
Acta Horticulturae Sinica ›› 2022, Vol. 49 ›› Issue (2): 304-318.doi: 10.16420/j.issn.0513-353x.2021-0044
• Research Papers • Previous Articles Next Articles
WANG Guangpeng, LIU Tongkun, XU Xinfeng, LI Zhubo, GAO Zhanyuan, HOU Xilin( )
)
Received:2021-04-06
Revised:2021-10-14
Online:2022-02-25
Published:2022-02-28
Contact:
HOU Xilin
E-mail:hxl@njau.edu.cn
CLC Number:
WANG Guangpeng, LIU Tongkun, XU Xinfeng, LI Zhubo, GAO Zhanyuan, HOU Xilin. Identification of LEA Family and Expression Analysis of Some Members Under Low-temperature Stress in Chinese Cabbage[J]. Acta Horticulturae Sinica, 2022, 49(2): 304-318.
Add to citation manager EndNote|Ris|BibTeX
URL: https://www.ahs.ac.cn/EN/10.16420/j.issn.0513-353x.2021-0044
| 基因 ID Gene ID | 基因名称 Gene name | 引物序列(5′-3′) Primer sequence |
|---|---|---|
| Bra008242 | BrDehydrin2 | F:GGAATCATCAGCGGAGGTCA;R:CCTCGTGTTTAACCTCAGGCA |
| Bra025819 | BrDehydrin4 | F:TTCCGGTGAAGGAGGAAACG;R:CCGTGTCGACCACTTGAGAA |
| Bra012230 | BrDehydrin5 | F:TTCACCGATCCAACAGCTCC;R:CCTCCGTGTTGACGACTTGA |
| Bra015779 | BrDehydrin6 | F:AGGAATCATCAACGGCCACC;R:CACCGGCTCTGAGATGTGAA |
| Bra003732 | BrDehydrin7 | F:AGGACACGGAAAGAAACCCG;R:CCTCAACGGTCTTTGAGTGG |
| Bra013992 | BrDehydrin8 | F:TGCAGCTTCCAAAGGTTGTG;R:CGTCGCGTCAGTCACATAGA |
| Bra035491 | BrLEA_1-3 | F:GGGCACGGACTAAAGAGGAG;R:TCACAGGTGTGTGATGAGGC |
| Bra000414 | BrLEA_2-2 | F:AAGGACGATAGCGTCGGGTA;R:CCGCTATGCTATAGGCCACC |
| Bra026541 | BrLEA_4-3 | F:GAATGCGACCATTACACCGC;R:CCACTTCACTCCCCATGTCC |
| Bra000263 | BrLEA_4-6 | F:GGCCTTGAAAAATGGCGAGA;R:TTTGTGGCGTCCTTAGCCTC |
| Bra000265 | BrLEA_4-7 | F:CTCAGGAGCTGTTCTCAGTGG;R:ATCAACGACTTCTTGCGCTG |
| Bra000173 | BrLEA_5-1 | F:GCCGAGGAGGGAATACAAGG;R:TGGGTTCGTCCATCTCCTCT |
| Bra022950 | BrLEA_6-1 | F:GCCACTAGAAACGAGTCCGTA;R:GGCCATGCTTAACCTGTTGG |
| Actin | F:AGGCTACACGTTCGGACAAG;R:TGGGGCACTAAACACAGTCA |
Table 1 Primers used in this study
| 基因 ID Gene ID | 基因名称 Gene name | 引物序列(5′-3′) Primer sequence |
|---|---|---|
| Bra008242 | BrDehydrin2 | F:GGAATCATCAGCGGAGGTCA;R:CCTCGTGTTTAACCTCAGGCA |
| Bra025819 | BrDehydrin4 | F:TTCCGGTGAAGGAGGAAACG;R:CCGTGTCGACCACTTGAGAA |
| Bra012230 | BrDehydrin5 | F:TTCACCGATCCAACAGCTCC;R:CCTCCGTGTTGACGACTTGA |
| Bra015779 | BrDehydrin6 | F:AGGAATCATCAACGGCCACC;R:CACCGGCTCTGAGATGTGAA |
| Bra003732 | BrDehydrin7 | F:AGGACACGGAAAGAAACCCG;R:CCTCAACGGTCTTTGAGTGG |
| Bra013992 | BrDehydrin8 | F:TGCAGCTTCCAAAGGTTGTG;R:CGTCGCGTCAGTCACATAGA |
| Bra035491 | BrLEA_1-3 | F:GGGCACGGACTAAAGAGGAG;R:TCACAGGTGTGTGATGAGGC |
| Bra000414 | BrLEA_2-2 | F:AAGGACGATAGCGTCGGGTA;R:CCGCTATGCTATAGGCCACC |
| Bra026541 | BrLEA_4-3 | F:GAATGCGACCATTACACCGC;R:CCACTTCACTCCCCATGTCC |
| Bra000263 | BrLEA_4-6 | F:GGCCTTGAAAAATGGCGAGA;R:TTTGTGGCGTCCTTAGCCTC |
| Bra000265 | BrLEA_4-7 | F:CTCAGGAGCTGTTCTCAGTGG;R:ATCAACGACTTCTTGCGCTG |
| Bra000173 | BrLEA_5-1 | F:GCCGAGGAGGGAATACAAGG;R:TGGGTTCGTCCATCTCCTCT |
| Bra022950 | BrLEA_6-1 | F:GCCACTAGAAACGAGTCCGTA;R:GGCCATGCTTAACCTGTTGG |
| Actin | F:AGGCTACACGTTCGGACAAG;R:TGGGGCACTAAACACAGTCA |
| 基因号 Gene number | 基因名称 Gene name | 氨基酸长度 Amino acid length | 等电点 pI | 分子量/D MW | 不稳定性 指数 Instability index | 脂肪族指数 Aliphatic index | 亲水性 GRAVY | 亚细胞定位 Subcellular localization |
|---|---|---|---|---|---|---|---|---|
| Bra011874 | BrDehydrin-1 | 149 | 5.65 | 15 928.40 | 23.71 | 58.93 | -0.863 | 细胞核 Nucleus |
| Bra036843 | BrDehydrin-10 | 134 | 9.19 | 13 834.22 | 42.98 | 49.55 | -0.887 | 细胞核 Nucleus |
| Bra037177 | BrDehydrin-11 | 144 | 8.81 | 15 139.46 | 45.61 | 33.26 | -1.360 | 细胞核 Nucleus |
| Bra008242 | BrDehydrin-2 | 194 | 5.52 | 21 840.38 | 62.43 | 53.71 | -1.352 | 细胞核 Nucleus |
| Bra031809 | BrDehydrin-3 | 192 | 7.14 | 18 844.17 | 33.09 | 32.14 | -1.021 | 细胞核 Nucleus |
| Bra025819 | BrDehydrin-4 | 271 | 5.00 | 31 018.16 | 61.44 | 47.49 | -1.508 | 细胞核 Nucleus |
| Bra012230 | BrDehydrin-5 | 220 | 5.11 | 24 837.39 | 62.24 | 55.41 | -1.399 | 细胞核 Nucleus |
| Bra015779 | BrDehydrin-6 | 195 | 5.45 | 22 044.52 | 63.15 | 50.41 | -1.455 | 细胞核 Nucleus |
| Bra003732 | BrDehydrin-7 | 194 | 5.47 | 21 755.35 | 56.38 | 53.25 | -1.308 | 细胞核 Nucleus |
| Bra013992 | BrDehydrin-8 | 257 | 6.17 | 28 978.00 | 34.53 | 35.33 | -1.645 | 细胞核 Nucleus |
| Bra031192 | BrDehydrin-9 | 183 | 6.49 | 19 183.88 | 36.64 | 49.18 | -1.016 | 细胞核 Nucleus |
| Bra005911 | BrLEA_1-1 | 159 | 9.22 | 16 286.99 | 17.53 | 43.71 | -0.789 | 细胞核 Nucleus |
| Bra005353 | BrLEA_1-2 | 98 | 9.22 | 10 666.89 | 42.18 | 49.18 | -1.115 | 细胞核 Nucleus |
| Bra035491 | BrLEA_1-3 | 132 | 9.68 | 14 933.95 | 56.61 | 51.89 | -1.109 | 线粒体 Mitochondrion |
| Bra023278 | BrLEA_1-4 | 133 | 9.24 | 14 507.53 | 59.47 | 48.65 | -0.878 | 线粒体 Mitochondrion |
| Bra009225 | BrLEA_1-5 | 159 | 8.93 | 16 180.83 | 16.45 | 38.81 | -0.818 | 细胞核 Nucleus |
| Bra000339 | BrLEA_2-1 | 777 | 9.02 | 84 314.53 | 32.33 | 96.22 | -0.135 | 叶绿体 Chloroplast |
| Bra000414 | BrLEA_2-2 | 166 | 4.81 | 17 823.38 | 17.04 | 96.81 | 0.030 | 胞外Extracellular |
| Bra039288 | BrLEA_2-3 | 166 | 4.50 | 18 119.88 | 16.96 | 100.36 | 0.013 | 叶绿体 Chloroplast |
| Bra037669 | BrLEA_2-4 | 506 | 8.85 | 55 465.06 | 27.06 | 85.89 | -0.399 | 细胞核 Nucleus |
| Bra004565 | BrLEA_2-5 | 166 | 4.81 | 17 725.34 | 20.39 | 106.20 | 0.093 | 细胞膜 Cell membrane |
| Bra030494 | BrLEA_2-6 | 151 | 4.72 | 16 404.92 | 10.57 | 106.42 | 0.075 | 细胞膜 Cell membrane |
| Bra033520 | BrLEA_3-1 | 99 | 9.72 | 10 582.01 | 50.01 | 70.10 | -0.400 | 叶绿体 Chloroplast |
| Bra000884 | BrLEA_3-2 | 94 | 9.85 | 10 095.53 | 39.42 | 78.94 | -0.349 | 叶绿体 Chloroplast |
| Bra014864 | BrLEA_3-3 | 122 | 9.20 | 14 152.16 | 50.98 | 63.20 | -0.598 | 叶绿体 Chloroplast |
| Bra018556 | BrLEA_3-4 | 93 | 9.99 | 10 112.56 | 52.38 | 72.47 | -0.482 | 叶绿体 Chloroplast |
| Bra038061 | BrLEA_3-5 | 57 | 4.94 | 6 614.40 | 49.22 | 56.67 | -0.912 | 线粒体 Mitochondrion |
| Bra036272 | BrLEA_3-6 | 94 | 9.99 | 10 088.55 | 43.90 | 72.77 | -0.299 | 叶绿体 Chloroplast |
| Bra033350 | BrLEA_3-7 | 98 | 8.03 | 10 298.59 | 40.91 | 76.84 | -0.229 | 叶绿体 Chloroplast |
| Bra021126 | BrLEA_4-1 | 226 | 9.02 | 24 598.55 | 28.79 | 22.79 | -1.475 | 线粒体 Mitochondrion |
| Bra022221 | BrLEA_4-10 | 298 | 5.45 | 32 431.59 | 30.10 | 54.50 | -1.126 | 胞外Extracellular |
| Bra027219 | BrLEA_4-11 | 229 | 8.83 | 24 810.86 | 27.04 | 22.97 | -1.414 | 线粒体 Mitochondrion |
| Bra005256 | BrLEA_4-12 | 399 | 5.74 | 43 565.34 | 20.90 | 52.93 | -0.970 | 细胞膜 Cell membrane |
| Bra019628 | BrLEA_4-13 | 451 | 5.20 | 48 554.47 | 12.26 | 47.98 | -1.141 | 细胞核 Nucleus |
| Bra025130 | BrLEA_4-14 | 201 | 5.40 | 22 054.03 | 38.33 | 77.21 | -0.429 | 叶绿体 Chloroplast |
| Bra039616 | BrLEA_4-15 | 480 | 6.47 | 52 652.67 | 18.82 | 47.04 | -0.921 | 叶绿体 Chloroplast |
| Bra010561 | BrLEA_4-16 | 487 | 5.63 | 52 431.94 | 23.72 | 46.65 | -0.923 | 叶绿体 Chloroplast |
| Bra020879 | BrLEA_4-17 | 218 | 5.12 | 24 376.73 | 15.32 | 38.72 | -1.441 | 细胞核 Nucleus |
| Bra027542 | BrLEA_4-18 | 311 | 7.60 | 35 005.70 | 23.66 | 43.79 | -1.371 | 叶绿体 Chloroplast |
| Bra037225 | BrLEA_4-19 | 722 | 8.58 | 80 928.05 | 43.18 | 68.66 | -0.722 | 叶绿体 Chloroplast |
| Bra013489 | BrLEA_4-2 | 253 | 8.55 | 27 837.93 | 20.67 | 47.31 | -1.202 | 叶绿体 Chloroplast |
| Bra036021 | BrLEA_4-20 | 253 | 5.72 | 28 430.30 | 32.23 | 43.72 | -1.355 | 叶绿体 Chloroplast |
| Bra041061 | BrLEA_4-21 | 241 | 6.17 | 26 330.13 | 20.46 | 50.83 | -1.111 | 叶绿体 Chloroplast |
| Bra026541 | BrLEA_4-3 | 192 | 8.57 | 20 399.00 | 33.96 | 62.55 | -0.669 | 叶绿体 Chloroplast |
| Bra008006 | BrLEA_4-4 | 462 | 9.03 | 50 701.68 | 32.68 | 78.53 | -0.390 | 胞外Extracellular |
| Bra001666 | BrLEA_4-5 | 276 | 5.54 | 30 311.37 | 25.62 | 53.22 | -1.093 | 叶绿体 Chloroplast |
| Bra000263 | BrLEA_4-6 | 142 | 8.65 | 14 760.62 | 19.57 | 77.75 | -0.430 | 叶绿体 Chloroplast |
| Bra000265 | BrLEA_4-7 | 129 | 5.31 | 13 894.51 | 22.81 | 71.86 | -0.692 | 叶绿体 Chloroplast |
| Bra017229 | BrLEA_4-8 | 398 | 6.02 | 43 386.25 | 20.77 | 55.63 | -1.065 | 细胞核 Nucleus |
| Bra016868 | BrLEA_4-9 | 679 | 5.98 | 72 972.61 | 33.88 | 45.77 | -1.088 | 细胞核 Nucleus |
| Bra000173 | BrLEA_5-1 | 88 | 5.88 | 9 630.38 | 37.14 | 25.57 | -1.659 | 线粒体 Mitochondrion |
| Bra004981 | BrLEA_5-2 | 84 | 6.74 | 9 264.98 | 49.89 | 27.96 | -1.704 | 线粒体 Mitochondrion |
| Bra040894 | BrLEA_5-3 | 172 | 6.17 | 18 997.70 | 42.85 | 30.64 | -1.635 | 细胞核 Nucleus |
| Bra022950 | BrLEA_6-1 | 71 | 6.03 | 7 581.23 | 52.98 | 40.00 | -1.238 | 细胞核 Nucleus |
| Bra021869 | BrLEA_6-2 | 72 | 5.21 | 7 627.30 | 58.73 | 40.83 | -1.174 | 细胞核 Nucleus |
| Bra039946 | BrLEA_6-3 | 82 | 4.72 | 8 433.11 | 44.84 | 37.07 | -1.055 | 线粒体 Mitochondrion |
| Bra022642 | BrSMP-1 | 173 | 5.88 | 18 379.46 | 40.43 | 86.36 | -0.357 | 细胞膜 Cell membrane |
| Bra020604 | BrSMP-2 | 181 | 4.76 | 18 366.26 | 31.03 | 71.44 | -0.481 | 细胞核 Nucleus |
| Bra001869 | BrSMP-3 | 239 | 5.70 | 25 385.45 | 46.14 | 76.78 | -0.380 | 细胞膜 Cell membrane |
| Bra029085 | BrSMP-4 | 177 | 5.43 | 18 691.81 | 41.22 | 82.20 | -0.351 | 细胞膜 Cell membrane |
| Bra039956 | BrSMP-5 | 262 | 4.71 | 26 692.50 | 34.19 | 77.21 | -0.239 | 细胞膜 Cell membrane |
| Bra009983 | BrSMP-6 | 192 | 4.76 | 19 473.41 | 23.22 | 69.32 | -0.515 | 细胞膜 Cell membrane |
| Bra036112 | BrSMP-7 | 191 | 4.99 | 19 600.77 | 26.99 | 75.81 | -0.414 | 细胞核 Nucleus |
| Bra033375 | BrSMP-8 | 177 | 4.53 | 18 185.98 | 30.21 | 71.24 | -0.540 | 叶绿体 Chloroplast |
| Bra033377 | BrSMP-9 | 283 | 8.89 | 32 586.25 | 52.20 | 75.72 | -0.543 | 叶绿体 Chloroplast |
Table 2 Physical and chemical properties analysis
| 基因号 Gene number | 基因名称 Gene name | 氨基酸长度 Amino acid length | 等电点 pI | 分子量/D MW | 不稳定性 指数 Instability index | 脂肪族指数 Aliphatic index | 亲水性 GRAVY | 亚细胞定位 Subcellular localization |
|---|---|---|---|---|---|---|---|---|
| Bra011874 | BrDehydrin-1 | 149 | 5.65 | 15 928.40 | 23.71 | 58.93 | -0.863 | 细胞核 Nucleus |
| Bra036843 | BrDehydrin-10 | 134 | 9.19 | 13 834.22 | 42.98 | 49.55 | -0.887 | 细胞核 Nucleus |
| Bra037177 | BrDehydrin-11 | 144 | 8.81 | 15 139.46 | 45.61 | 33.26 | -1.360 | 细胞核 Nucleus |
| Bra008242 | BrDehydrin-2 | 194 | 5.52 | 21 840.38 | 62.43 | 53.71 | -1.352 | 细胞核 Nucleus |
| Bra031809 | BrDehydrin-3 | 192 | 7.14 | 18 844.17 | 33.09 | 32.14 | -1.021 | 细胞核 Nucleus |
| Bra025819 | BrDehydrin-4 | 271 | 5.00 | 31 018.16 | 61.44 | 47.49 | -1.508 | 细胞核 Nucleus |
| Bra012230 | BrDehydrin-5 | 220 | 5.11 | 24 837.39 | 62.24 | 55.41 | -1.399 | 细胞核 Nucleus |
| Bra015779 | BrDehydrin-6 | 195 | 5.45 | 22 044.52 | 63.15 | 50.41 | -1.455 | 细胞核 Nucleus |
| Bra003732 | BrDehydrin-7 | 194 | 5.47 | 21 755.35 | 56.38 | 53.25 | -1.308 | 细胞核 Nucleus |
| Bra013992 | BrDehydrin-8 | 257 | 6.17 | 28 978.00 | 34.53 | 35.33 | -1.645 | 细胞核 Nucleus |
| Bra031192 | BrDehydrin-9 | 183 | 6.49 | 19 183.88 | 36.64 | 49.18 | -1.016 | 细胞核 Nucleus |
| Bra005911 | BrLEA_1-1 | 159 | 9.22 | 16 286.99 | 17.53 | 43.71 | -0.789 | 细胞核 Nucleus |
| Bra005353 | BrLEA_1-2 | 98 | 9.22 | 10 666.89 | 42.18 | 49.18 | -1.115 | 细胞核 Nucleus |
| Bra035491 | BrLEA_1-3 | 132 | 9.68 | 14 933.95 | 56.61 | 51.89 | -1.109 | 线粒体 Mitochondrion |
| Bra023278 | BrLEA_1-4 | 133 | 9.24 | 14 507.53 | 59.47 | 48.65 | -0.878 | 线粒体 Mitochondrion |
| Bra009225 | BrLEA_1-5 | 159 | 8.93 | 16 180.83 | 16.45 | 38.81 | -0.818 | 细胞核 Nucleus |
| Bra000339 | BrLEA_2-1 | 777 | 9.02 | 84 314.53 | 32.33 | 96.22 | -0.135 | 叶绿体 Chloroplast |
| Bra000414 | BrLEA_2-2 | 166 | 4.81 | 17 823.38 | 17.04 | 96.81 | 0.030 | 胞外Extracellular |
| Bra039288 | BrLEA_2-3 | 166 | 4.50 | 18 119.88 | 16.96 | 100.36 | 0.013 | 叶绿体 Chloroplast |
| Bra037669 | BrLEA_2-4 | 506 | 8.85 | 55 465.06 | 27.06 | 85.89 | -0.399 | 细胞核 Nucleus |
| Bra004565 | BrLEA_2-5 | 166 | 4.81 | 17 725.34 | 20.39 | 106.20 | 0.093 | 细胞膜 Cell membrane |
| Bra030494 | BrLEA_2-6 | 151 | 4.72 | 16 404.92 | 10.57 | 106.42 | 0.075 | 细胞膜 Cell membrane |
| Bra033520 | BrLEA_3-1 | 99 | 9.72 | 10 582.01 | 50.01 | 70.10 | -0.400 | 叶绿体 Chloroplast |
| Bra000884 | BrLEA_3-2 | 94 | 9.85 | 10 095.53 | 39.42 | 78.94 | -0.349 | 叶绿体 Chloroplast |
| Bra014864 | BrLEA_3-3 | 122 | 9.20 | 14 152.16 | 50.98 | 63.20 | -0.598 | 叶绿体 Chloroplast |
| Bra018556 | BrLEA_3-4 | 93 | 9.99 | 10 112.56 | 52.38 | 72.47 | -0.482 | 叶绿体 Chloroplast |
| Bra038061 | BrLEA_3-5 | 57 | 4.94 | 6 614.40 | 49.22 | 56.67 | -0.912 | 线粒体 Mitochondrion |
| Bra036272 | BrLEA_3-6 | 94 | 9.99 | 10 088.55 | 43.90 | 72.77 | -0.299 | 叶绿体 Chloroplast |
| Bra033350 | BrLEA_3-7 | 98 | 8.03 | 10 298.59 | 40.91 | 76.84 | -0.229 | 叶绿体 Chloroplast |
| Bra021126 | BrLEA_4-1 | 226 | 9.02 | 24 598.55 | 28.79 | 22.79 | -1.475 | 线粒体 Mitochondrion |
| Bra022221 | BrLEA_4-10 | 298 | 5.45 | 32 431.59 | 30.10 | 54.50 | -1.126 | 胞外Extracellular |
| Bra027219 | BrLEA_4-11 | 229 | 8.83 | 24 810.86 | 27.04 | 22.97 | -1.414 | 线粒体 Mitochondrion |
| Bra005256 | BrLEA_4-12 | 399 | 5.74 | 43 565.34 | 20.90 | 52.93 | -0.970 | 细胞膜 Cell membrane |
| Bra019628 | BrLEA_4-13 | 451 | 5.20 | 48 554.47 | 12.26 | 47.98 | -1.141 | 细胞核 Nucleus |
| Bra025130 | BrLEA_4-14 | 201 | 5.40 | 22 054.03 | 38.33 | 77.21 | -0.429 | 叶绿体 Chloroplast |
| Bra039616 | BrLEA_4-15 | 480 | 6.47 | 52 652.67 | 18.82 | 47.04 | -0.921 | 叶绿体 Chloroplast |
| Bra010561 | BrLEA_4-16 | 487 | 5.63 | 52 431.94 | 23.72 | 46.65 | -0.923 | 叶绿体 Chloroplast |
| Bra020879 | BrLEA_4-17 | 218 | 5.12 | 24 376.73 | 15.32 | 38.72 | -1.441 | 细胞核 Nucleus |
| Bra027542 | BrLEA_4-18 | 311 | 7.60 | 35 005.70 | 23.66 | 43.79 | -1.371 | 叶绿体 Chloroplast |
| Bra037225 | BrLEA_4-19 | 722 | 8.58 | 80 928.05 | 43.18 | 68.66 | -0.722 | 叶绿体 Chloroplast |
| Bra013489 | BrLEA_4-2 | 253 | 8.55 | 27 837.93 | 20.67 | 47.31 | -1.202 | 叶绿体 Chloroplast |
| Bra036021 | BrLEA_4-20 | 253 | 5.72 | 28 430.30 | 32.23 | 43.72 | -1.355 | 叶绿体 Chloroplast |
| Bra041061 | BrLEA_4-21 | 241 | 6.17 | 26 330.13 | 20.46 | 50.83 | -1.111 | 叶绿体 Chloroplast |
| Bra026541 | BrLEA_4-3 | 192 | 8.57 | 20 399.00 | 33.96 | 62.55 | -0.669 | 叶绿体 Chloroplast |
| Bra008006 | BrLEA_4-4 | 462 | 9.03 | 50 701.68 | 32.68 | 78.53 | -0.390 | 胞外Extracellular |
| Bra001666 | BrLEA_4-5 | 276 | 5.54 | 30 311.37 | 25.62 | 53.22 | -1.093 | 叶绿体 Chloroplast |
| Bra000263 | BrLEA_4-6 | 142 | 8.65 | 14 760.62 | 19.57 | 77.75 | -0.430 | 叶绿体 Chloroplast |
| Bra000265 | BrLEA_4-7 | 129 | 5.31 | 13 894.51 | 22.81 | 71.86 | -0.692 | 叶绿体 Chloroplast |
| Bra017229 | BrLEA_4-8 | 398 | 6.02 | 43 386.25 | 20.77 | 55.63 | -1.065 | 细胞核 Nucleus |
| Bra016868 | BrLEA_4-9 | 679 | 5.98 | 72 972.61 | 33.88 | 45.77 | -1.088 | 细胞核 Nucleus |
| Bra000173 | BrLEA_5-1 | 88 | 5.88 | 9 630.38 | 37.14 | 25.57 | -1.659 | 线粒体 Mitochondrion |
| Bra004981 | BrLEA_5-2 | 84 | 6.74 | 9 264.98 | 49.89 | 27.96 | -1.704 | 线粒体 Mitochondrion |
| Bra040894 | BrLEA_5-3 | 172 | 6.17 | 18 997.70 | 42.85 | 30.64 | -1.635 | 细胞核 Nucleus |
| Bra022950 | BrLEA_6-1 | 71 | 6.03 | 7 581.23 | 52.98 | 40.00 | -1.238 | 细胞核 Nucleus |
| Bra021869 | BrLEA_6-2 | 72 | 5.21 | 7 627.30 | 58.73 | 40.83 | -1.174 | 细胞核 Nucleus |
| Bra039946 | BrLEA_6-3 | 82 | 4.72 | 8 433.11 | 44.84 | 37.07 | -1.055 | 线粒体 Mitochondrion |
| Bra022642 | BrSMP-1 | 173 | 5.88 | 18 379.46 | 40.43 | 86.36 | -0.357 | 细胞膜 Cell membrane |
| Bra020604 | BrSMP-2 | 181 | 4.76 | 18 366.26 | 31.03 | 71.44 | -0.481 | 细胞核 Nucleus |
| Bra001869 | BrSMP-3 | 239 | 5.70 | 25 385.45 | 46.14 | 76.78 | -0.380 | 细胞膜 Cell membrane |
| Bra029085 | BrSMP-4 | 177 | 5.43 | 18 691.81 | 41.22 | 82.20 | -0.351 | 细胞膜 Cell membrane |
| Bra039956 | BrSMP-5 | 262 | 4.71 | 26 692.50 | 34.19 | 77.21 | -0.239 | 细胞膜 Cell membrane |
| Bra009983 | BrSMP-6 | 192 | 4.76 | 19 473.41 | 23.22 | 69.32 | -0.515 | 细胞膜 Cell membrane |
| Bra036112 | BrSMP-7 | 191 | 4.99 | 19 600.77 | 26.99 | 75.81 | -0.414 | 细胞核 Nucleus |
| Bra033375 | BrSMP-8 | 177 | 4.53 | 18 185.98 | 30.21 | 71.24 | -0.540 | 叶绿体 Chloroplast |
| Bra033377 | BrSMP-9 | 283 | 8.89 | 32 586.25 | 52.20 | 75.72 | -0.543 | 叶绿体 Chloroplast |
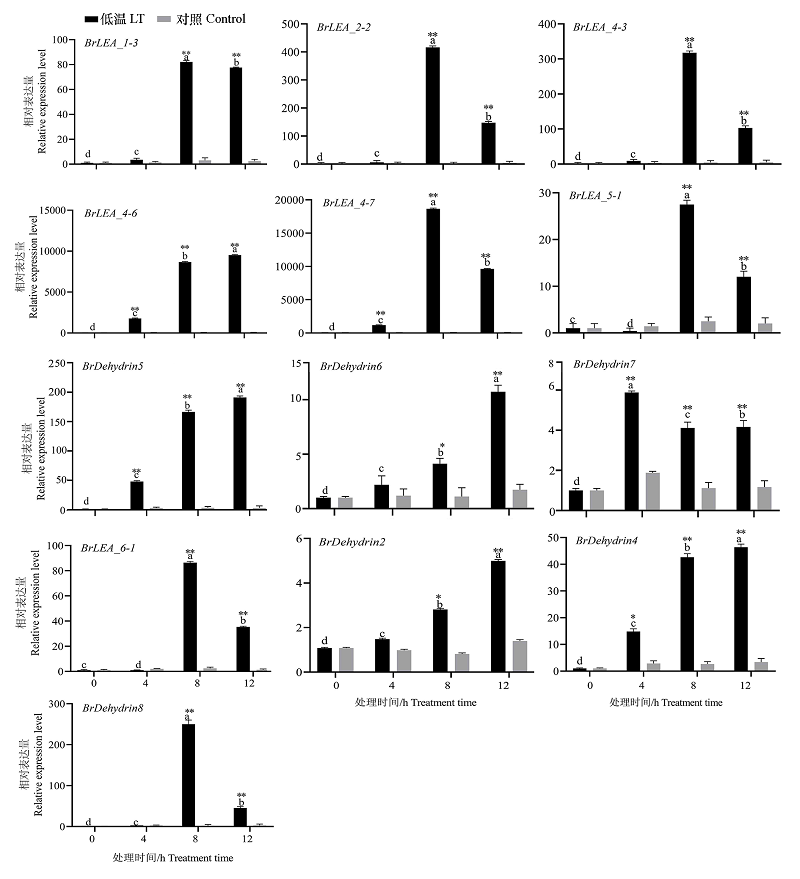
Fig. 6 Expression analysis of 13 BrLEA genes under low-temperature treatment Different lowercase letters indicate significant differences in low temperature treatment at 0.05 level at different times. *,** indicated significant difference between treatment and control at 0.05 and 0.01 levels.
| [1] |
Amara I, Odena A, Oliveira E, Moreno A, Masmoudi K, Pagès M, Goday A. 2012. Insights into maize LEA proteins:from proteomics to functional approaches. Plant Cell Physiol, 53 (2):312-329.
doi: 10.1093/pcp/pcr183 pmid: 22199372 |
| [2] |
Baker J, Van Dennsteele C, Dure L. 1988. Sequence and characterization of 6 LEA proteins and their genes from cotton. Plant Mol Biol, 11 (3):277-291.
doi: 10.1007/BF00027385 pmid: 24272341 |
| [3] |
Battaglia M, Olvera-Carrillo Y, Garciarrubio A, Campos F, Covarrubias A A. 2008. The enigmatic LEA proteins and other hydrophilins. Plant Physiol, 148 (1):6-24.
doi: 10.1104/pp.108.120725 pmid: 18772351 |
| [4] |
Bies-Ethève N, Gaubier-Comella P, Debures A, Lasserre E, Jobet E, Raynal M, Cooke R, Delseny M. 2008. Inventory,evolution and expression profiling diversity of the LEA(late embryogenesis abundant)protein gene family in Arabidopsis thaliana. Plant Mol Biol, 67 (1-2):107-124.
doi: 10.1007/s11103-008-9304-x pmid: 18265943 |
| [5] |
Candat A, Paszkiewicz G, Neveu M, Gautier R, Logan D C, Avelange-Macherel M H, Macherel D. 2014. The ubiquitous distribution of late embryogenesis abundant proteins across cell compartments in Arabidopsis offers tailored protection against abiotic stress. Plant Cell, 26 (7):3148-3166.
doi: 10.1105/tpc.114.127316 URL |
| [6] |
Cao J, Li X. 2015. Identification and phylogenetic analysis of late embryogenesis abundant proteins family in tomato(Solanum lycopersicum). Planta, 241 (3):757-772.
doi: 10.1007/s00425-014-2215-y URL |
| [7] |
Chen C, Chen H, Zhang Y, Thomas H R, Frank M H, He Y, Xia R. 2020. TBtools:an integrative toolkit developed for interactive analyses of big biological data. Mol Plant, 13 (8):1194-1202.
doi: 10.1016/j.molp.2020.06.009 URL |
| [8] | Chen Fang, Gu Xiaoping, Liang Ping, Yu Fei. 2017. Effects of low temperature freezing injury on morphological changes of Chinese cabbage. Meteorological and Environmental Sciences, 40 (2):55-59. (in Chinese) |
| 陈芳, 谷晓平, 梁平, 于飞. 2017. 低温冻害对大白菜形态变化的影响. 气象与环境科学, 40 (2):55-59. | |
| [9] | Chen Yiyin. 2007. Study on cold resistance of tobacco and transgenic plants induced by low temperature gene transformation of Amongillus amurensis[M. D. Dissertation]. BeiJing: Beijing Forestry University. (in Chinese) |
| 陈奕吟. 2007. 沙冬青低温诱导基因转化烟草及转基因植物的抗寒性研究[硕士论文]. 北京: 北京林业大学. | |
| [10] |
Daniel C J, Christopher J P, Jürg B. 2008. Rapidly regulated genes are intron poor. Trends Genet, 24 (8):375-378.
doi: 10.1016/j.tig.2008.05.006 URL |
| [11] | Du Yongting, Wang Tiejuan, Du Junrui, Chen Jing. 2018. Expression and sequence analysis of LEA gene of Artemisia argyi under drought stress. Biotechnology, 28 (4):318-322,371. (in Chinese) |
| 杜永婷, 王铁娟, 杜俊瑞, 陈静. 2018. 干旱胁迫下白沙蒿LEA基因的表达及序列分析. 生物技术, 28 (4):318-322,371. | |
| [12] |
Dure L Ⅲ. 1993. A repeating 11-mer amino acid motif and plant desiccation. Plant J, 3 (3):363-369.
pmid: 8220448 |
| [13] |
Dure L Ⅲ, Crouch M, Harada J, Ho T H, Mundy J, Quatrano R, Thomas T, Sung Z R. 1989. Common amino acid sequence domains among the LEA proteins of higher plants. Plant Mol Biol, 12 (5):475-486.
doi: 10.1007/BF00036962 pmid: 24271064 |
| [14] |
Dure L Ⅲ, Greenway S C, Galau G A. 1981. Developmental biochemistry of cottonseed embryogenesis and germination:changing messenger ribonucleic acid populations as shown by in vitro and in vivo protein synthesis. Biochemistry, 20 (14):4162-4168.
pmid: 7284317 |
| [15] |
Espelund M, Saebøe-Larssen S, Hughes D W, Galau G A, Larsen F, Jakobsen K S. 1992. Late embryogenesis-abundant genes encoding proteins with different numbers of hydrophilic repeats are regulated differentially by abscisic acid and osmotic stress. Plant J, 2 (2):241-252.
pmid: 1302052 |
| [16] | Finn R D, Mistry J, Tate J, Coggill P, Heger A, Pollington J E, Gavin O L, Gunasekaran P, Ceric G, Forslund K, Holm L, Sonnhammer E L, Eddy R, Bateman A. 2000. The Pfam protein families database. Nucleic Acids Res, 38 (Database issue):D211-D222. |
| [17] |
Gao T, Mo Y X, Huang H Y, Yu J M, Wang Y, Wang W D. 2021. Heterologous expression of Camellia sinensis late embryogenesis abundant protein gene 1(CsLEA1)confers cold stress tolerance in Escherichia coli and yeast. Horticultural Plant Journal, 7 (1):89-96.
doi: 10.1016/j.hpj.2020.09.005 URL |
| [18] |
Garay-Arroyo A, Colmener-Flores J M, Garciarrubio A, Covarrubias A A. 2000. Highly hydrophilic proteins in prokaryotes and eukaryotes are common during conditions of water deficit. J Biol Chem, 275 (8):5668-5674.
doi: 10.1074/jbc.275.8.5668 pmid: 10681550 |
| [19] |
Hand S C, Menze M A, Toner M, Boswell L, Moore D. 1996. LEA proteins during water stress:not just for plants anymore. Annu Rev Physiol, 73:115-134.
doi: 10.1146/physiol.2011.73.issue-1 URL |
| [20] |
Hundertmark M, Hincha D K. 2008. LEA(late embryogenesis abundant)proteins and their encoding genes in Arabidopsis thaliana. BMC Genomics, 9 (1):118.
doi: 10.1186/1471-2164-9-118 URL |
| [21] |
Imai R, Chang L, Ohta A, Bray E A, Takagi M. 1996. A lea-class gene of tomato confers salt and freezing tolerance when expressed in Saccharomyces cerevisiae. Gene, 170 (2):243-248.
pmid: 8666253 |
| [22] |
Jeffares D C, Penkett C J, Bähler J. 2008. Rapidly regulated genes are intron poor. Trends Genet, 24 (8):375-378.
doi: 10.1016/j.tig.2008.05.006 pmid: 18586348 |
| [23] |
Kosová K, Vítámvás Prášil, Prasil I. 2007. The role of dehydrins in plant response to cold. Biol Plant, 51 (4):601-617.
doi: 10.1007/s10535-007-0133-6 URL |
| [24] |
Lysak M A, Koch M A, Pecinka A, Schubert I. 2005. Chromosome triplication found across the tribe Brassiceae. Genome Res, 15 (4):516-525.
doi: 10.1101/gr.3531105 URL |
| [25] |
Liu Y, Xie L, Liang X, Zhang S. 2015. pLEA5,the Late Embryogenesis Abundant Protein Gene from Chimonanthus praecox,possesses low temperature and osmotic resistances in prokaryote and eukaryotes. Int J Mol Sci, 16 (11):26978-26990.
doi: 10.3390/ijms161126006 URL |
| [26] |
Morran S, Eini O, Pyvovarenko T, Parent B, Singh R, Ismagul A, Eliby S, Shirley N, Langridge P, Lopato S. 2011. Improvement of stress tolerance of wheat and barley by modulation of expression of DREB/CBF factors. Plant Biotechnol J, 9 (2):230-249.
doi: 10.1111/j.1467-7652.2010.00547.x pmid: 20642740 |
| [27] |
Mowla S B, Cuypers A, Driscoll S P, Kiddle G, Thomson J, Foyer C H, Theodoulou F L. 2006. Yeast complementation reveals a role for an Arabidopsis thaliana late embryogenesis abundant(LEA)-like protein in oxidative stress tolerance. Plant J, 48 (5):743-756.
doi: 10.1111/tpj.2006.48.issue-5 URL |
| [28] |
Nagaraju M, Kumar S A, Reddy P S, Kumar A, Rao D M, Kavi Kishor P B. 2019. Genome-scale identification,classification,and tissue specific expression analysis of late embryogenesis abundant(LEA)genes under abiotic stress conditions in Sorghum bicolor L. PLoS ONE, 14 (1):e0209980.
doi: 10.1371/journal.pone.0209980 URL |
| [29] |
Olvera-Carrillo Y, Luis Reyes J, Covarrubias A A. 2011. Late embryogenesis abundant proteins:versatile players in the plant adaptation to water limiting environments. Plant Signal Behav, 6 (4):586-589.
doi: 10.4161/psb.6.4.15042 pmid: 21447997 |
| [30] |
Park S C, Kim Y H, Jeong J C, Kim C Y, Lee H S, Bang J W, Kwak S S. 2011. Sweetpotato late embryogenesis abundant 14(IbLEA14)gene influences lignification and increases osmotic- and salt stress-tolerance of transgenic calli. Planta, 233 (3):621-634.
doi: 10.1007/s00425-010-1326-3 URL |
| [31] |
Tong C, Wang X, Yu J, Wu J, Li W, Huang J, Dong C, Hua W, Liu S. 2013. Comprehensive analysis of RNA-seq data reveals the complexity of the transcriptome in Brassica rapa. BMC Genomics, 14:689.
doi: 10.1186/1471-2164-14-689 URL |
| [32] |
Tunnacliffe A, Wise M J. 2007. The continuing conundrum of the LEA proteins. Naturwissenschaften, 94 (10):791-812.
doi: 10.1007/s00114-007-0254-y URL |
| [33] | Wang Ying, Zhang Changwei, Lv Shanwu, Hou Xilin. 2018. Genome identification and expression analysis BrCNGC Chinese cabbage. Journal of Nanjing Agricultural University, 41 (6):994-1002. (in Chinese) |
| 汪影, 张昌伟, 吕善武, 侯喜林. 2018. 大白菜BrCNGC全基因组鉴定及其表达分析. 南京农业大学学报, 41 (6):994-1002. | |
| [34] |
Wang X S, Zhu H B, Jin G L, Liu H L, Wu W R, Zhu J. 2006. Genome-scale identification and analysis of LEA genes in rice ( Oryza sativa L.). Plant Science, 172 (2):414-420.
doi: 10.1016/j.plantsci.2006.10.004 URL |
| [35] |
Wang Y D, Chen G J, Lei J J, Cao B H, Chen C M. 2020. Identification and characterization of a LEA-like gene,CaMF5,specifically expressed in the anthers of male-fertile Capsicum annuum. Horticultural Plant Journal, 6 (1):39-48.
doi: 10.1016/j.hpj.2019.07.004 URL |
| [36] |
Wise M J, Tunnacliffe A. 2004. POPP the question:what do LEA proteins do? Trends Plant Sci, 9 (1):13-17.
doi: 10.1016/j.tplants.2003.10.012 URL |
| [37] |
Yu J, Lai Y, Wu X, Wu G, Guo C. 2016. Overexpression of OsEm1 encoding a group I LEA protein confers enhanced drought tolerance in rice. Biochem Biophys Res Commun, 478 (2):703-709.
doi: 10.1016/j.bbrc.2016.08.010 URL |
| [38] | Yu J N, Zhang J S, Shan L, Chen S Y. 2005. Two new group 3 LEA genes of wheat and their functional analysis in yeast. Journal of Integrative Plant Biology Formerly Aacta Botanica Sinica, 47 (11):1372-1381. |
| [39] | Zhang L, Cai X, Wu J, Liu M, Grob S, Cheng F, Liang J, Cai C, Liu Z, Liu B, Wang F, Li S, Li X, Cheng L, Yang W, Li M H, Grossniklaus U, Zheng H, Wang X. 2019. Improved Brassica rapa reference genome by single-molecule sequencing and chromosome conformation capture technologies. Hortic Res, 13 (6):124. |
| [40] | Zhang L S, Zhao W M. 2003. LEA protein functions to tolerance drought of the plant. Plant Physiol Comm, 39:61-66. |
| [41] | Zhang Mei, Zhang Hui. 2017. Research progress in late embryogenesis abundant protein(LEA protein)and plant stress resistance. Biotic Resources, 39 (3):155-161. (in Chinese) |
| 张美, 张会. 2017. 胚胎发育晚期丰富蛋白(LEA蛋白)与植物抗逆性研究进展. 生物资源, 39 (3):155-161. | |
| [42] | Zong Mei, Cai Yongping. 2005. Correlation between seed dehydration tolerance and protective system. Acta Horticulturae Sinica, 46 (2):342-347. (in Chinese) |
| 宗梅, 蔡永萍. 2005. 种子脱水耐性与保护系统的相关性. 园艺学报, 46 (2):342-347. |
| [1] | YU Tingting, LI Huan, NING Yuansheng, SONG Jianfei, PENG Lulin, JIA Junqi, ZHANG Weiwei, and YANG Hongqiang. Genome-wide Identification of GRAS Gene Family in Apple and Expression Analysis of Its Response to Auxin [J]. Acta Horticulturae Sinica, 2023, 50(2): 397-409. |
| [2] | LIN Haijiao, LIANG Yuchen, LI Ling, MA Jun, ZHANG Lu, LAN Zhenying, YUAN Zening. Exploration and Regulation Network Analysis of CBF Pathway Related Cold Tolerance Genes in Lavandula angustifolia [J]. Acta Horticulturae Sinica, 2023, 50(1): 131-144. |
| [3] | HAN Shuhui, HAN Caifeng, HAN Shurong, HAN Caimei, and HAN Xu. A New Autumn Chinese Cabbage Hybrid‘Jiaoyan Qiubao’ [J]. Acta Horticulturae Sinica, 2022, 49(S2): 83-84. |
| [4] | WANG Weihong, ZHANG Fenglan, YU Yangjun, ZHANG Deshuang, ZHAO Xiuyun, YU Shuancang, SU Tongbing, LI Peirong, and XIN Xiaoyun. A New Chinese Cabbage Cultivar‘Jingqiu 1518’for Autumn Cultivation [J]. Acta Horticulturae Sinica, 2022, 49(S2): 85-86. |
| [5] | YU Yangjun, WANG Weihong, SU Tongbing, ZHANG Fenglan, ZHANG Deshuang, ZHAO Xiuyun, YU Shuancang, LI Peirong, XIN Xiaoyun, and WANG Jiao. A New Chinese Cabbage Cultivar‘Jingchun CR3’with Clubroot Resistance and Bolting Tolerance [J]. Acta Horticulturae Sinica, 2022, 49(S2): 87-88. |
| [6] | WANG Lili, WANG Xin, WU Haidong, WEN Qiang, and Yang Xiaofei. A New Chinese Cabbage Cultivar‘Liaobai 28’with Resistance to Clubroot Disease [J]. Acta Horticulturae Sinica, 2022, 49(S2): 89-90. |
| [7] | YU Yangjun, SU Tongbing, ZHANG Fenglan, ZHANG Deshuang, ZHAO Xiuyun, YU Shuancang, WANG Weihong, LI Peirong, XIN Xiaoyun, WANG Jiao, and WU Changjian. A New Purple Seedling-Edible Chinese Cabbage F1 Hybrid‘Jingyan Zikuaicai’ [J]. Acta Horticulturae Sinica, 2022, 49(S2): 91-92. |
| [8] | HUANG Li, CHEN Caizhi, YU Xiaolin, YAO Xiangtan, and CAO Jiashu, . A New Early-mid Maturing Chinese Cabbage Pak-choi Cultivar‘Zhedaqing’ [J]. Acta Horticulturae Sinica, 2022, 49(S2): 93-94. |
| [9] | XU Ligong, HAN Taili, SUN Jifeng, YANG Xiaodong, and TAN Jinxia. A New Seedling-edible Chinese Cabbage Cultivar‘Jinlü 2’ [J]. Acta Horticulturae Sinica, 2022, 49(S1): 67-68. |
| [10] | GAO Yanlong, WU Yuxia, ZHANG Zhongxing, WANG Shuangcheng, ZHANG Rui, ZHANG De, WANG Yanxiu. Bioinformatics Analysis of Apple ELO Gene Family and Its Expression Analysis Under Low Temperature Stress [J]. Acta Horticulturae Sinica, 2022, 49(8): 1621-1636. |
| [11] | WANG Yu, ZHANG Xue, ZHANG Xueying, ZHANG Siyu, WEN Tingting, WANG Yingjun, GAN Caixia, PANG Wenxing. The Effect of Camalexin on Chinese Cabbage Resistance to Clubroot [J]. Acta Horticulturae Sinica, 2022, 49(8): 1689-1698. |
| [12] | QIAN Jieyu, JIANG Lingli, ZHENG Gang, CHEN Jiahong, LAI Wuhao, XU Menghan, FU Jianxin, ZHANG Chao. Identification and Expression Analysis of MYB Transcription Factors Regulating the Anthocyanin Biosynthesis in Zinnia elegans and Function Research of ZeMYB9 [J]. Acta Horticulturae Sinica, 2022, 49(7): 1505-1518. |
| [13] | ZHANG Lugang, LU Qianqian, HE Qiong, XUE Yihua, MA Xiaomin, MA Shuai, NIE Shanshan, YANG Wenjing. Creation of Novel Germplasm of Purple-orange Heading Chinese Cabbage [J]. Acta Horticulturae Sinica, 2022, 49(7): 1582-1588. |
| [14] | CHEN Jianxin, WEI Yuqian, TANG Jie, ZHU Yongjin, MA Huancheng, WU Jianrong. Molecular Detection and Sequence Analysis of ORSV and CymMV in Cultivated Cymbidium goeringii in Dali,Yunnan [J]. Acta Horticulturae Sinica, 2022, 49(3): 655-662. |
| [15] | JIANG Cuicui, FANG Zhizhen, ZHOU Danrong, LIN Yanjuan, YE Xinfu. Identification and Expression Analysis of Sugar Transporter Family Genes in‘Furongli’(Prunus salicina) [J]. Acta Horticulturae Sinica, 2022, 49(2): 252-264. |
| Viewed | ||||||
|
Full text |
|
|||||
|
Abstract |
|
|||||
Copyright © 2012 Acta Horticulturae Sinica 京ICP备10030308号-2 国际联网备案号 11010802023439
Tel: 010-82109523 E-Mail: yuanyixuebao@126.com
Support by: Beijing Magtech Co.Ltd