崔丽红, 高小宁, 张迪, 黄丽丽, 黄蔚, 陈继富. 2019. 湘西地区猕猴桃细菌性溃疡病抗性资源筛选及其抗性机理研究. 植物保护, 45 (3):158-164.
|
|
| [3] |
|
| [4] |
|
|
方炎祖, 王宇道. 1990. 湖南猕猴桃病害调查研究初报. 四川果树科技, 18 (1):28-29.
|
|
| [5] |
|
| [6] |
|
| [7] |
|
|
顾霞, 陈庆红, 何华平, 徐爱春. 2009. ‘金农’和‘金阳’猕猴桃优良性状评价. 中国南方果树, 38 (6):30-31.
|
|
| [8] |
|
| [9] |
|
|
井赵斌. 2021. 猕猴桃抗溃疡病转录组分析和基因功能注释. 分子植物育种, 19 (6):1830-1838.
|
|
| [10] |
doi: 10.1016/s1369-5266(02)00275-3 pmid: 12179966 |
| [11] |
|
| [12] |
|
|
李黎, 潘慧, 李文艺, 汪祖鹏, 钟彩虹. 2022. 中国野生猕猴桃资源的溃疡病抗性种质筛选. 植物科学学报, 40 (6):801-809.
|
|
| [13] |
|
| [14] |
doi: 10.1016/j.tim.2012.01.003 pmid: 22341410 |
| [15] |
|
|
刘健, 王彦昌, 吴世权, 何仕松, 向敬宗. 2016. 红肉型猕猴桃新品种红昇的选育及栽培技术. 落叶果树, 48 (4):26-29.
|
|
| [16] |
doi: 10.16420/j.issn.0513-353x.2017-0687 |
|
刘秀霞, 梁宇宁, 张伟伟, 霍艳红, 冯守千, 邱化荣, 何晓文, 吴树敬, 陈学森. 2018. 超表达MdFLS2的拟南芥fls2突变体识别细菌鞭毛蛋白,提高对轮纹病菌的抗性. 园艺学报, 45 (5):827-844.
doi: 10.16420/j.issn.0513-353x.2017-0687 |
|
| [17] |
|
| [18] |
|
| [19] |
|
| [20] |
doi: 10.1093/gbe/evx055 pmid: 28369338 |
| [21] |
|
| [22] |
|
| [23] |
doi: 10.1038/nchembio.164 pmid: 19377457 |
| [24] |
|
| [25] |
|
| [26] |
|
| [27] |
|
|
宋雅林. 2019. 猕猴桃应答溃疡病菌侵染的转录组研究及抗性相关基因挖掘[博士论文]. 武汉: 华中农业大学.
|
|
| [28] |
|
| [29] |
|
| [30] |
|
| [31] |
|
|
王发明, 齐贝贝, 叶开玉, 龚弘娟, 莫权辉, 蒋桥生, 刘平平, 李洁维. 2021. 九个中华类红肉猕猴桃品种的亲缘关系及其溃疡病抗性分析. 分子植物育种, 19 (1):193-199.
|
|
| [32] |
|
| [33] |
|
|
王睿, 韩烈保. 2022. CRISPR/Cas9介导的二穗短柄草bdfls2敲除突变体的获得. 生物技术通报, 38 (1):70-76.
doi: 10.13560/j.cnki.biotech.bull.1985.2021-0326 |
|
| [34] |
|
| [35] |
|
| [36] |
|
| [37] |
|
|
温欣, 秦红艳, 艾军, 王月, 杨义明, 范书田, 张宝香, 舒楠. 2020. 不同抗性猕猴桃材料接种溃疡病菌后防御酶活性变化研究. 特产研究, 42 (3):6-11.
|
|
| [38] |
|
| [39] |
|
| [40] |
|
|
张迪, 高小宁, 赵志博, 秦虎强, 黄丽丽. 2019. 不同猕猴桃品种对溃疡病的抗性差异及其机制研究. 果树学报, 36 (11):1549-1557.
|
|
| [41] |
|
| [42] |
|
| [1] | ZHANG Wenhao, ZHANG Hui, LIU Yuting, WANG Yan, ZHANG Yingying, WANG Xinman, WANG Quanhua, ZHU Weimin, YANG Xuedong. Preliminary Transcriptome Analysis in Two Tomato Fruits Materials with Different Sugar Content [J]. Acta Horticulturae Sinica, 2024, 51(2): 281-294. |
| [2] | WANG Hong, MIAO Chen, DING Xiaotao, SHEN Haibin, ZHU Weimin. Growth,Development and Transcriptome Analysis of Tomato Cultivars with Different Low Light Tolerances Under Low Light Condition [J]. Acta Horticulturae Sinica, 2024, 51(2): 335-345. |
| [3] | BU Wenxuan, YAO Yiping, HUANG Yu, YANG Xingyu, LUO Xiaoning, ZHANG Minhuan, LEI Weiqun, WANG Zheng, TIAN Jianing, CHEN Lujie, QIN Liping. Transcriptome Analysis of the Response of Tree Peony‘Hu Hong’Under High Temperature Stress [J]. Acta Horticulturae Sinica, 2024, 51(12): 2800-2816. |
| [4] | FENG Yisi, TIAN Xiaoyu, YANG Chongshan, CHEN Xiangling, DENG Xiuxin, XIE Kaidong, GUO Wenwu, XIE Zongzhou, CHAI Lijun, and YE Junli, . Exploitation and Genetic Identification of Triploid Plants from Seedling Populations of Orah Mandarin(Citrus reticulata) [J]. Acta Horticulturae Sinica, 2024, 51(10): 2231-2242. |
| [5] | LU Yanqing, LIN Yanjin, LU Xinkun. Cell Wall Material Metabolism and the Response to High Temperature and Water Deficiency Stresses in Pericarp are Associated with Fruit Cracking of‘Duwei’Pummelo [J]. Acta Horticulturae Sinica, 2023, 50(8): 1747-1768. |
| [6] | RUAN Ruoxin, LUO Huifeng, ZHANG Chen, HUANG Kangkang, XI Dujun, PEI Jiabo, XING Mengyun, LIU Hui. Transcriptome Analysis of Flower Buds of Sweet Cherry Cultivars with Different Chilling Requirements During Dormancy Stages in Hangzhou [J]. Acta Horticulturae Sinica, 2023, 50(6): 1187-1202. |
| [7] | XU Yue, LI Zixiong, CHEN Jie, SUN Liang. Transcriptional Bases of the Regulation of 2,4-D on Tomato Fruit Shape [J]. Acta Horticulturae Sinica, 2023, 50(4): 802-814. |
| [8] | WANG Zehan, YU Wentao, WANG Pengjie, LIU Caiguo, FAN Xiaojing, GU Mengya, CAI Chunping, WANG Pan, YE Naixing. WGCNA Analysis of Differentially Expressed Genes in Floral Organs of Tea Germplasms with Ovary-glabrous and Ovary-trichome [J]. Acta Horticulturae Sinica, 2023, 50(3): 620-634. |
| [9] | JIANG Yu, TU Xunliang, HE Junrong. Analysis of Differential Expression Genes in Leaves of Leaf Color Mutant of Chinese Orchid [J]. Acta Horticulturae Sinica, 2023, 50(2): 371-381. |
| [10] | XIE Qian, ZHANG Shiyan, JIANG Lai, DING Mingyue, LIU Lingling, WU Rujian, CHEN Qingxi. Analysis of Canarium album Transcriptome SSR Information and Molecular Marker Development and Application [J]. Acta Horticulturae Sinica, 2023, 50(11): 2350-2364. |
| [11] | ZHAO Xueyan, WANG Qi, WANG Li, WANG Fangyuan, WANG Qing, LI Yan. Comparative Transcriptome Analysis of Differential Expression in Different Tissues of Corydalis yanhusuo [J]. Acta Horticulturae Sinica, 2023, 50(1): 177-187. |
| [12] | WANG Yan, LIU Zhenshan, ZHANG Jing, YANG Pengfei, MA Lan, WANG Zhiyi, TU Hongxia, YANG Shaofeng, WANG Hao, CHEN Tao, WANG Xiaorong. Inheritance Trend of Flower and Fruit Traits in F1 Progenies of Chinese Cherry [J]. Acta Horticulturae Sinica, 2022, 49(9): 1853-1865. |
| [13] | ZHOU Xuzixin, YANG Wei, MAO Meiqin, XUE Yanbin, MA Jun. Identification of Pigment Components and Key Genes in Carotenoid Pathway in Mutants of Chimeric Ananas comosus var. bracteatus [J]. Acta Horticulturae Sinica, 2022, 49(5): 1081-1091. |
| [14] | LI Li, FENG Dandan, PAN Hui, LI Wenyi, DENG Lei, WANG Zupeng, ZHONG Caihong. Comparison of Sterilization Methods for Kiwifruit Pollen and Its Effect on Fruit Quality [J]. Acta Horticulturae Sinica, 2022, 49(4): 769-777. |
| [15] | SHEN Nan, ZHANG Jingcheng, WANG Chengchen, BIAN Yinbing, XIAO Yang. Studies on Transcriptome During Fruiting Body Development of Lentinula edodes [J]. Acta Horticulturae Sinica, 2022, 49(4): 801-815. |
| Viewed | ||||||
|
Full text |
|
|||||
|
Abstract |
|
|||||
Copyright © 2012 Acta Horticulturae Sinica 京ICP备10030308号-2 国际联网备案号 11010802023439
Tel: 010-82109523 E-Mail: yuanyixuebao@126.com
Support by: Beijing Magtech Co.Ltd