
园艺学报 ›› 2022, Vol. 49 ›› Issue (5): 1031-1046.doi: 10.16420/j.issn.0513-353x.2021-0003
肖学宸1, 刘梦雨1, 蒋梦琦1, 陈燕1, 薛晓东1, 周承哲1, 吴兴健1, 吴君楠1, 郭寅生1, 叶开温2, 赖钟雄1, 林玉玲1,*( )
)
收稿日期:2021-08-01
修回日期:2021-09-07
出版日期:2022-05-25
发布日期:2022-05-25
通讯作者:
林玉玲
E-mail:buliang84@163.com
基金资助:
XIAO Xuechen1, LIU Mengyu1, JIANG Mengqi1, CHEN Yan1, XUE Xiaodong1, ZHOU Chengzhe1, WU Xingjian1, WU Junnan1, GUO Yinsheng1, YEH Kaiwen2, LAI Zhongxiong1, LIN Yuling1,*( )
)
Received:2021-08-01
Revised:2021-09-07
Online:2022-05-25
Published:2022-05-25
Contact:
LIN Yuling
E-mail:buliang84@163.com
摘要:
基于第3代龙眼(Dimocarpus longan)基因组及转录组数据库,对褪黑素合成路径上可能的限速基因SNAT(serotonin N-acetyltransferase)、ASMT(N-acetylserotonin methyltransferase)和COMT(caffeic acid O-methyltransferase)进行基因家族成员鉴定,利用实时荧光定量PCR技术研究其在龙眼体胚早期的表达谱,并研究外源IAA、GA3、MeJA、SA和ABA对龙眼胚性愈伤组织中褪黑素含量的影响,以及褪黑素合成途径相关基因的表达模式。结果显示:龙眼SNAT(DlSNAT)、ASMT(DlASMT)和COMT(DlCOMT)基因家族分别含有2、18和7个成员,蛋白质结构域保守,相同家族的成员之间保守基序相差极小,DlASMT和DlCOMT家族成员高度相似。基于氨基酸序列进行系统进化树分析,龙眼、拟南芥、水稻、小麦、番茄、辣椒和卷柏的SNAT家族可分为3个亚组;ASMT和COMT家族具有高度同源性,共同建树并分为3个亚组。DlSNAT、DlASMT和DlCOMT家族成员的启动子中含有大量响应生长素、赤霉素、茉莉酸甲酯、水杨酸和脱落酸的顺式作用元件。表达谱分析显示,DlASMT和DlCOMT家族成员主要在龙眼不完全胚性紧实结构和球形胚阶段高表达,DlSNAT1和DlSNAT2在龙眼体胚发生早期始终处于高水平表达。用0.1 mmol · L-1IAA处理龙眼胚性愈伤组织,内源褪黑素含量在24 h内显著降低,GA3、MeJA、SA和ABA处理则显著提高了褪黑素含量;同时分析DlSNAT、DlASMT、DlCOMT成员表达情况,内源褪黑素含量变化趋势和DlSNAT表达基本一致。综上,DlSNAT、DlASMT和DlCOMT可能通过影响内源褪黑素的合成参与调控龙眼早期体胚发生。
中图分类号:
肖学宸, 刘梦雨, 蒋梦琦, 陈燕, 薛晓东, 周承哲, 吴兴健, 吴君楠, 郭寅生, 叶开温, 赖钟雄, 林玉玲. 龙眼褪黑素合成途径SNAT、ASMT和COMT家族基因鉴定及表达分析[J]. 园艺学报, 2022, 49(5): 1031-1046.
XIAO Xuechen, LIU Mengyu, JIANG Mengqi, CHEN Yan, XUE Xiaodong, ZHOU Chengzhe, WU Xingjian, WU Junnan, GUO Yinsheng, YEH Kaiwen, LAI Zhongxiong, LIN Yuling. Whole-genome Identification and Expression Analysis of SNAT,ASMT and COMT Families of Melatonin Synthesis Pathway in Dimocarpus longan[J]. Acta Horticulturae Sinica, 2022, 49(5): 1031-1046.
| 引物名称 Primer name | 引物序列 Primer sequence | 引物名称 Primer name | 引物序列 Primer sequence |
|---|---|---|---|
| DlSNAT1-F | TTCATTCTCCACCGCAC | DlASMT16-F | CCCTCTGATGCCTCTCTAAG |
| DlSNAT1-R | CTCCATCCAAAGCAGACTG | DlASMT16-R | TCACGAACGGCTCTGAT |
| DlSNAT2-F | GCCACATTGCATTCTGTTAG | DlASMT12-F | CAAGCAATGACCAGTGACTC |
| DlSNAT2-R | GACCTTGACCCTGATAGCTAG | DlASMT12-R | CTTCCTCCTACATCAACCAATG |
| DlASMT6-F | TTGTTGGTTGCTCTGTGG | DlCOMT6-F | CGCCTTTGATTCTGATACAG |
| DlASMT6-R | CTCCTTCAAGAACGGCAT | DlCOMT6-R | GCCATCGTTGAACATCTTG |
| DlASMT4-F | CGAACGCATTTGAGTATCC | DlCOMT1-F | TCGGATGGGAAAGTTGAG |
| DlASMT4-R | TGTGAGGGTATTTGGCAGT | DlCOMT1-R | TGCCTTGTTGAATGGGA |
| DlASMT2-F | GTTCAACCACACAACCATAGTC | DlCOMT7-F | CCATTTGTGCGTCTCATTC |
| DlASMT2-R | CCAACATCCACAATCTGCT | DlCOMT7-R | AGTCAGAGTCAGTGTTCATTGC |
| DlASMT11-F | CAGGGCTTATGAGACAACACT | DlCOMT3-F | TTACCCTTTCAGAGTTGGCT |
| DlASMT11-R | CCACATCAACCAGGGAAT | DlCOMT3-R | AGAAGTTGGTGGTGGCAT |
表1 DlSNAT、DlASMT和DlCOMT家族成员引物序列
Table. 1 The primer sequence of D1SNAT,D1ASMT and D1COMT family members
| 引物名称 Primer name | 引物序列 Primer sequence | 引物名称 Primer name | 引物序列 Primer sequence |
|---|---|---|---|
| DlSNAT1-F | TTCATTCTCCACCGCAC | DlASMT16-F | CCCTCTGATGCCTCTCTAAG |
| DlSNAT1-R | CTCCATCCAAAGCAGACTG | DlASMT16-R | TCACGAACGGCTCTGAT |
| DlSNAT2-F | GCCACATTGCATTCTGTTAG | DlASMT12-F | CAAGCAATGACCAGTGACTC |
| DlSNAT2-R | GACCTTGACCCTGATAGCTAG | DlASMT12-R | CTTCCTCCTACATCAACCAATG |
| DlASMT6-F | TTGTTGGTTGCTCTGTGG | DlCOMT6-F | CGCCTTTGATTCTGATACAG |
| DlASMT6-R | CTCCTTCAAGAACGGCAT | DlCOMT6-R | GCCATCGTTGAACATCTTG |
| DlASMT4-F | CGAACGCATTTGAGTATCC | DlCOMT1-F | TCGGATGGGAAAGTTGAG |
| DlASMT4-R | TGTGAGGGTATTTGGCAGT | DlCOMT1-R | TGCCTTGTTGAATGGGA |
| DlASMT2-F | GTTCAACCACACAACCATAGTC | DlCOMT7-F | CCATTTGTGCGTCTCATTC |
| DlASMT2-R | CCAACATCCACAATCTGCT | DlCOMT7-R | AGTCAGAGTCAGTGTTCATTGC |
| DlASMT11-F | CAGGGCTTATGAGACAACACT | DlCOMT3-F | TTACCCTTTCAGAGTTGGCT |
| DlASMT11-R | CCACATCAACCAGGGAAT | DlCOMT3-R | AGAAGTTGGTGGTGGCAT |
| 基因ID Gene-ID | 基因名称 Gene name | 染色体定位 Chromosome location | 氨基酸数 Amino acid number | 分子量/kD Molecular weight | 理论等电点 Theoretical pI | 不稳定系数 The Instabi- lity index | 亲水性 GRAVY | 亚细胞定位 Subcellular location | 信号肽 Signal peptide |
|---|---|---|---|---|---|---|---|---|---|
| Dlo018311 | DlSNAT1 | 8 | 187 | 20.81 | 9.73 | 33.60 | 0.064 | 过氧化物酶体Peroxisome | 无No |
| Dlo030488 | DlSNAT2 | 14 | 225 | 24.68 | 4.89 | 44.33 | -0.070 | 叶绿体Chloroplast | 无No |
| Dlo004250 | DlASMT1 | 2 | 347 | 38.76 | 5.22 | 39.04 | 0.077 | 细胞质Cytoplasm | 无No |
| Dlo011828 | DlASMT2 | 5 | 389 | 43.13 | 6.02 | 35.12 | 0.000 | 细胞质Cytoplasm | 无No |
| Dlo011829 | DlASMT3 | 5 | 442 | 49.48 | 5.77 | 41.16 | -0.223 | 细胞质Cytoplasm | 无No |
| 基因ID Gene-ID | 基因名称 Gene name | 染色体定位 Chromosome location | 氨基酸数 Amino acid number | 分子量/kD Molecular weight | 理论等电点 Theoretical pI | 不稳定系数 The Instabi- Lity index | 亲水性 GRAVY | 亚细胞定位 Subcellular location | 信号肽 Signal peptide |
| Dlo011833 | DlASMT4 | 5 | 365 | 40.35 | 5.39 | 33.80 | -0.099 | 细胞质Cytoplasm | 无No |
| Dlo011834 | DlASMT5 | 5 | 355 | 38.89 | 5.55 | 32.36 | 0.090 | 细胞质Cytoplasm | 无No |
| Dlo011835 | DlASMT6 | 5 | 354 | 39.07 | 5.55 | 35.08 | 0.007 | 细胞质Cytoplasm | 无No |
| Dlo017622 | DlASMT7 | 8 | 244 | 27.16 | 5.59 | 29.58 | -0.029 | 细胞骨架Cytoskeleton | 无No |
| Dlo017625 | DlASMT8 | 8 | 367 | 41.17 | 5.20 | 36.62 | -0.096 | 细胞骨架Cytoskeleton | 无No |
| Dlo017687 | DlASMT9 | 8 | 365 | 40.99 | 5.45 | 32.12 | -0.074 | 细胞质Cytoplasm | 无No |
| Dlo017690 | DlASMT10 | 8 | 364 | 40.91 | 5.52 | 31.91 | -0.074 | 叶绿体Chloroplast | 无No |
| Dlo020696 | DlASMT11 | 9 | 370 | 41.10 | 5.68 | 32.47 | -0.144 | 细胞质Cytoplasm | 无No |
| Dlo021675 | DlASMT12 | 10 | 396 | 44.11 | 5.27 | 36.38 | 0.051 | 细胞质Cytoplasm | 无No |
| Dlo021676 | DlASMT13 | 10 | 163 | 18.04 | 4.80 | 35.40 | -0.007 | 叶绿体Chloroplast | 无No |
| Dlo027273 | DlASMT14 | 13 | 502 | 55.16 | 6.22 | 35.67 | -0.084 | 细胞质Cytoplasm | 无No |
| Dlo027379 | DlASMT15 | 13 | 361 | 40.58 | 5.66 | 22.65 | -0.063 | 细胞质Cytoplasm | 无No |
| Dlo029539 | DlASMT16 | 14 | 359 | 39.66 | 5.64 | 41.86 | -0.133 | 细胞质Cytoplasm | 无No |
| Dlo032559 | DlASMT17 | 15 | 370 | 41.19 | 5.79 | 32.94 | -0.108 | 细胞质Cytoplasm | 无No |
| Dlo032561 | DlASMT18 | 15 | 374 | 41.28 | 5.32 | 32.59 | -0.103 | 细胞质Cytoplasm | 无No |
| Dlo010102 | DlCOMT1 | 4 | 364 | 39.73 | 5.59 | 27.48 | 0.048 | 细胞质Cytoplasm | 无No |
| Dlo010103 | DlCOMT2 | 4 | 375 | 41.29 | 5.41 | 27.60 | 0.139 | 细胞质Cytoplasm | 无No |
| Dlo021674 | DlCOMT3 | 10 | 357 | 40.12 | 5.35 | 34.99 | -0.173 | 细胞质Cytoplasm | 无No |
| Dlo023091 | DlCOMT4 | 11 | 357 | 39.29 | 5.93 | 43.69 | 0.018 | 细胞质Cytoplasm | 无No |
| Dlo027274 | DlCOMT5 | 13 | 351 | 38.88 | 5.43 | 48.29 | 0.027 | 细胞骨架Cytoskeleton | 无No |
| Dlo027275 | DlCOMT6 | 13 | 349 | 38.48 | 6.20 | 39.43 | -0.080 | 细胞骨架Cytoskeleton | 无No |
| Dlo027378 | DlCOMT7 | 13 | 360 | 40.09 | 5.58 | 35.85 | 0.034 | 细胞质Cytoplasm | 无No |
表2 DlSNAT、DlASMT和DlCOMT家族成员蛋白质理化性质
Table 2 The physicochemical properties of the proteins of D1SNAT,D1ASMT and D1COMT family members
| 基因ID Gene-ID | 基因名称 Gene name | 染色体定位 Chromosome location | 氨基酸数 Amino acid number | 分子量/kD Molecular weight | 理论等电点 Theoretical pI | 不稳定系数 The Instabi- lity index | 亲水性 GRAVY | 亚细胞定位 Subcellular location | 信号肽 Signal peptide |
|---|---|---|---|---|---|---|---|---|---|
| Dlo018311 | DlSNAT1 | 8 | 187 | 20.81 | 9.73 | 33.60 | 0.064 | 过氧化物酶体Peroxisome | 无No |
| Dlo030488 | DlSNAT2 | 14 | 225 | 24.68 | 4.89 | 44.33 | -0.070 | 叶绿体Chloroplast | 无No |
| Dlo004250 | DlASMT1 | 2 | 347 | 38.76 | 5.22 | 39.04 | 0.077 | 细胞质Cytoplasm | 无No |
| Dlo011828 | DlASMT2 | 5 | 389 | 43.13 | 6.02 | 35.12 | 0.000 | 细胞质Cytoplasm | 无No |
| Dlo011829 | DlASMT3 | 5 | 442 | 49.48 | 5.77 | 41.16 | -0.223 | 细胞质Cytoplasm | 无No |
| 基因ID Gene-ID | 基因名称 Gene name | 染色体定位 Chromosome location | 氨基酸数 Amino acid number | 分子量/kD Molecular weight | 理论等电点 Theoretical pI | 不稳定系数 The Instabi- Lity index | 亲水性 GRAVY | 亚细胞定位 Subcellular location | 信号肽 Signal peptide |
| Dlo011833 | DlASMT4 | 5 | 365 | 40.35 | 5.39 | 33.80 | -0.099 | 细胞质Cytoplasm | 无No |
| Dlo011834 | DlASMT5 | 5 | 355 | 38.89 | 5.55 | 32.36 | 0.090 | 细胞质Cytoplasm | 无No |
| Dlo011835 | DlASMT6 | 5 | 354 | 39.07 | 5.55 | 35.08 | 0.007 | 细胞质Cytoplasm | 无No |
| Dlo017622 | DlASMT7 | 8 | 244 | 27.16 | 5.59 | 29.58 | -0.029 | 细胞骨架Cytoskeleton | 无No |
| Dlo017625 | DlASMT8 | 8 | 367 | 41.17 | 5.20 | 36.62 | -0.096 | 细胞骨架Cytoskeleton | 无No |
| Dlo017687 | DlASMT9 | 8 | 365 | 40.99 | 5.45 | 32.12 | -0.074 | 细胞质Cytoplasm | 无No |
| Dlo017690 | DlASMT10 | 8 | 364 | 40.91 | 5.52 | 31.91 | -0.074 | 叶绿体Chloroplast | 无No |
| Dlo020696 | DlASMT11 | 9 | 370 | 41.10 | 5.68 | 32.47 | -0.144 | 细胞质Cytoplasm | 无No |
| Dlo021675 | DlASMT12 | 10 | 396 | 44.11 | 5.27 | 36.38 | 0.051 | 细胞质Cytoplasm | 无No |
| Dlo021676 | DlASMT13 | 10 | 163 | 18.04 | 4.80 | 35.40 | -0.007 | 叶绿体Chloroplast | 无No |
| Dlo027273 | DlASMT14 | 13 | 502 | 55.16 | 6.22 | 35.67 | -0.084 | 细胞质Cytoplasm | 无No |
| Dlo027379 | DlASMT15 | 13 | 361 | 40.58 | 5.66 | 22.65 | -0.063 | 细胞质Cytoplasm | 无No |
| Dlo029539 | DlASMT16 | 14 | 359 | 39.66 | 5.64 | 41.86 | -0.133 | 细胞质Cytoplasm | 无No |
| Dlo032559 | DlASMT17 | 15 | 370 | 41.19 | 5.79 | 32.94 | -0.108 | 细胞质Cytoplasm | 无No |
| Dlo032561 | DlASMT18 | 15 | 374 | 41.28 | 5.32 | 32.59 | -0.103 | 细胞质Cytoplasm | 无No |
| Dlo010102 | DlCOMT1 | 4 | 364 | 39.73 | 5.59 | 27.48 | 0.048 | 细胞质Cytoplasm | 无No |
| Dlo010103 | DlCOMT2 | 4 | 375 | 41.29 | 5.41 | 27.60 | 0.139 | 细胞质Cytoplasm | 无No |
| Dlo021674 | DlCOMT3 | 10 | 357 | 40.12 | 5.35 | 34.99 | -0.173 | 细胞质Cytoplasm | 无No |
| Dlo023091 | DlCOMT4 | 11 | 357 | 39.29 | 5.93 | 43.69 | 0.018 | 细胞质Cytoplasm | 无No |
| Dlo027274 | DlCOMT5 | 13 | 351 | 38.88 | 5.43 | 48.29 | 0.027 | 细胞骨架Cytoskeleton | 无No |
| Dlo027275 | DlCOMT6 | 13 | 349 | 38.48 | 6.20 | 39.43 | -0.080 | 细胞骨架Cytoskeleton | 无No |
| Dlo027378 | DlCOMT7 | 13 | 360 | 40.09 | 5.58 | 35.85 | 0.034 | 细胞质Cytoplasm | 无No |
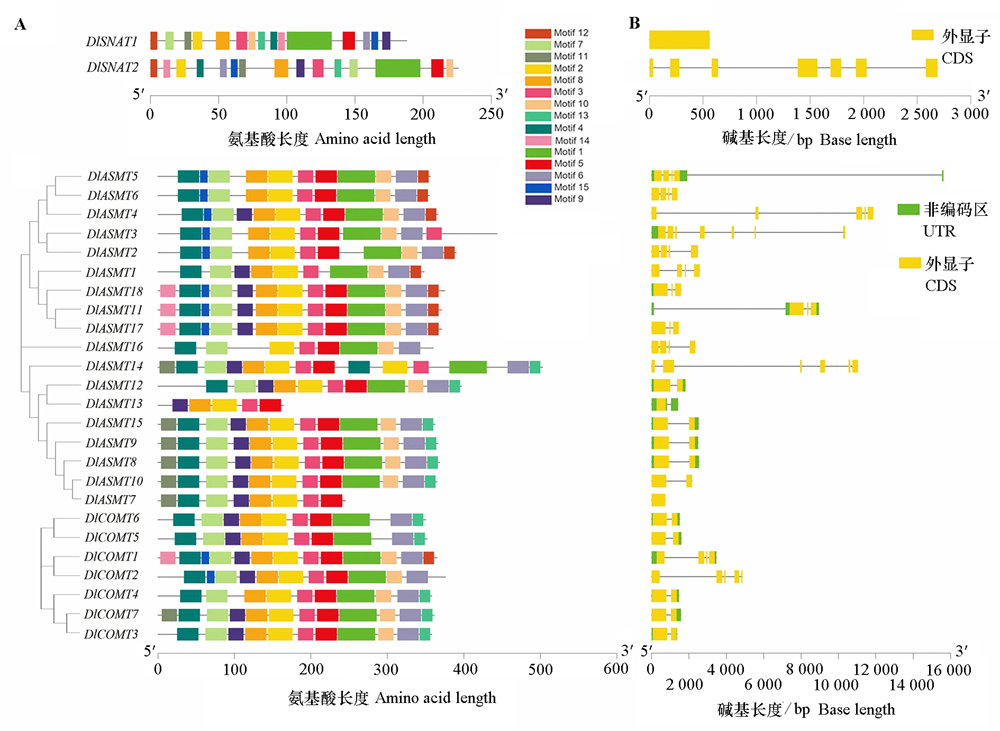
图2 DlSNAT、DlASMT和DlCOMT家族成员保守motif分布(A)和基因结构(B)
Fig.2 The conservative motif distribution(A)and gene structures(B)of DlSNAT,DlASMT and DlCOMT family members

图3 龙眼(Dl)、拟南芥(At)、水稻(Os)、小麦(Ta)、番茄(Sl)、辣椒(Ca)和卷柏(St)SNAT、ASMT和COMT家族成员进化树
Fig.3 The evolutionary tree of SNAT,ASMT and COMT family members of longan(Dl),Arabidopsis(At),rice(Os),wheat(Ta),tomato(Sl),pepper(Ca)and Selaginella(St)

图5 DlSNAT、DlASMT和DlCOMT家族成员在龙眼体胚发生早期不同阶段特异性表达的FPKM值(A)和qRT-PCR检测值(B) EC:愈伤组织;ICpEC:不完全胚性紧实结构;GE:球形胚。
Fig. 5 FPKM value(A)and the value detected by qRT-PCR(B)of specific expression of DlSNAT,DlASMT and DlCOMT family members in the early stages of somatic embryogenesis in longan EC:Embryonic callus;ICpEC:Incomplete embryonic compact structure;GE:Globular embryo.
| 基因Gene | P | Pearson相关系数 Pearson correlation |
|---|---|---|
| DlASMT4 | 0.029 | 0.999 |
| DlASMT11 | 0.010 | 1.000 |
| DlCOMT3 | 0.048 | 0.997 |
表3 DlASMT4、DlASMT11、DlCOMT3在龙眼体胚发生早期的FPKM值和相对表达量变化的相关性检测
Table 3 The Pearson correlation between the change of the FPKM and the relative expression of DlASMT4,DlASMT11 and DlCOMT3 in the early stages of somatic embryogenesis in longan
| 基因Gene | P | Pearson相关系数 Pearson correlation |
|---|---|---|
| DlASMT4 | 0.029 | 0.999 |
| DlASMT11 | 0.010 | 1.000 |
| DlCOMT3 | 0.048 | 0.997 |
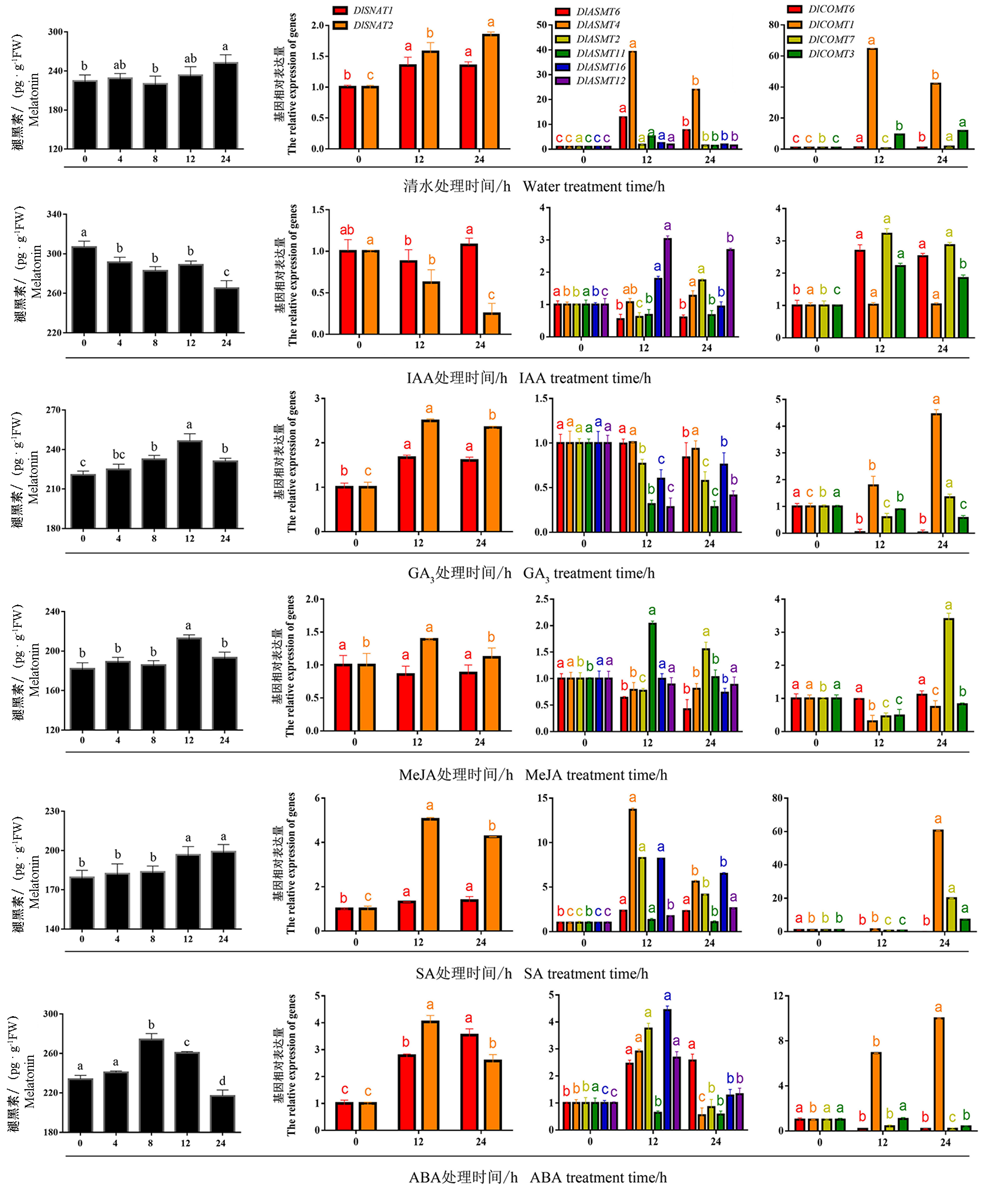
图6 外源IAA、GA3、MeJA、SA和ABA处理龙眼EC内源褪黑素含量及合成相关基因表达变化 不同小写字母代表处理间显著差异(P ≤ 0.05)。
Fig. 6 The endogenous melatonin content changes(A1-F1)and the expression changes of related synthetic genes(A2-F2) in longan EC treated with IAA,GA3,MeJA,SA and ABA treatments Different lowercase letters in the picture represent significant differences between different treatments(P ≤ 0.05).
| 处理Treatment | P | Pearson相关系数 Pearson correlation |
|---|---|---|
| IAA | 0.050 | 0.997 |
| MeJA | 0.045 | 0.997 |
表4 IAA和MeJA处理下的龙眼愈伤组织DlSNAT2相对表达量和褪黑素含量变化的相关性
Table 4 The Pearson correlation between the change of relative expression of DlSNAT2 and the melatonin content under the IAA and MeJA treatments in longan callus
| 处理Treatment | P | Pearson相关系数 Pearson correlation |
|---|---|---|
| IAA | 0.050 | 0.997 |
| MeJA | 0.045 | 0.997 |
| [1] |
Arnao M B, Hernández-Ruiz, Josefa. 2015. Functions of melatonin in plants:a review. Journal of Pineal Research, 59 (2):133-150.
doi: 10.1111/jpi.12253 URL |
| [2] |
Arnao M B, Hernández-Ruiz, Josefa. 2014. Melatonin:plant growth regulator and/or biostimulator during stress? Trends in Plant Science, 19 (12):789-797.
doi: 10.1016/j.tplants.2014.07.006 URL |
| [3] |
Back K, Tan D X, Reiter R J. 2016. Melatonin biosynthesis in plants:multiple pathways catalyze tryptophan to melatonin in the cytoplasm or chloroplasts. Journal of Pineal Research, 61 (4):426-437.
doi: 10.1111/jpi.12364 URL |
| [4] |
Byeon Y, Back K. 2015. Molecular cloning of melatonin 2-hydroxylase responsible for 2-hydroxymelatonin production in rice(Oryza sativa). Journal of Pineal Research, 58 (3):343-351.
doi: 10.1111/jpi.12220 URL |
| [5] |
Byeon Y, Lee H Y, Lee K, Park S, Back K. 2014. Caffeic acid O-methyltransferase is involved in the synthesis of melatonin by methylating N-acetylserotonin in Arabidopsis. Journal of Pineal Research, 57 (2):219-227.
doi: 10.1111/jpi.12160 URL |
| [6] |
Byeon Y, Lee H Y, Hwang O J, Lee H J, Lee K, Back K. 2015. Coordinated regulation of melatonin synthesis and degradation genes in rice leaves in response to cadmium treatment. Journal of Pineal Research, 58 (4):470-478.
doi: 10.1111/jpi.12232 URL |
| [7] |
Chen C J, Chen H, Zhang Y, Hannah R T, Margaret H F, He Y H, Xia R. 2020. Tbtools:an integrative toolkit developed for interactive analyses of big biological data. Molecular Plant,DOI: 10.1016/j.molp.2020.06.009.
doi: 10.1016/j.molp.2020.06.009 URL |
| [8] |
Debnath B, Islam W, Li M, Sun Y T, Lu X C, Mitra S. 2019. Melatonin mediates enhancement of stress tolerance in plants. International Journal of Molecular Sciences, 20 (5):1040.
doi: 10.3390/ijms20051040 URL |
| [9] | Dodd A, Salathia N, Hall A, Kevei E, Toth R, Nagy F, Hibberd J, Millar A, Webb A A R. 2005. Plant circadian clocks increase photosynthesis,growth,survival,and competitive advantage. Science, 309:633. |
| [10] |
Galano A, Tan D X, Reiter R J. 2011. Melatonin as a natural ally against oxidative stress:a physicochemical examination. Journal of Pineal Research, 51 (1):1-16.
doi: 10.1111/j.1600-079X.2011.00916.x pmid: 21752095 |
| [11] |
Ganguly S, Coon S L, Klein D C. 2002. Control of melatonin synthesis in the mammalian pineal gland:the critical role of serotonin acetylation. Cell and Tissue Research, 309:127-137.
doi: 10.1007/s00441-002-0579-y URL |
| [12] |
Hernandez-Ruiz J, Cano A, Arnao M B. 2010. Melatonin acts as a growth-stimulating compound in some monocot species. Journal of Pineal Research, 39:137-142.
doi: 10.1111/j.1600-079X.2005.00226.x URL |
| [13] | Kang K, Lee K, Park S, Kim Y S, Back K. 2010. Enhanced production of melatonin by ectopic overexpression of human serotonin N-acetyltransferase plays a role in cold resistance in transgenic rice seedlings. Journal of Pineal Research, 49 (2). |
| [14] |
Kang K, Kong K, Park S, Natsagdorj U, Kim Y S, Back K. 2011. Molecular cloning of a plant N-acetylserotonin methyltransferase and its expression characteristics in rice. Journal of Pineal Research, 50:304-309.
doi: 10.1111/j.1600-079X.2010.00841.x URL |
| [15] | Kolar J, Johnson C H, Machackova I. 1999. Presence and possible role of melatonin in a short-day flowering plant, Chenopodium rubrum. Advances in Experimental Medicine and Biology, 460:391-393. |
| [16] | Lai Zhong-xiong, Chen Chun-ling, Huang Su-hua, Sang Qing-liang, Pan Dong-ming, Chen Zhen-guang. 2001. Long-term subculture of embryogenic calli and their genetic variation of thechromosome number in longan. Journal of Fujian Agricultural University,(1):30-33. (in Chinese) |
| 赖钟雄, 陈春玲, 黄素华, 桑庆亮, 潘东明, 陈振光. 2001. 龙眼胚性愈伤组织长期继代培养及其染色体数目变异. 福建农业大学学报,(1):30-33. | |
| [17] |
Lee Y J, Kim B G, Chong Y, Lim Y, Ahn J H. 2008. Cation dependent O-methyltransferase from rice. Planta, 227:641-647.
doi: 10.1007/s00425-007-0646-4 URL |
| [18] | Liang D, Shen Y, Ni Z, Wang Q, Lei Z, Xu N, Deng Q, Lin L, Jin W, Lv X. 2018. Exogenous melatonin application delays senescence of kiwifruit leaves by regulating the antioxidant capacity and biosynthesis of flavonoids. Frontiers in Plant ence, 9:426. |
| [19] |
Li M Q, Hasan M K, Li C X, Ahammed G J, Xia X J, Shi K, Zhou Y H, Reiter R J, Yu J Q, Xu M X, Zhou J. 2016. Melatonin mediates selenium-induced tolerance to cadmium stress in tomato plants. Journal of Pineal Research, 61:291-302.
doi: 10.1111/jpi.12346 URL |
| [20] |
Lin Y L, Lai Z X. 2010. Reference gene selection for qPCR analysis during somatic embryogenesis in longan tree. Plant Science, 178 (4):359-365.
doi: 10.1016/j.plantsci.2010.02.005 URL |
| [21] |
Liu T, Borjigin J. 2005. N-acetyltransferase is not the rate-limiting enzyme of melatonin synthesis at night. Journal of Pineal Research, 39:91-96.
doi: 10.1111/j.1600-079X.2005.00223.x URL |
| [22] |
Liu W, Zhao D, Zheng C, Chen C, Xin P, Yuan C, Wan H. 2017. Genomic analysis of the ASMT gene family in Solanum lycopersicum. Molecules, 22 (11):1984.
doi: 10.3390/molecules22111984 URL |
| [23] |
Mandal M K, Suren H, Ward B, Boroujerdi A, Kousik C. 2018. Differential roles of melatonin in plant-host resistance and pathogen suppression in cucurbits. Journal of Pineal Research, 65:12505.
doi: 10.1111/jpi.12505 pmid: 29766569 |
| [24] |
Mauriz J L, Collado P S, Veneroso C, Reiter R J, Gonzalez-Gallego J. 2013. A review of the molecular aspects of melatonin’s anti-inflammatory actions:recent insights and new perspectives. Journal of Pineal Research, 54 (1):1-14.
doi: 10.1111/j.1600-079X.2012.01014.x pmid: 22725668 |
| [25] |
Morteza S A, Luo Z S, Jannatizadeh A, Sheikh-Assadi M, Sharafi Y, Farmani B, Fard J R, Razavi F. 2019. Employing exogenous melatonin applying confers chilling tolerance in tomato fruits by upregulating ZAT2/6/12 giving rise to promoting endogenous polyamines,proline,and nitric oxide accumulation by triggering arginine pathway activity. Food Chemistry, 275:549-556.
doi: 10.1016/j.foodchem.2018.09.157 URL |
| [26] |
Murch S J, KrishnaRaj S, Saxena P K. 2000. Tryptophan is a precursor for melatonin and serotonin biosynthesis in in vitro regenerated St. John’s wort(Hypericum perforatum L. cv. Anthos)plants. Plant Cell Reports, 19 (7):698-704.
doi: 10.1007/s002990000206 pmid: 30754808 |
| [27] |
Park S, Byeon Y, Back K. 2013. Functional analyses of three ASMT gene family members in rice plants. Journal of Pineal Research, 55 (4):409-415.
doi: 10.1111/jpi.12088 URL |
| [28] | Pan L Z, Zheng J Q, Liu J, Guo J, Fawan, Lecheng. 2019. Analysis of the ASMT gene family in pepper(Capsicum annuum L.):identification,phylogeny,and expression profiles. International Journal of Genomics. |
| [29] |
Rajniak J, Barco B, Clay N K, Sattely E S. 2015. A new cyanogenic metabolite in Arabidopsis required for inducible pathogen defence. Nature, 525:376-395.
doi: 10.1038/nature14907 URL |
| [30] |
Tan D X, Hardeland R, Back K, Manchester L C, Alatorre-Jimenez M A, Reiter R J. 2016. On the significance of an alternate pathway of melatonin synthesis via 5-methoxytryptamine:comparisons across species. Journal of Pineal Research, 61:27-40.
doi: 10.1111/jpi.12336 URL |
| [31] |
Tan D X, Manchester L C, Esteban-Zubero E, Zhou Z, Reiter R J. 2015. Melatonin as a potent and inducible endogenous antioxidant:synthesis and metabolism. Molecules, 20 (10):18886-18906.
doi: 10.3390/molecules201018886 URL |
| [32] |
Wei W, Li Q T, Chu Y N, Reiter R J, Yu X M, Zhu D H, Zhang W K, Ma B, Lin Q, Zhang J S, Chen S Y. 2015. Melatonin enhances plant growth and abiotic stress tolerance in soybean plants. Journal of Experimental Botany, 66:695-707.
doi: 10.1093/jxb/eru392 pmid: 25297548 |
| [33] |
Xie T, Chen C J, Li C H, Liu J R, Liu C Y, He Y H. 2018. Genome-wide investigation of WRKY gene family in pineapple:evolution and expression profiles during development and stress. BMC Genomics, 19 (1):490.
doi: 10.1186/s12864-018-4880-x URL |
| [34] | Ya Rong, Xu Weirong, Zhang Ying, Xia Siqi, Zhang Ningbo. 2020. Investigation of melatonin on somatic embryo induction for‘Thompson Seedless’grapevine. Acta Horticulturae Sinica, 47 (5):142-151. (in Chinese) |
| 雅蓉, 徐伟荣, 张莹, 夏思琪, 张宁波. 2020. 褪黑素对‘无核白’葡萄体细胞胚的诱导作用. 园艺学报, 47 (5):142-151. | |
| [35] | Zhang Lai-jun, Jia Jing-fen. 2013. Effect of melatonin on the proliferation and differentiation of calli of Scutellaria amoena. Northern Horticulture,(8):106-109. (in Chinese) |
| 张来军, 贾敬芬. 2013. 外源褪黑素对滇黄芩愈伤组织增殖和分化的影响. 北方园艺,(8):106-109. |
| [1] | 赵雪艳, 王琪, 王莉, 王方圆, 王庆, 李艳. 基于比较转录组的延胡索组织差异性表达分析[J]. 园艺学报, 2023, 50(1): 177-187. |
| [2] | 邓朝军, 许奇志, 蒋际谋, 胡文舜, 郑少泉, 陈秀萍, 姜 帆, 许家辉, 苏文炳, 张雅玲, 黄敬峰. 浓香型龙眼新品种‘醇香’[J]. 园艺学报, 2022, 49(S2): 75-76. |
| [3] | 邓朝军, 陈秀萍, 许奇志, 蒋际谋, 郑少泉, 胡文舜, 姜 帆, 许家辉, 苏文炳, 张雅玲, 黄敬峰. 浓香型龙眼新品种‘福香’[J]. 园艺学报, 2022, 49(S2): 77-78. |
| [4] | 徐小萍, 曹清影, 蔡柔荻, 官庆栩, 张梓浩, 陈裕坤, 徐涵, 林玉玲, 赖钟雄. 龙眼miR408与DlLAC12克隆及其在球形胚发生和非生物胁迫下的表达分析[J]. 园艺学报, 2022, 49(9): 1866-1882. |
| [5] | 高彦龙, 吴玉霞, 张仲兴, 王双成, 张瑞, 张德, 王延秀. 苹果ELO家族基因鉴定及其在低温胁迫下的表达分析[J]. 园艺学报, 2022, 49(8): 1621-1636. |
| [6] | 邱子文, 刘林敏, 林永盛, 林晓洁, 李永裕, 吴少华, 杨超. 千层金MbEGS基因的克隆与功能分析[J]. 园艺学报, 2022, 49(8): 1747-1760. |
| [7] | 郑林, 王帅, 刘语诺, 杜美霞, 彭爱红, 何永睿, 陈善春, 邹修平. 柑橘响应黄龙病菌侵染的NAC基因的克隆及表达分析[J]. 园艺学报, 2022, 49(7): 1441-1457. |
| [8] | 马维峰, 李艳梅, 马宗桓, 陈佰鸿, 毛娟. 苹果POD家族基因的鉴定与MdPOD15的功能分析[J]. 园艺学报, 2022, 49(6): 1181-1199. |
| [9] | 张凯, 麻明英, 王萍, 李益, 金燕, 盛玲, 邓子牛, 马先锋. 柑橘HSP20家族基因鉴定及其响应溃疡病菌侵染表达分析[J]. 园艺学报, 2022, 49(6): 1213-1232. |
| [10] | 梁晨, 孙如意, 向锐, 孙艺萌, 师校欣, 杜国强, 王莉. 葡萄生长调控因子GRF家族基因的鉴定及表达分析[J]. 园艺学报, 2022, 49(5): 995-1007. |
| [11] | 向妙莲, 吴帆, 李树成, 马巧利, 王印宝, 肖刘华, 陈金印, 陈明. 外源褪黑素调控活性氧代谢诱导梨果实抗采后黑斑病[J]. 园艺学报, 2022, 49(5): 1102-1110. |
| [12] | 潘鑫峰, 叶方婷, 毛志君, 李兆伟, 范凯. 睡莲WRKY家族的全基因组鉴定和分子进化分析[J]. 园艺学报, 2022, 49(5): 1121-1135. |
| [13] | 高玮林, 张力曼, 薛超玲, 张垚, 刘孟军, 赵锦. 枣E类MADS基因在花和果中的表达及其蛋白互作研究[J]. 园艺学报, 2022, 49(4): 739-748. |
| [14] | 夏铭, 李经纬, 罗章瑞, 祖贵东, 王娅, 张万萍. 外源褪黑素影响萝卜生长及对链格孢菌抗性的机理研究[J]. 园艺学报, 2022, 49(3): 548-560. |
| [15] | 刘梦雨, 蒋梦琦, 陈燕, 张舒婷, 薛晓东, 肖学宸, 赖钟雄, 林玉玲. 龙眼GDSL酯酶/脂肪酶基因的全基因组鉴定及表达分析[J]. 园艺学报, 2022, 49(3): 597-612. |
| 阅读次数 | ||||||
|
全文 |
|
|||||
|
摘要 |
|
|||||
版权所有 © 2012 《园艺学报》编辑部 京ICP备10030308号-2 国际联网备案号 11010802023439
编辑部地址: 北京市海淀区中关村南大街12号中国农业科学院蔬菜花卉研究所 邮编: 100081
电话: 010-82109523 E-Mail: yuanyixuebao@126.com
技术支持:北京玛格泰克科技发展有限公司