
Acta Horticulturae Sinica ›› 2021, Vol. 48 ›› Issue (8): 1485-1503.doi: 10.16420/j.issn.0513-353x.2020-0595
• Research Papers • Previous Articles Next Articles
ZHANG Zhiqiang, LU Shixiong, MA Zonghuan, LI Yanbiao, GAO Caixia, CHEN Baihong, MAO Juan( )
)
Received:2021-01-22
Revised:2021-04-06
Online:2021-08-25
Published:2021-09-06
Contact:
MAO Juan
E-mail:maojuan-81@163.com
CLC Number:
ZHANG Zhiqiang, LU Shixiong, MA Zonghuan, LI Yanbiao, GAO Caixia, CHEN Baihong, MAO Juan. Bioinformatic Identification and Expression Analysis of Candidate Genes of LIM Protein Family in Strawberry Under Abiotic Stresses[J]. Acta Horticulturae Sinica, 2021, 48(8): 1485-1503.
Add to citation manager EndNote|Ris|BibTeX
URL: https://www.ahs.ac.cn/EN/10.16420/j.issn.0513-353x.2020-0595
| 基因 Gene | 上游引物(5′-3′) Forward primer | 下游引物(5′-3′) Reverse primer |
|---|---|---|
| FvLIM1 | ATATGTGGCTTGACGCAGAGACG | GAGGACGCTGAGGAGGAAGAGG |
| FvLIM2 | AAACACCACCACATCCAGCTCTTC | GGCTTGCTCGTTCGTTGATTGC |
| FvLIM3 | ATATGTGGCTTGACGCAGAGACG | GAGGACGCTGAGGAGGAAGAGG |
| FvLIM4 | GCACCACCACACCCAACTCATC | GCAACTGCTGTGGCTTTCTCATTG |
| FvLIM5 | TTGCCCAGTTGTTCAAGGAGAAGG | TGCTGCGTCTGGCTTTGGTTC |
| FvLIM6 | TGCAAGGCATGTGAGAAGACTGTG | AGTGGTGGCATCTGAAGCAAGC |
| FvLIM7 | GAATAGGGCACCCAGCAAACTCTC | TGAGCACACCTGAAGCATGACTTG |
| FvLIM8 | TTAGACCATGCAATCGCGCTCTC | GCTCGTTATCCTTCTCCTGCCATC |
| FvLIM9 | CTGGTATGTTTTCTGGCACGCAAG | TGGCTCTCCACTGTCACCTTCTC |
| FaLIM1 | ATATGTGGCTTGACGCAGAGACG | AAGAGGACGCTGAGGAGGAAGAC |
| FaLIM2 | GCCGAAGATTGGAGCAGATTACCG | ACGAGAATTGCCGTCACTTCACAG |
| FaLIM3 | ATATGTGGCTTGACGCAGAGACG | GAGGACGCTGAGGAGGAAGAGG |
| FaLIM4 | ATATGTGGCTTGACGCAGAGACG | AGGACGCTGAGAAGGAAGAGGAAG |
| FaLIM5 | GCACCACCACACCCAACTCATC | GCAACTGCTGTGGCTTTCTCATTG |
| FaLIM6 | GATAGGACTCCACGGTGTTGTAGC | TTCAAGGTAAAGGGGCTGGCATTC |
| FaLIM7 | GCTAGATGCCGAGCTTATGTCTGG | GCGTGCCCTTCTTTGATGAATTGG |
| FaLIM8 | AAACACCACCACATCCAGCTCTTC | GGCTTGCTCGTTCGTTGATTGC |
| FaLIM9 | TTGCCCAGTTGTTCAAGGAGAAGG | TGCTGCGTCTGGCTTTGGTTG |
| FaLIM10 | TTGCCCAGTTGTTCAAGGAGAAGG | TGCTGCGTCTGGCTTTGGTTC |
| FaLIM11 | CACCGCCTCTACTGTAAGCATCAC | TACTCCGCCACTGTGCTCTCAG |
| FaLIM12 | TTGCCCAGTTGTTCAAGGAGAAGG | TGCTGCGTCTGGCTTTGGTTC |
| FaLIM13 | TGCAAGGCATGTGAGAAGACTGTG | AGTGGTGGCATCTGAAGCAAGC |
| FaLIM14 | GAATAGGGCACCCAGCAAACTCTC | TGAGCACACCTGAAGCATGACTTG |
| FaLIM15 | AGTGCTTCTGTTGTCGCTCTTGTC | GGATGGGCCAGCTCCTTGAAAC |
| FaLIM16 | AGGAGAAGGGGAGCTACAACCATC | TGGAATGCTGGCAACTGCTACTG |
| FaLIM17 | GAATAGGGCACCCAGCAAACTCTC | TGAGCACACCTGAAGCATGACTTG |
| FaLIM18 | GAATAGGGCACCCAGCAAACTCTC | TGAGCACACCTGAAGCATGACTTG |
| FaLIM19 | TTAGACCATGCAATCGCGCTCTC | GCTCGTTATCCTTCTCCTGCCATC |
| FaLIM20 | CGACTAGGTCACCCAGCAAAGC | CGACTTGTGATATGCCTGGCTCTC |
| FaLIM21 | TTAGACCATGCAATCGCGCTCTC | GCTCGTTATCCTTCTCCTGCCATC |
| FaLIM22 | TCAAGGAGAAGGGGAGCTACAACC | TTAGGCTTCTGGAATGCTGGCAAC |
| FaLIM23 | TCAAGGAGAAGGGGAGCTACAACC | TTAGGCTTCTGGAATGCTGGCAAC |
| FaLIM24 | AGTGCTTCTGTTGTCGCTCTTGTC | GGATGGGCCAGCTCCTTGAAAC |
| FaLIM25 | AAGGTCCATTTCGCTCAGCTCTTC | CTTCTGGTTGTGGCTCTGCTTCTG |
| FaLIM26 | CTGGTATGTTTTCTGGCACGCAAG | TGGCTCTCCACTGTCACCTTCTC |
| FaLIM27 | AGTGCTTCTGTTGTCGCTCTTGTG | GGATGGGCCAGCTCCTTGAAAC |
| FaLIM28 | GAATAGGGCACCCAGCAAACTCTC | TGAGCACACCTGAAGCATGACTTG |
| GAPDH | CATTCATCACCACCGACTACA | GAAGGGTCTTCTCATCCTTGAC |
Table 1 Real-time PCR primers for strawberry LIM family candidate genes
| 基因 Gene | 上游引物(5′-3′) Forward primer | 下游引物(5′-3′) Reverse primer |
|---|---|---|
| FvLIM1 | ATATGTGGCTTGACGCAGAGACG | GAGGACGCTGAGGAGGAAGAGG |
| FvLIM2 | AAACACCACCACATCCAGCTCTTC | GGCTTGCTCGTTCGTTGATTGC |
| FvLIM3 | ATATGTGGCTTGACGCAGAGACG | GAGGACGCTGAGGAGGAAGAGG |
| FvLIM4 | GCACCACCACACCCAACTCATC | GCAACTGCTGTGGCTTTCTCATTG |
| FvLIM5 | TTGCCCAGTTGTTCAAGGAGAAGG | TGCTGCGTCTGGCTTTGGTTC |
| FvLIM6 | TGCAAGGCATGTGAGAAGACTGTG | AGTGGTGGCATCTGAAGCAAGC |
| FvLIM7 | GAATAGGGCACCCAGCAAACTCTC | TGAGCACACCTGAAGCATGACTTG |
| FvLIM8 | TTAGACCATGCAATCGCGCTCTC | GCTCGTTATCCTTCTCCTGCCATC |
| FvLIM9 | CTGGTATGTTTTCTGGCACGCAAG | TGGCTCTCCACTGTCACCTTCTC |
| FaLIM1 | ATATGTGGCTTGACGCAGAGACG | AAGAGGACGCTGAGGAGGAAGAC |
| FaLIM2 | GCCGAAGATTGGAGCAGATTACCG | ACGAGAATTGCCGTCACTTCACAG |
| FaLIM3 | ATATGTGGCTTGACGCAGAGACG | GAGGACGCTGAGGAGGAAGAGG |
| FaLIM4 | ATATGTGGCTTGACGCAGAGACG | AGGACGCTGAGAAGGAAGAGGAAG |
| FaLIM5 | GCACCACCACACCCAACTCATC | GCAACTGCTGTGGCTTTCTCATTG |
| FaLIM6 | GATAGGACTCCACGGTGTTGTAGC | TTCAAGGTAAAGGGGCTGGCATTC |
| FaLIM7 | GCTAGATGCCGAGCTTATGTCTGG | GCGTGCCCTTCTTTGATGAATTGG |
| FaLIM8 | AAACACCACCACATCCAGCTCTTC | GGCTTGCTCGTTCGTTGATTGC |
| FaLIM9 | TTGCCCAGTTGTTCAAGGAGAAGG | TGCTGCGTCTGGCTTTGGTTG |
| FaLIM10 | TTGCCCAGTTGTTCAAGGAGAAGG | TGCTGCGTCTGGCTTTGGTTC |
| FaLIM11 | CACCGCCTCTACTGTAAGCATCAC | TACTCCGCCACTGTGCTCTCAG |
| FaLIM12 | TTGCCCAGTTGTTCAAGGAGAAGG | TGCTGCGTCTGGCTTTGGTTC |
| FaLIM13 | TGCAAGGCATGTGAGAAGACTGTG | AGTGGTGGCATCTGAAGCAAGC |
| FaLIM14 | GAATAGGGCACCCAGCAAACTCTC | TGAGCACACCTGAAGCATGACTTG |
| FaLIM15 | AGTGCTTCTGTTGTCGCTCTTGTC | GGATGGGCCAGCTCCTTGAAAC |
| FaLIM16 | AGGAGAAGGGGAGCTACAACCATC | TGGAATGCTGGCAACTGCTACTG |
| FaLIM17 | GAATAGGGCACCCAGCAAACTCTC | TGAGCACACCTGAAGCATGACTTG |
| FaLIM18 | GAATAGGGCACCCAGCAAACTCTC | TGAGCACACCTGAAGCATGACTTG |
| FaLIM19 | TTAGACCATGCAATCGCGCTCTC | GCTCGTTATCCTTCTCCTGCCATC |
| FaLIM20 | CGACTAGGTCACCCAGCAAAGC | CGACTTGTGATATGCCTGGCTCTC |
| FaLIM21 | TTAGACCATGCAATCGCGCTCTC | GCTCGTTATCCTTCTCCTGCCATC |
| FaLIM22 | TCAAGGAGAAGGGGAGCTACAACC | TTAGGCTTCTGGAATGCTGGCAAC |
| FaLIM23 | TCAAGGAGAAGGGGAGCTACAACC | TTAGGCTTCTGGAATGCTGGCAAC |
| FaLIM24 | AGTGCTTCTGTTGTCGCTCTTGTC | GGATGGGCCAGCTCCTTGAAAC |
| FaLIM25 | AAGGTCCATTTCGCTCAGCTCTTC | CTTCTGGTTGTGGCTCTGCTTCTG |
| FaLIM26 | CTGGTATGTTTTCTGGCACGCAAG | TGGCTCTCCACTGTCACCTTCTC |
| FaLIM27 | AGTGCTTCTGTTGTCGCTCTTGTG | GGATGGGCCAGCTCCTTGAAAC |
| FaLIM28 | GAATAGGGCACCCAGCAAACTCTC | TGAGCACACCTGAAGCATGACTTG |
| GAPDH | CATTCATCACCACCGACTACA | GAAGGGTCTTCTCATCCTTGAC |
| 名称 Name | 染色体定位 Chromosome location | 氨基酸数 Amino acids | 分子量 Molecular weight | 等电点 pI | 不稳定指数 Instability index | 亲水性 Hydrophilic | 亚细胞定位 Subcellular localization |
|---|---|---|---|---|---|---|---|
| FvLIM1 | Fvb1:7945940..7952121 | 538 | 60 510.36 | 5.20 | 53.78 | -0.718 | 细胞核 Nucleus |
| FvLIM2 | Fvb2:2982770..2998244 | 197 | 22 232.22 | 8.89 | 42.90 | -0.703 | 细胞核 Nucleus |
| FvLIM3 | Fvb2:5932861..5935114 | 527 | 59 323.18 | 5.33 | 52.19 | -0.698 | 细胞核 Nucleus |
| FvLIM4 | Fvb2:24843809..24848493 | 191 | 21 205.29 | 9.05 | 29.01 | -0.539 | 细胞核 Nucleus |
| FvLIM5 | Fvb5:1235177..1237221 | 212 | 23 391.44 | 7.52 | 45.56 | -0.62 | 细胞核 Nucleus |
| FvLIM6 | Fvb5:9381785..9385122 | 188 | 21 211.15 | 8.86 | 28.77 | -0.586 | 细胞核 Nucleus |
| FvLIM7 | Fvb7:12341249..12343536 | 217 | 23 962.13 | 6.50 | 45.65 | -0.567 | 细胞核 Nucleus |
| FvLIM8 | Fvb7:18342947..18346948 | 504 | 57 014.81 | 8.26 | 52.67 | -0.541 | 细胞核Nucleus |
| FvLIM9 | Fvb7:18517083..18519868 | 200 | 21 721.91 | 9.08 | 37.48 | -0.362 | 细胞核 Nucleus |
| FaLIM1 | Fvb1-1:20431547..20437334 | 534 | 59 915.92 | 5.22 | 52.12 | -0.668 | 细胞核 Nucleus |
| FaLIM2 | Fvb1-2:9164763..9171264 | 533 | 60 168.24 | 5.39 | 51.21 | -0.737 | 细胞核 Nucleus |
| FaLIM3 | Fvb1-3:8145568..8151824 | 539 | 60 522.40 | 5.20 | 54.39 | -0.719 | 细胞核 Nucleus |
| FaLIM4 | Fvb1-4:6945308..6951056 | 538 | 60 571.45 | 5.17 | 53.06 | -0.712 | 细胞核 Nucleus |
| FaLIM5 | Fvb2-2:728488..730564 | 191 | 21 205.29 | 9.05 | 29.01 | -0.539 | 细胞核 Nucleus |
| FaLIM6 | Fvb2-2:18290991..18293017 | 231 | 27 087.33 | 7.80 | 45.08 | -0.512 | 细胞核 Nucleus |
| FaLIM7 | Fvb2-3:2848239..2854332 | 491 | 55 136.60 | 5.75 | 44.43 | -0.671 | 细胞核 Nucleus |
| FaLIM8 | Fvb2-4:1747334..1750087 | 176 | 19 899.54 | 8.83 | 44.58 | -0.618 | 细胞核 Nucleus |
| FaLIM9 | Fvb5-1:1118664..1120389 | 213 | 23 551.67 | 7.92 | 45.67 | -0.623 | 细胞核 Nucleus |
| FaLIM10 | Fvb5-2:1037693..1039462 | 216 | 23 835.04 | 8.18 | 44.28 | -0.569 | 细胞核 Nucleus |
| FaLIM11 | Fvb5-3:18813760..18816438 | 188 | 21 172.07 | 8.86 | 31.15 | -0.599 | 细胞核 Nucleus |
| FaLIM12 | Fvb5-4:1027068..1028886 | 211 | 23 297.40 | 7.52 | 43.54 | -0.607 | 细胞核 Nucleus |
| FaLIM13 | Fvb5-4:7252377..7255038 | 188 | 21 199.10 | 8.86 | 29.68 | -0.613 | 细胞核 Nucleus |
| FaLIM14 | Fvb7-1:16915680..16917768 | 217 | 23 964.10 | 6.50 | 45.95 | -0.579 | 细胞核 Nucleus |
| FaLIM15 | Fvb7-1:25526059..25530050 | 506 | 57 362.23 | 8.54 | 52.15 | -0.549 | 细胞核Nucleus |
| FaLIM16 | Fvb7-1:25665993..25670535 | 200 | 21 818.02 | 9.08 | 37.58 | -0.360 | 细胞核 Nucleus |
| FaLIM17 | Fvb7-2:11429297..11431530 | 217 | 23 962.13 | 6.50 | 45.65 | -0.567 | 细胞核 Nucleus |
| FaLIM18 | Fvb7-2:17869656..17871925 | 217 | 23 949.13 | 6.50 | 45.65 | -0.554 | 细胞核 Nucleus |
| FaLIM19 | Fvb7-2:23292270..23296225 | 493 | 55 877.59 | 8.37 | 53.86 | -0.540 | 细胞核 Nucleus |
| FaLIM20 | Fvb7-2:23449921..23452679 | 200 | 21 691.82 | 9.08 | 37.63 | -0.376 | 细胞核 Nucleus |
| FaLIM21 | Fvb7-2:23655808..23659719 | 430 | 49 337.54 | 8.63 | 48.07 | -0.525 | 细胞核Nucleus |
| FaLIM22 | Fvb7-2:23849991..23852934 | 200 | 21 721.91 | 9.08 | 37.48 | -0.362 | 细胞核 Nucleus |
| FaLIM23 | Fvb7-3:5169640..5172605 | 200 | 21 704.86 | 9.01 | 38.00 | -0.353 | 细胞核 Nucleus |
| FaLIM24 | Fvb7-3:6773453..6777457 | 506 | 57 325.21 | 8.45 | 52.87 | -0.541 | 细胞核Nucleus |
| FaLIM25 | Fvb7-3:12069028..12071004 | 215 | 23 793.94 | 6.50 | 44.19 | -0.573 | 细胞核 Nucleus |
| FaLIM26 | Fvb7-4:5056752..5059611 | 200 | 21 721.91 | 9.08 | 37.48 | -0.362 | 细胞核 Nucleus |
| FaLIM27 | Fvb7-4:5292185..5296170 | 506 | 57 277.11 | 8.45 | 51.10 | -0.545 | 细胞核Nucleus |
| FaLIM28 | Fvb7-4:11548941..11551124 | 214 | 23 743.92 | 6.54 | 42.18 | -0.534 | 细胞核 Nucleus |
Table 2 Physical and chemical properties for strawberry LIM family candidate genes
| 名称 Name | 染色体定位 Chromosome location | 氨基酸数 Amino acids | 分子量 Molecular weight | 等电点 pI | 不稳定指数 Instability index | 亲水性 Hydrophilic | 亚细胞定位 Subcellular localization |
|---|---|---|---|---|---|---|---|
| FvLIM1 | Fvb1:7945940..7952121 | 538 | 60 510.36 | 5.20 | 53.78 | -0.718 | 细胞核 Nucleus |
| FvLIM2 | Fvb2:2982770..2998244 | 197 | 22 232.22 | 8.89 | 42.90 | -0.703 | 细胞核 Nucleus |
| FvLIM3 | Fvb2:5932861..5935114 | 527 | 59 323.18 | 5.33 | 52.19 | -0.698 | 细胞核 Nucleus |
| FvLIM4 | Fvb2:24843809..24848493 | 191 | 21 205.29 | 9.05 | 29.01 | -0.539 | 细胞核 Nucleus |
| FvLIM5 | Fvb5:1235177..1237221 | 212 | 23 391.44 | 7.52 | 45.56 | -0.62 | 细胞核 Nucleus |
| FvLIM6 | Fvb5:9381785..9385122 | 188 | 21 211.15 | 8.86 | 28.77 | -0.586 | 细胞核 Nucleus |
| FvLIM7 | Fvb7:12341249..12343536 | 217 | 23 962.13 | 6.50 | 45.65 | -0.567 | 细胞核 Nucleus |
| FvLIM8 | Fvb7:18342947..18346948 | 504 | 57 014.81 | 8.26 | 52.67 | -0.541 | 细胞核Nucleus |
| FvLIM9 | Fvb7:18517083..18519868 | 200 | 21 721.91 | 9.08 | 37.48 | -0.362 | 细胞核 Nucleus |
| FaLIM1 | Fvb1-1:20431547..20437334 | 534 | 59 915.92 | 5.22 | 52.12 | -0.668 | 细胞核 Nucleus |
| FaLIM2 | Fvb1-2:9164763..9171264 | 533 | 60 168.24 | 5.39 | 51.21 | -0.737 | 细胞核 Nucleus |
| FaLIM3 | Fvb1-3:8145568..8151824 | 539 | 60 522.40 | 5.20 | 54.39 | -0.719 | 细胞核 Nucleus |
| FaLIM4 | Fvb1-4:6945308..6951056 | 538 | 60 571.45 | 5.17 | 53.06 | -0.712 | 细胞核 Nucleus |
| FaLIM5 | Fvb2-2:728488..730564 | 191 | 21 205.29 | 9.05 | 29.01 | -0.539 | 细胞核 Nucleus |
| FaLIM6 | Fvb2-2:18290991..18293017 | 231 | 27 087.33 | 7.80 | 45.08 | -0.512 | 细胞核 Nucleus |
| FaLIM7 | Fvb2-3:2848239..2854332 | 491 | 55 136.60 | 5.75 | 44.43 | -0.671 | 细胞核 Nucleus |
| FaLIM8 | Fvb2-4:1747334..1750087 | 176 | 19 899.54 | 8.83 | 44.58 | -0.618 | 细胞核 Nucleus |
| FaLIM9 | Fvb5-1:1118664..1120389 | 213 | 23 551.67 | 7.92 | 45.67 | -0.623 | 细胞核 Nucleus |
| FaLIM10 | Fvb5-2:1037693..1039462 | 216 | 23 835.04 | 8.18 | 44.28 | -0.569 | 细胞核 Nucleus |
| FaLIM11 | Fvb5-3:18813760..18816438 | 188 | 21 172.07 | 8.86 | 31.15 | -0.599 | 细胞核 Nucleus |
| FaLIM12 | Fvb5-4:1027068..1028886 | 211 | 23 297.40 | 7.52 | 43.54 | -0.607 | 细胞核 Nucleus |
| FaLIM13 | Fvb5-4:7252377..7255038 | 188 | 21 199.10 | 8.86 | 29.68 | -0.613 | 细胞核 Nucleus |
| FaLIM14 | Fvb7-1:16915680..16917768 | 217 | 23 964.10 | 6.50 | 45.95 | -0.579 | 细胞核 Nucleus |
| FaLIM15 | Fvb7-1:25526059..25530050 | 506 | 57 362.23 | 8.54 | 52.15 | -0.549 | 细胞核Nucleus |
| FaLIM16 | Fvb7-1:25665993..25670535 | 200 | 21 818.02 | 9.08 | 37.58 | -0.360 | 细胞核 Nucleus |
| FaLIM17 | Fvb7-2:11429297..11431530 | 217 | 23 962.13 | 6.50 | 45.65 | -0.567 | 细胞核 Nucleus |
| FaLIM18 | Fvb7-2:17869656..17871925 | 217 | 23 949.13 | 6.50 | 45.65 | -0.554 | 细胞核 Nucleus |
| FaLIM19 | Fvb7-2:23292270..23296225 | 493 | 55 877.59 | 8.37 | 53.86 | -0.540 | 细胞核 Nucleus |
| FaLIM20 | Fvb7-2:23449921..23452679 | 200 | 21 691.82 | 9.08 | 37.63 | -0.376 | 细胞核 Nucleus |
| FaLIM21 | Fvb7-2:23655808..23659719 | 430 | 49 337.54 | 8.63 | 48.07 | -0.525 | 细胞核Nucleus |
| FaLIM22 | Fvb7-2:23849991..23852934 | 200 | 21 721.91 | 9.08 | 37.48 | -0.362 | 细胞核 Nucleus |
| FaLIM23 | Fvb7-3:5169640..5172605 | 200 | 21 704.86 | 9.01 | 38.00 | -0.353 | 细胞核 Nucleus |
| FaLIM24 | Fvb7-3:6773453..6777457 | 506 | 57 325.21 | 8.45 | 52.87 | -0.541 | 细胞核Nucleus |
| FaLIM25 | Fvb7-3:12069028..12071004 | 215 | 23 793.94 | 6.50 | 44.19 | -0.573 | 细胞核 Nucleus |
| FaLIM26 | Fvb7-4:5056752..5059611 | 200 | 21 721.91 | 9.08 | 37.48 | -0.362 | 细胞核 Nucleus |
| FaLIM27 | Fvb7-4:5292185..5296170 | 506 | 57 277.11 | 8.45 | 51.10 | -0.545 | 细胞核Nucleus |
| FaLIM28 | Fvb7-4:11548941..11551124 | 214 | 23 743.92 | 6.54 | 42.18 | -0.534 | 细胞核 Nucleus |
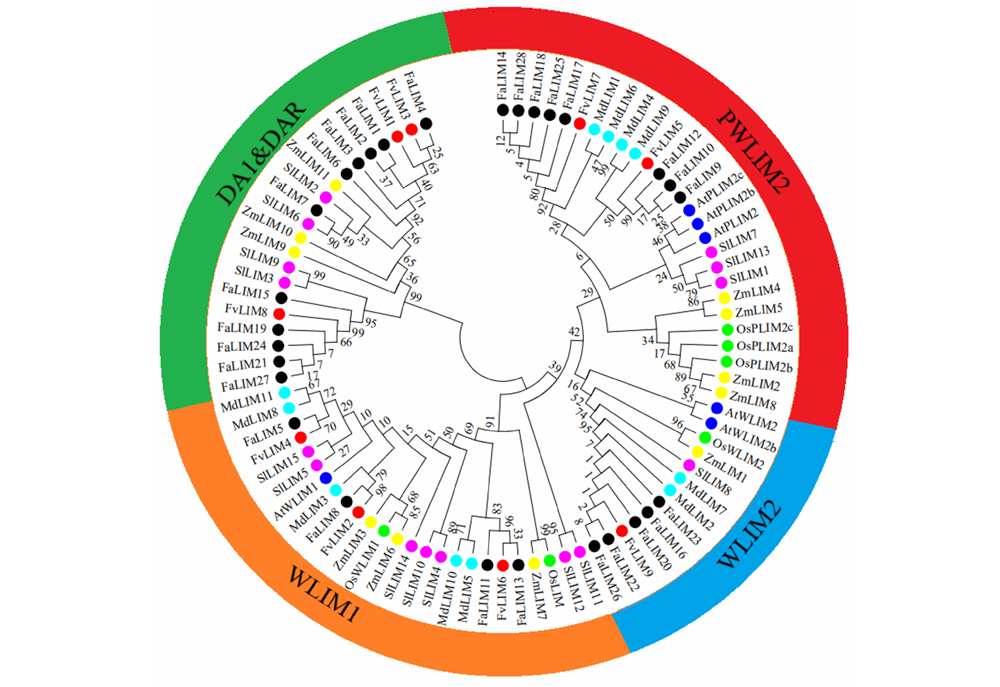
Fig. 1 The phylogenetic tree for strawberry LIM family candidate genes Fa:Fragaria ananassa;Fv:Fragaria vesca;Md:Malus × domestica;At:Arabidopsis thaliana;Sl:Solanum lycopersicum;Zm:Zea mays;Os:Oryza sativa.
| 基序编号 Conserved motif | 氨基酸数 Number of amino acids | 氨基酸保守序列 Amino acid conserved sequence |
|---|---|---|
| motif1 | 50 | GAVYHKSCFKCHHCKGPJKLSNYSSSEGVLYCKPHFEZLFKETGNVCKSF |
| motif2 | 50 | VYPLEKVTVEGESYHKSCFRCTHGGCPJTPSNYAALEGILYCKHHFAQLF |
| motif3 | 50 | AIMDTHECQPLYLEIQEFYEGLNMKVEQQVPMLLVERQALNEAMVGEKNG |
| motif4 | 50 | IDMRTQPQKLTRRCEVTAILVLYGLPRLLTGSILAHEMMHAWLRLKGYPN |
| motif5 | 50 | IPTNSAGLIEYRAHPFWLQKYCPSHERDRTPRCCSCERMEPKDTKYJLLD |
| motif6 | 21 | FPGTRQKCKACEKTVYLVDKL |
| motif7 | 50 | EKKLGEFFKHQIESDSSPAYGEGFRAGNQAVLKYGLRRTLDHIRITGSFP |
| motif8 | 21 | PSKLSSMFSGTQDKCATCKKT |
Table 3 Conserved motifs for strawberry LIM family candidate genes
| 基序编号 Conserved motif | 氨基酸数 Number of amino acids | 氨基酸保守序列 Amino acid conserved sequence |
|---|---|---|
| motif1 | 50 | GAVYHKSCFKCHHCKGPJKLSNYSSSEGVLYCKPHFEZLFKETGNVCKSF |
| motif2 | 50 | VYPLEKVTVEGESYHKSCFRCTHGGCPJTPSNYAALEGILYCKHHFAQLF |
| motif3 | 50 | AIMDTHECQPLYLEIQEFYEGLNMKVEQQVPMLLVERQALNEAMVGEKNG |
| motif4 | 50 | IDMRTQPQKLTRRCEVTAILVLYGLPRLLTGSILAHEMMHAWLRLKGYPN |
| motif5 | 50 | IPTNSAGLIEYRAHPFWLQKYCPSHERDRTPRCCSCERMEPKDTKYJLLD |
| motif6 | 21 | FPGTRQKCKACEKTVYLVDKL |
| motif7 | 50 | EKKLGEFFKHQIESDSSPAYGEGFRAGNQAVLKYGLRRTLDHIRITGSFP |
| motif8 | 21 | PSKLSSMFSGTQDKCATCKKT |
| 基因Gene | T3s | C3s | A3s | G3s | CAI | Fop | ENC | GC3s | GC |
|---|---|---|---|---|---|---|---|---|---|
| FvLIM1 | 0.48 | 0.18 | 0.41 | 0.23 | 0.211 | 0.37 | 51.41 | 0.31 | 0.43 |
| FvLIM3 | 0.48 | 0.18 | 0.41 | 0.23 | 0.210 | 0.37 | 51.69 | 0.31 | 0.43 |
| FvLIM8 | 0.48 | 0.20 | 0.37 | 0.22 | 0.192 | 0.36 | 51.25 | 0.32 | 0.44 |
| FaLIM4 | 0.48 | 0.18 | 0.41 | 0.23 | 0.212 | 0.37 | 51.64 | 0.31 | 0.43 |
| FaLIM2 | 0.48 | 0.18 | 0.41 | 0.24 | 0.214 | 0.37 | 51.73 | 0.32 | 0.43 |
| FaLIM3 | 0.47 | 0.19 | 0.41 | 0.23 | 0.212 | 0.37 | 51.89 | 0.31 | 0.44 |
| FaLIM1 | 0.48 | 0.18 | 0.41 | 0.23 | 0.209 | 0.36 | 51.19 | 0.31 | 0.44 |
| FaLIM6 | 0.49 | 0.20 | 0.42 | 0.24 | 0.201 | 0.31 | 56.42 | 0.32 | 0.42 |
| FaLIM7 | 0.43 | 0.22 | 0.40 | 0.25 | 0.169 | 0.34 | 54.45 | 0.35 | 0.44 |
| FaLIM15 | 0.48 | 0.20 | 0.37 | 0.22 | 0.195 | 0.37 | 51.76 | 0.32 | 0.44 |
| FaLIM19 | 0.48 | 0.20 | 0.37 | 0.22 | 0.191 | 0.36 | 50.99 | 0.32 | 0.44 |
| FaLIM27 | 0.48 | 0.20 | 0.37 | 0.22 | 0.193 | 0.36 | 51.75 | 0.32 | 0.44 |
| FaLIM24 | 0.47 | 0.21 | 0.37 | 0.23 | 0.192 | 0.37 | 52.14 | 0.33 | 0.45 |
| FvLIM21 | 0.48 | 0.20 | 0.36 | 0.24 | 0.181 | 0.35 | 50.30 | 0.33 | 0.44 |
| FvLIM7 | 0.38 | 0.34 | 0.38 | 0.24 | 0.266 | 0.42 | 43.11 | 0.43 | 0.46 |
| FaLIM17 | 0.38 | 0.34 | 0.38 | 0.23 | 0.268 | 0.43 | 43.14 | 0.43 | 0.46 |
| FaLIM18 | 0.38 | 0.34 | 0.37 | 0.24 | 0.270 | 0.43 | 42.82 | 0.43 | 0.46 |
| FaLIM14 | 0.42 | 0.33 | 0.34 | 0.25 | 0.262 | 0.42 | 43.15 | 0.43 | 0.46 |
| FaLIM28 | 0.41 | 0.33 | 0.34 | 0.25 | 0.260 | 0.43 | 44.11 | 0.44 | 0.46 |
| FaLIM25 | 0.41 | 0.32 | 0.37 | 0.24 | 0.273 | 0.44 | 41.34 | 0.42 | 0.46 |
| FvLIM5 | 0.33 | 0.38 | 0.35 | 0.28 | 0.271 | 0.45 | 52.09 | 0.49 | 0.48 |
| 基因Gene | T3s | C3s | A3s | G3s | CAI | Fop | ENC | GC3s | GC |
| FaLIM10 | 0.32 | 0.39 | 0.34 | 0.28 | 0.273 | 0.46 | 60.52 | 0.50 | 0.48 |
| FaLIM12 | 0.32 | 0.39 | 0.34 | 0.28 | 0.272 | 0.45 | 55.80 | 0.50 | 0.48 |
| FaLIM9 | 0.32 | 0.38 | 0.35 | 0.29 | 0.265 | 0.44 | 51.91 | 0.50 | 0.47 |
| FaLIM22 | 0.39 | 0.32 | 0.29 | 0.32 | 0.334 | 0.45 | 56.12 | 0.48 | 0.47 |
| FaLIM23 | 0.39 | 0.31 | 0.28 | 0.33 | 0.348 | 0.47 | 52.97 | 0.48 | 0.47 |
| FaLIM20 | 0.39 | 0.31 | 0.29 | 0.32 | 0.333 | 0.45 | 55.85 | 0.48 | 0.47 |
| FvLIM9 | 0.39 | 0.32 | 0.29 | 0.32 | 0.332 | 0.44 | 55.51 | 0.48 | 0.47 |
| FaLIM26 | 0.39 | 0.32 | 0.29 | 0.32 | 0.332 | 0.44 | 55.51 | 0.48 | 0.47 |
| FaLIM16 | 0.39 | 0.31 | 0.29 | 0.32 | 0.351 | 0.46 | 52.26 | 0.47 | 0.47 |
| FvLIM2 | 0.29 | 0.42 | 0.34 | 0.30 | 0.266 | 0.46 | 58.30 | 0.53 | 0.47 |
| FaLIM8 | 0.27 | 0.44 | 0.34 | 0.31 | 0.242 | 0.44 | 59.77 | 0.55 | 0.48 |
| FvLIM4 | 0.29 | 0.41 | 0.38 | 0.24 | 0.254 | 0.48 | 51.73 | 0.50 | 0.47 |
| FaLIM5 | 0.29 | 0.42 | 0.38 | 0.23 | 0.259 | 0.48 | 51.62 | 0.50 | 0.47 |
| FvLIM6 | 0.40 | 0.35 | 0.28 | 0.33 | 0.318 | 0.48 | 59.48 | 0.50 | 0.45 |
| FaLIM13 | 0.41 | 0.34 | 0.29 | 0.32 | 0.316 | 0.47 | 56.22 | 0.48 | 0.45 |
| FaLIM11 | 0.40 | 0.36 | 0.28 | 0.31 | 0.320 | 0.48 | 55.92 | 0.49 | 0.45 |
| 平均 Average | 0.41 | 0.29 | 0.35 | 0.26 | 0.256 | 0.42 | 52.10 | 0.42 | 0.45 |
Table 4 Main parameters for strawberry LIM family candidate genes
| 基因Gene | T3s | C3s | A3s | G3s | CAI | Fop | ENC | GC3s | GC |
|---|---|---|---|---|---|---|---|---|---|
| FvLIM1 | 0.48 | 0.18 | 0.41 | 0.23 | 0.211 | 0.37 | 51.41 | 0.31 | 0.43 |
| FvLIM3 | 0.48 | 0.18 | 0.41 | 0.23 | 0.210 | 0.37 | 51.69 | 0.31 | 0.43 |
| FvLIM8 | 0.48 | 0.20 | 0.37 | 0.22 | 0.192 | 0.36 | 51.25 | 0.32 | 0.44 |
| FaLIM4 | 0.48 | 0.18 | 0.41 | 0.23 | 0.212 | 0.37 | 51.64 | 0.31 | 0.43 |
| FaLIM2 | 0.48 | 0.18 | 0.41 | 0.24 | 0.214 | 0.37 | 51.73 | 0.32 | 0.43 |
| FaLIM3 | 0.47 | 0.19 | 0.41 | 0.23 | 0.212 | 0.37 | 51.89 | 0.31 | 0.44 |
| FaLIM1 | 0.48 | 0.18 | 0.41 | 0.23 | 0.209 | 0.36 | 51.19 | 0.31 | 0.44 |
| FaLIM6 | 0.49 | 0.20 | 0.42 | 0.24 | 0.201 | 0.31 | 56.42 | 0.32 | 0.42 |
| FaLIM7 | 0.43 | 0.22 | 0.40 | 0.25 | 0.169 | 0.34 | 54.45 | 0.35 | 0.44 |
| FaLIM15 | 0.48 | 0.20 | 0.37 | 0.22 | 0.195 | 0.37 | 51.76 | 0.32 | 0.44 |
| FaLIM19 | 0.48 | 0.20 | 0.37 | 0.22 | 0.191 | 0.36 | 50.99 | 0.32 | 0.44 |
| FaLIM27 | 0.48 | 0.20 | 0.37 | 0.22 | 0.193 | 0.36 | 51.75 | 0.32 | 0.44 |
| FaLIM24 | 0.47 | 0.21 | 0.37 | 0.23 | 0.192 | 0.37 | 52.14 | 0.33 | 0.45 |
| FvLIM21 | 0.48 | 0.20 | 0.36 | 0.24 | 0.181 | 0.35 | 50.30 | 0.33 | 0.44 |
| FvLIM7 | 0.38 | 0.34 | 0.38 | 0.24 | 0.266 | 0.42 | 43.11 | 0.43 | 0.46 |
| FaLIM17 | 0.38 | 0.34 | 0.38 | 0.23 | 0.268 | 0.43 | 43.14 | 0.43 | 0.46 |
| FaLIM18 | 0.38 | 0.34 | 0.37 | 0.24 | 0.270 | 0.43 | 42.82 | 0.43 | 0.46 |
| FaLIM14 | 0.42 | 0.33 | 0.34 | 0.25 | 0.262 | 0.42 | 43.15 | 0.43 | 0.46 |
| FaLIM28 | 0.41 | 0.33 | 0.34 | 0.25 | 0.260 | 0.43 | 44.11 | 0.44 | 0.46 |
| FaLIM25 | 0.41 | 0.32 | 0.37 | 0.24 | 0.273 | 0.44 | 41.34 | 0.42 | 0.46 |
| FvLIM5 | 0.33 | 0.38 | 0.35 | 0.28 | 0.271 | 0.45 | 52.09 | 0.49 | 0.48 |
| 基因Gene | T3s | C3s | A3s | G3s | CAI | Fop | ENC | GC3s | GC |
| FaLIM10 | 0.32 | 0.39 | 0.34 | 0.28 | 0.273 | 0.46 | 60.52 | 0.50 | 0.48 |
| FaLIM12 | 0.32 | 0.39 | 0.34 | 0.28 | 0.272 | 0.45 | 55.80 | 0.50 | 0.48 |
| FaLIM9 | 0.32 | 0.38 | 0.35 | 0.29 | 0.265 | 0.44 | 51.91 | 0.50 | 0.47 |
| FaLIM22 | 0.39 | 0.32 | 0.29 | 0.32 | 0.334 | 0.45 | 56.12 | 0.48 | 0.47 |
| FaLIM23 | 0.39 | 0.31 | 0.28 | 0.33 | 0.348 | 0.47 | 52.97 | 0.48 | 0.47 |
| FaLIM20 | 0.39 | 0.31 | 0.29 | 0.32 | 0.333 | 0.45 | 55.85 | 0.48 | 0.47 |
| FvLIM9 | 0.39 | 0.32 | 0.29 | 0.32 | 0.332 | 0.44 | 55.51 | 0.48 | 0.47 |
| FaLIM26 | 0.39 | 0.32 | 0.29 | 0.32 | 0.332 | 0.44 | 55.51 | 0.48 | 0.47 |
| FaLIM16 | 0.39 | 0.31 | 0.29 | 0.32 | 0.351 | 0.46 | 52.26 | 0.47 | 0.47 |
| FvLIM2 | 0.29 | 0.42 | 0.34 | 0.30 | 0.266 | 0.46 | 58.30 | 0.53 | 0.47 |
| FaLIM8 | 0.27 | 0.44 | 0.34 | 0.31 | 0.242 | 0.44 | 59.77 | 0.55 | 0.48 |
| FvLIM4 | 0.29 | 0.41 | 0.38 | 0.24 | 0.254 | 0.48 | 51.73 | 0.50 | 0.47 |
| FaLIM5 | 0.29 | 0.42 | 0.38 | 0.23 | 0.259 | 0.48 | 51.62 | 0.50 | 0.47 |
| FvLIM6 | 0.40 | 0.35 | 0.28 | 0.33 | 0.318 | 0.48 | 59.48 | 0.50 | 0.45 |
| FaLIM13 | 0.41 | 0.34 | 0.29 | 0.32 | 0.316 | 0.47 | 56.22 | 0.48 | 0.45 |
| FaLIM11 | 0.40 | 0.36 | 0.28 | 0.31 | 0.320 | 0.48 | 55.92 | 0.49 | 0.45 |
| 平均 Average | 0.41 | 0.29 | 0.35 | 0.26 | 0.256 | 0.42 | 52.10 | 0.42 | 0.45 |
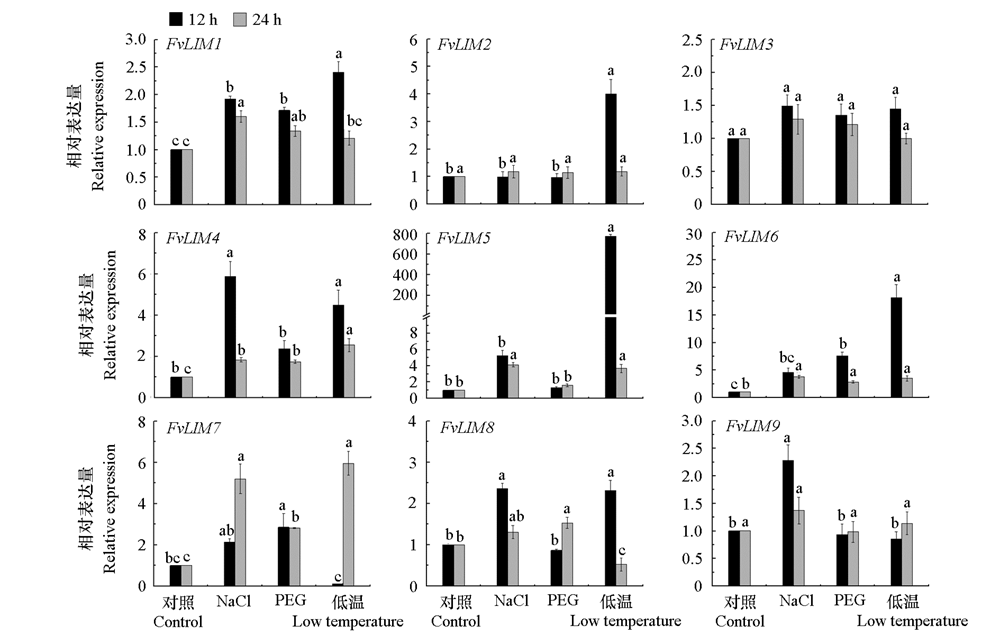
Fig. 9 Quantitative expression of LIM family candidate genes in Fragaria vesca Different lowercase letters indicate that there are significant differences between the treatment and the control.
| [1] |
Agulnick A D, Taira M, Breen J J, Tanaka T, Dawid I B, Westphal H. 1996. Interactions of the LIM-domain-binding factor Ldb 1 with LIM homeodomain proteins. Nature, 384 (6606):270-272.
doi: 10.1038/384270a0 URL |
| [2] | Arnaud D, Dejardin A, Leple J C, Lesage-Descauses M C, Boizot N, Villar M, Benedetti H, Pilate G. 2012. Expression analysis of LIM gene family in poplar,toward an updated phylogenetic classifification. BMC Research, 5 (1):102. |
| [3] |
Arnaud D, Déjardin A, Leplé J C, Lesage-descauses M C, Pilate G. 2007. Genome-wide analysis of LIM gene family in Populus trichocarpa,Arabidopsis thaliana,and Oryza sativa. DNA Research, 14 (3):103-116.
doi: 10.1093/dnares/dsm013 URL |
| [4] | Bailey T L, Boden M, Buske F A, Frith M, Grant C E, Clementi L, Ren J Y, LI W W, Noble W S. 2009. MEME SUITE:tools for motif discovery and searching. Nucleic Acids Research, 37 (Web Server issue):W202. |
| [5] |
Cheng X, Li G H, Muhammad A, Zhang J Y, Jiang T S, Jin Q, Zhao H, Cai Y P, Lin Y. 2019. Molecular identification,phylogenomic characterization and expression patterns analysis of the LIM(LIN-11,Isl1 and MEC-3 domains)gene family in pear(Pyrus bretschneideri)reveal its potential role in lignin metabolism. Gene, 686:237-249.
doi: S0378-1119(18)31207-1 pmid: 30468911 |
| [6] |
Chenna R, Sugawara H, Koike T, Lopez R, Gibson T J, Higgins D G, Thompson J D. 2003. Multiple sequence alignment with the clustal series of programs. Nucleic Acids Research, 31 (13):3497-3500.
pmid: 12824352 |
| [7] |
Dixon R A, Paiva N L. 1995. Stress-induced phenylpropanoid metabolism. The Plant Cell, 7 (7):1085-1097.
pmid: 12242399 |
| [8] |
Feng L, Xia R, Liu Y L. 2019. Comprehensive characterization of miRNA and PHAS loci in the diploid strawberry(Fragaria vesca)genome. Horticultural Plant Journal, 5 (6):255-267.
doi: 10.1016/j.hpj.2019.11.004 |
| [9] | Gao Ya. 2019. Genetic transformation of Gossypium hirsutum and function of LIM gene in offspring of transgenic cotton[M. D. Dissertation]. Wuhan: Central China Normal University. (in Chinese) |
| 高雅. 2019. 棉花(Gossypium hirsutum)遗传转化及LIM基因在转基因棉花后代中的功能研究[硕士论文]. 武汉: 华中师范大学. | |
| [10] | He Hong-hong, Ma Zong-huan, Zhang Yuan-xia, Zhang Juan, Lu Shi-xiong, Zhang Zhi-qiang, Zhao Xin, Wu Yu-xia, Mao Juan. 2018. Identification and expression analysis of LBD gene family in grape. Scientia Agricultura Sinica, 51 (21):4102-4118. (in Chinese) |
| 何红红, 马宗桓, 张元霞, 张娟, 卢世雄, 张志强, 赵鑫, 吴玉霞, 毛娟. 2018. 葡萄LBD基因家族的鉴定与表达分析. 中国农业科学, 51 (21):4102-4118. | |
| [11] |
Jurata L W, Gill G N. 1997. Functional analysis of the nuclear LIM domain interactor NLI. Molecular and Cellular Biology, 17 (10):5688-5698.
doi: 10.1128/MCB.17.10.5688 pmid: 9315627 |
| [12] | Kadrmas J L, Beckerle M C. 2004. The LIM domain:from the cytoskeleton to the nucleus. Nature Reviews. Molecular Cell Biology, 5 (11):920-931. |
| [13] |
Kawaoka A, Ebinuma H. 2001. Transcriptional control of lignin biosynthesis by tobacco LIM protein. Phytochemistry, 57 (7):1149-1157.
pmid: 11430987 |
| [14] |
Khadiza K, Arif H K R, Jong-In P, Nasar U A, Chang K K, Ki-Byung L, Min-Bae K, Do-Jin L, Ill S N, Mi-Young C. 2016. Genome-wide identification,characterization and expression profiling of LIM family genes in Solanum lycopersicum L. Plant Physiology and Biochemistry, 108:177-190.
doi: S0981-9428(16)30273-X pmid: 27439220 |
| [15] | Li Tong. 2017. Transcriptional regulation of sorghum SbLIM1 on lignin synthesis and recognition of interacting proteins[M. D. Dissertation]. Jinan: Shandong University. (in Chinese) |
| 李彤. 2017. 高梁SbLIM1对木质素合成的转录调控及互作蛋白识别研究[硕士论文]. 济南:山东大学. | |
| [16] |
Li Ya, Yue Xiao-feng, Que Ya-wei, Yan Xia, Ma Zhong-hua, Talbot Nicholas J, Wang Zheng-yi. 2014. Characterisation of four LIM protein-encoding genes involved in infection-related development and pathogenicity by the rice blast fungus Magnaporthe oryzae. PLoS One, 9 (2):e88246.
doi: 10.1371/journal.pone.0088246 URL |
| [17] | Lian Peng, Du Meng-qing, Wang Li-juan. 2019. Tissue culture and disinfection of strawberry stem tips and optimization of hormone ratio. Journal of Tianjin Agricultural College, 26 (3):21-25. (in Chinese) |
| 连朋, 杜梦卿, 王丽娟. 2019. 草莓茎尖组培消毒方法及激素配比的优选. 天津农学院学报, 26 (3):21-25. | |
| [18] | Liu Tao, Wang Ping-ping, He Hong-hong, Liang Guo-ping, Lu Shi-xiong, Chen Bai-hong, Mao Juan. 2020. Identification and expression analysis of CIPK gene family in strawberry. Acta Horticulturae Sinica, 47 (1):127-142. (in Chinese) |
| 刘涛, 王萍萍, 何红红, 梁国平, 卢世雄, 陈佰鸿, 毛娟. 2020. 草莓CIPK基因家族的鉴定与表达分析. 园艺学报, 47 (1):127-142. | |
| [19] | Liu Wan-da, Zhang Bing-xiu, Wei Yuan-yuan, Wang Yu, Dong Chao, Zhu Yan-ming. 2014. Analysis of codon preference of strawberry DHAR gene. Northern Horticulture,(14):92-97. (in Chinese) |
| 刘万达, 张丙秀, 魏媛媛, 王禹, 董超, 朱延明. 2014. 草莓DHAR基因密码子偏好性分析. 北方园艺,(14):92-97. | |
| [20] | Liu Zhi. 2011. Functional analysis of Oslim transcription factor involved in non-host resistance of rice[M. D. Dissertation]. Beijing:Chinese Academy of Agricultural Sciences. (in Chinese) |
| 刘峙. 2011. LIM转录因子(Oslim)参与水稻非寄主抗性的功能分析[硕士论文]. 北京:中国农业科学院. | |
| [21] |
Livak K J, Schmittgen T D. 2001. Analysis of relative gene expression data using real-time quantitative and the 2-ΔΔCT method. Methods, 25 (4):402-408.
pmid: 11846609 |
| [22] | Ma Zong-huan, Mao Juan, Li Wen-fang, Yang Shi-mao, Wu Jin-hong, Chen Bai-hong. 2016. Identification and expression analysis of SnRK 2 family genes in grape. Acta Horticulturae Sinica, 43 (10):1891-1903. (in Chinese) |
| 马宗桓, 毛娟, 李文芳, 杨世茂, 吴金红, 陈佰鸿. 2016. 葡萄SnRK2家族基因的鉴定与表达分析. 园艺学报, 43 (10):1891-1903. | |
| [23] | Mao Juan, Lu Shi-xiong, Wang Ping, Ma Zong-huan, Liu Tao, He Hong-hong, Nai Guo-jie, Zhang Zhi-qiang, Chen Bai-hong. 2020. Analysis of gene characteristic and expression pattern of LIM family in grape. Acta Horticulturae Sinica, 47 (7):1277-1288. (in Chinese) |
| 毛娟, 卢世雄, 王萍, 马宗桓, 刘涛, 何红红, 乃国洁, 张志强, 陈佰鸿. 2020. 葡萄LIM家族基因特征及表达模式分析. 园艺学报, 47 (7):1277-1288. | |
| [24] | Michelsen J W, Schmeichel K L, Beckerle M C, Winge D R. 1993. The LIM motif defines a specific zinc-binding protein domain. Proceedings of the National Academy of Sciences of the United States of America, 90 (10):4404-4408. |
| [25] |
Rosane L C, Ellen C P, Isabel L V, Rafael W, Amilton S, Luciano D S P, César V R, Vanessa G. 2020. Genome-wide identification,and characterization of the CDPK gene family reveal their involvement in abiotic stress response in Fragaria ananassa. Scientific Reports, 10 (1):1-17.
doi: 10.1038/s41598-019-56847-4 URL |
| [26] |
Sánchez-García I, Rabbitts T H. 1994. The LIM domain:a new structural motif found in zinc-finger-like proteins. Trends in Genetics, 10 (9):315-320.
pmid: 7974745 |
| [27] | Schmeichel K L, Beckerle M C. 1994. The LIM domain is a modular protein-binding interface. MC Beckerle-Cell, 79 (2):211-219. |
| [28] |
Schmeichel K L, Beckerle M C. 1997. Molecular dissection of a LIM domain. Molecular Biology of the Cell, 8 (2):219-230.
pmid: 9190203 |
| [29] |
Taira M, Jamrich M, Good P J, Dawid I B. 1992. The LIM domain-containing homeo box gene Xlim-1 is expressed specifically in the organizer region of Xenopus gastrula embryos. Genes & Development, 6 (3):356-366.
doi: 10.1101/gad.6.3.356 URL |
| [30] | Wang Yong-qiang, He Xiao-liang, Liu Jian-guang, Du Shu-feng, Zhao Jun-li, Zhao Gui-yuan, Zhang Han-shan. 2014. Cloning and expression analysis of GhLIMa in cotton salt-tolerant gene. Acta Botanica Boreali-Occidentalis, 34 (12):2382-2387. (in Chinese) |
| 王永强, 何晓亮, 刘建光, 杜淑凤, 赵俊丽, 赵贵元, 张寒霜. 2014. 棉花耐盐相关基因GhLIMa的克隆与表达分析. 西北植物学报, 34 (12):2382-2387. | |
| [31] |
Wittkopp P J, Kalay G. 2011. Cis-regulatory elements:molecular mechanisms and evolutionary processes underlying divergence. Nature Reviews Genetics, 13 (1):59-69.
doi: 10.1038/nrg3095 URL |
| [32] | Xu Yang, Yuan Li-ping, Chen Jin-ling, Huang Xiao-hong. 2012. Research progress of actin depolymerizing factor/Cofilin function. Life Sciences, 24 (1):13-18. (in Chinese) |
| 徐阳, 原丽平, 陈金铃, 黄晓红. 2012. 肌动蛋白解聚因子/cofilin功能的研究进展. 生命科学, 24 (1):13-18. | |
| [33] |
Yang R, Chen M, Sun J C, Yu Y, Min D H, Chen J, Xu Z S, Zhou Y B, Ma Y Z, Zhang X H. 2019. Genome-wide analysis of LIM family genes in foxtail millet(Setaria italica L.)and characterization of the role of SiWLIM2b in drought tolerance. International Journal of Molecular Sciences, 20 (6):1303.
doi: 10.3390/ijms20061303 URL |
| [34] | Yang Rui. 2019. Effects of SiWLIM2b gene on drought resistance and low nitrogen stress tolerance in rice[M. D. Dissertation]. Xianyang:Northwest A & F University. (in Chinese) |
| 杨瑞. 2019. 谷子SiWLIM2b基因提高水稻抗旱及耐低氮胁迫的功能研究[硕士论文]. 咸阳: 西北农林科技大学. | |
| [35] | Li Bo, Hu wen-ran, Zhang Jing-bo, Fan Ling. 2015. Evolution and expression analysis of the LIM protein gene family in cotton. Plant Physiology Journal, 12:2133-2142. (in Chinese) |
| 杨洋, 李波, 胡文冉, 张经博, 范玲. 2015. 棉花LIM蛋白基因家族的进化及表达特征分析. 植物生理学报, 12:2133-2142. | |
| [36] |
Ye J R, Zhou L M, Xu M L. 2013. Arabidopsis LIM proteins PLIM2a and PLIM2b regulate actin configuration during pollen tube growth. Biologia Plantarum, 57 (3):433-441.
doi: 10.1007/s10535-013-0323-3 URL |
| [37] | Yi F, Guo J, Dabbagh D, Spear M, He S J, Kehn-Hall K, Fontenot J, Yin Y, Bibian M, Park C M, Zheng K, Park H J, Soloveva V, Gharaibeh D, Retterer C, Zamani R, Naughton J, Jiang Y J, Shang H, Hakami R M, Ling B H, Young J A T, Bavari S, Xu X H, Feng Y B, Wu Y T. 2017. Discovery of novel small-molecule inhibitors of LIM domain kinase for inhibiting HIV-1. Journal of Virology, 91 (13):e02418-16. |
| [38] | Yuan Gao-peng, Han Xiao-lei, Bian Shu-xun, Zhang Li-yi, Tian Yi, Zhang Cai-xia, Cong Peihua. 2019. Bioinformatics and expression analysis of apple LIM gene family. Scientia Agricultura Sinica, 52 (23):4322-4332. (in Chinese) |
| 袁高鹏, 韩晓蕾, 卞书迅, 张利义, 田义, 张彩霞, 丛佩华. 2019. 苹果LIM基因家族生物信息学及表达分析. 中国农业科学, 52 (23):4322-4332. | |
| [39] | Zhang Hai-yan, Li Zuo-tong, Zhao Chang-jiang, Yang Ke-jun, Wang Yu-feng, Hu Xue-wei, Zhao Ying. 2013. Analysis of LIM domain protein gene family in maize. Maize Science, 21 (3):40-47. (in Chinese) |
| 张海燕, 李佐同, 赵长江, 杨克军, 王玉凤, 胡雪微, 赵莹. 2013. 玉米LIM结构域蛋白基因家族分析. 玉米科学, 21 (3):40-47. | |
| [40] |
Zhang X, Zhen J B, Li Z H, Kang D M, Yang Y M, Kong J, Hua J P. 2011. Expression profile of early responsive genes under salt stress in upland cotton ( Gossypium hirsutum L.). Plant Molecular Biology Reporter, 29 (3):626-637.
doi: 10.1007/s11105-010-0269-y URL |
| [41] | Zhang Yi-han, LI Qing, Zhang Lei, Tan He-xin, Chen Xiang-hui. 2019. Bioinformatics analysis of the MADs-Box gene family in Crocus sativus. Genomics and Applied Biology, 38 (7):3140-3154. (in Chinese) |
| 张一菡, 李卿, 张磊, 谭何新, 陈祥慧. 2019. 西红花MADS-box基因家族的生物信息学分析. 基因组学与应用生物学, 38 (7):3140-3154. | |
| [42] |
Zhang Z B, Zhang J W, Chen Y J, Li R F, Wang H Z, Wei J H. 2012. Genome-wide analysis and identification of HAK potassium transporter gene family in maize(Zea mays L.). Molecular Biology Reports, 39 (8):8465-8473.
doi: 10.1007/s11033-012-1700-2 URL |
| [43] | Zhou Jian-ping. 2006. Phenotypic changes in cotton induced by excessive expression of LIM domain gene[M. D. Dissertation]. Chongqing:Southwest University. (in Chinese) |
| 周建平. 2006. 超量表达LIM结构域基因引起棉花的表型变化[硕士论文]. 重庆:西南大学. |
| [1] | YU Tingting, LI Huan, NING Yuansheng, SONG Jianfei, PENG Lulin, JIA Junqi, ZHANG Weiwei, and YANG Hongqiang. Genome-wide Identification of GRAS Gene Family in Apple and Expression Analysis of Its Response to Auxin [J]. Acta Horticulturae Sinica, 2023, 50(2): 397-409. |
| [2] | HE Chengyong, ZHAO Xiaoli, XU Tengfei, GAO Dehang, LI Shifang, WANG Hongqing. Genome Sequence Analysis of Strawberry Virus 1 from Shandong Province,China [J]. Acta Horticulturae Sinica, 2023, 50(1): 153-160. |
| [3] | YANG Lei, LI Li, DONG Hui, FENG Jia, ZHANG Jianjun, FAN Jingfang, YANG Qiuye, and YANG Li, . A New Strawberry Cultivar‘Shimei 11’ [J]. Acta Horticulturae Sinica, 2022, 49(S2): 79-80. |
| [4] | ZHAO Xia, LI Gang, LIU Lifeng, SONG Yanhong, and ZHOU Houcheng. A New Strawberry Cultivar‘Huashuo 1’ [J]. Acta Horticulturae Sinica, 2022, 49(S2): 81-82. |
| [5] | DONG Jing, CHANG Linlin, WANG Guixia, ZHONG Chuanfei, WEI Yongqing, SUN Jian, SUN Rui, ZHANG Hongli, GAO Yongshun, XU Liping, TAO Pang, LUO Zhiwei, and ZHANG Yuntao, . A New Ever-bearing Strawberry Cultivar‘Jinghong’ [J]. Acta Horticulturae Sinica, 2022, 49(S1): 61-62. |
| [6] | GAO Yanlong, WU Yuxia, ZHANG Zhongxing, WANG Shuangcheng, ZHANG Rui, ZHANG De, WANG Yanxiu. Bioinformatics Analysis of Apple ELO Gene Family and Its Expression Analysis Under Low Temperature Stress [J]. Acta Horticulturae Sinica, 2022, 49(8): 1621-1636. |
| [7] | QIAN Jieyu, JIANG Lingli, ZHENG Gang, CHEN Jiahong, LAI Wuhao, XU Menghan, FU Jianxin, ZHANG Chao. Identification and Expression Analysis of MYB Transcription Factors Regulating the Anthocyanin Biosynthesis in Zinnia elegans and Function Research of ZeMYB9 [J]. Acta Horticulturae Sinica, 2022, 49(7): 1505-1518. |
| [8] | ZHANG Qiuyue, LIU Changlai, YU Xiaojing, YANG Jiading, FENG Chaonian. Screening of Reference Genes for Differentially Expressed Genes in Pyrus betulaefolia Plant Under Salt Stress by qRT-PCR [J]. Acta Horticulturae Sinica, 2022, 49(7): 1557-1570. |
| [9] | WANG Hong, YANG Wangli, LIN Jing, YANG Qingsong, LI Xiaogang, SHENG Baolong, CHANG Youhong. Comparative Metabolic and Transcriptomic Analysis of Ripening Fruit in Pear Cultivars of‘Sucui 1’‘Cuiguan’and‘Huasu’ [J]. Acta Horticulturae Sinica, 2022, 49(3): 493-508. |
| [10] | CHEN Jianxin, WEI Yuqian, TANG Jie, ZHU Yongjin, MA Huancheng, WU Jianrong. Molecular Detection and Sequence Analysis of ORSV and CymMV in Cultivated Cymbidium goeringii in Dali,Yunnan [J]. Acta Horticulturae Sinica, 2022, 49(3): 655-662. |
| [11] | JIANG Cuicui, FANG Zhizhen, ZHOU Danrong, LIN Yanjuan, YE Xinfu. Identification and Expression Analysis of Sugar Transporter Family Genes in‘Furongli’(Prunus salicina) [J]. Acta Horticulturae Sinica, 2022, 49(2): 252-264. |
| [12] | WANG Guangpeng, LIU Tongkun, XU Xinfeng, LI Zhubo, GAO Zhanyuan, HOU Xilin. Identification of LEA Family and Expression Analysis of Some Members Under Low-temperature Stress in Chinese Cabbage [J]. Acta Horticulturae Sinica, 2022, 49(2): 304-318. |
| [13] | YANG Liyuan, WANG Qian, WANG Xuhui, XU Tongda, MA Jun. The Important Biological Function of T111,an Evolutional Conserved Amino Acid in FveTAA1,a Key Enzyme in Strawberry Auxin Biosynthesis [J]. Acta Horticulturae Sinica, 2021, 48(9): 1695-1705. |
| [14] | HONG Yanhong, YE Qinghua, LI Zekun, WANG Wei, XIE Qian, CHEN Qingxi, CHEN Jianqing. Accumulation of Anthocyanins in Red-flowered Strawberry‘Meihong’Petals and Expression Analysis of MYB Gene [J]. Acta Horticulturae Sinica, 2021, 48(8): 1470-1484. |
| [15] | HUANG Qianru, NIU Yonghao, WU Kuan, LIU Zhanjie, CAO Mengji, ZHAO Lei, WU Yunfeng. LncRNA Sequencing Identification of Viruses Infecting Strawberry in Shaanxi [J]. Acta Horticulturae Sinica, 2021, 48(8): 1589-1594. |
| Viewed | ||||||
|
Full text |
|
|||||
|
Abstract |
|
|||||
Copyright © 2012 Acta Horticulturae Sinica 京ICP备10030308号-2 国际联网备案号 11010802023439
Tel: 010-82109523 E-Mail: yuanyixuebao@126.com
Support by: Beijing Magtech Co.Ltd