
园艺学报 ›› 2022, Vol. 49 ›› Issue (4): 739-748.doi: 10.16420/j.issn.0513-353x.2021-0179
高玮林1, 张力曼1, 薛超玲1, 张垚1, 刘孟军2, 赵锦1,*( )
)
收稿日期:2021-11-18
修回日期:2022-01-18
出版日期:2022-04-25
发布日期:2022-04-24
通讯作者:
赵锦
E-mail:zhaojinbd@126.com
基金资助:
GAO Weilin1, ZHANG Liman1, XUE Chaoling1, ZHANG Yao1, LIU Mengjun2, ZHAO Jin1,*( )
)
Received:2021-11-18
Revised:2022-01-18
Online:2022-04-25
Published:2022-04-24
Contact:
ZHAO Jin
E-mail:zhaojinbd@126.com
摘要:
以‘金丝小枣’花和果实为试材进行E类MADS-box基因的生物信息、表达、亚细胞定位及蛋白互作研究。3个E-type MADS-box基因分别位于SEP1/2、SEP3和SEP4进化支。与其他物种的同源序列相比,E类蛋白在进化中非常保守。3个E类基因在枣的花和果实中均有表达,ZjSEP1/2在花不同发育时期表达比较稳定,ZjSEP3和ZjSEP4在花发育前期高表达,在感染枣疯病的花变叶中3个基因的表达均显著受抑制。3个蛋白均定位于细胞核。酵母单杂交结果表明ZjSEP1/2和ZjSEP3具有转录激活活性,可以作为转录因子调控下游基因,而ZjSEP4不具有此活性。酵母双杂交结果表明3个E类MADS蛋白都可以和A类、C/D类蛋白发生互作,ZjSEP4与ZjSEP1/2均可与自身互作,但ZjSEP3自身不发生互作。综上,3个E类MADS基因在枣花发育中具有重要作用,但功能和调控模式不同,ZjSEP3不能形成同源二聚体,ZjSEP1/2、ZjSEP4均可形成同源二聚体,3个E类蛋白均可与A、C/D类蛋白形成异源二聚体。
中图分类号:
高玮林, 张力曼, 薛超玲, 张垚, 刘孟军, 赵锦. 枣E类MADS基因在花和果中的表达及其蛋白互作研究[J]. 园艺学报, 2022, 49(4): 739-748.
GAO Weilin, ZHANG Liman, XUE Chaoling, ZHANG Yao, LIU Mengjun, ZHAO Jin. Expression of E-type MADS-box Genes in Flower and Fruits and Protein Interaction Analysis in Chinese Jujube[J]. Acta Horticulturae Sinica, 2022, 49(4): 739-748.
| 用途 Usage | 基因名称 Gene name | 正向序列(5′-3′) Forward primer sequence | 反向序列(5′-3′) Reverse primer sequence |
|---|---|---|---|
| q-RT-PCR Quantitative real-time PCR | ZjSEP1/2 | GGGGAGAGGTAAGGTGGAG | GGTTGGAGAAGATGATAAGAGC |
| ZjSEP3 | GGGGAAGACCTTGGACCTTT | AGCAAGTGTTCCTTGCGTTG | |
| ZjSEP4 | GGGGTTCTTCCAGGCCTTAG | ACTTGCTGGGCATGACTTGT | |
| 内参 Primers for internal control | ZjACT | AGCCTTCCTGCCAACGAGT | TTGCTTCTCACCCTTGATGC |
| 酵母双杂交 Yeast two- Hybrid(Y2H) | ZjBDZjSEP1/2 | ATGGCCATGGAGGCCGATGGGGAGAGGTAAGGTGGA | GGATCCCCGGGAATTTCAAAGCATCCACCCAGGG |
| ZjBDZjSEP3 | GGATCCCCGGGAATTTCAAAGCATCCACCCAGGG | GGATCCCCGGGAATTTCATGGTAACCATCCTGGCA | |
| ZjBDZjSEP4 | ATGGCCATGGAGGCCGATGGGGAGGGGTAGAGTTGA | GGATCCCCGGGAATTTCAAAGCATCCATCCAGGAA | |
| ZjADZjSEP1/2 | CCATGGAGGCCAGTGATGGGGAGAGGTAAGGTGGA | CACCCGGGTGGAATTTCAAAGCATCCACCCAGGG | |
| ZjADZjSEP3 | CCATGGAGGCCAGTGATGGGAAGAGGTAGAGTGGA | CACCCGGGTGGAATTTCATGGTAACCATCCTGGCA | |
| ZjADZjSEP4 | CCATGGAGGCCAGTGATGGGGAGGGGTAGAGTTGA | CACCCGGGTGGAATTTCAAAGCATCCATCCAGGAA | |
| ZjADZjMADS46 | CCATGGAGGCCAGTGATGGAGTTCCCAAATCAAGCA | CACCCGGGTGGAATTTCAAACAAGTTGGAGAGCTG | |
| ZjADMADS24 | CCATGGAGGCCAGTGATGGGGAGGGGAAGGGTTC | CACCCGGGTGGAATTGCATCCAAGCGGGCAGGAG | |
| 亚细胞定位 Subcellular localization | ZjSEP1/2 | AGCTCGGGTACCCGGGATGGGGAGAGGTAAGGTGGA | TCTAGAGGATCAAGCATCCACCCAGGG |
| ZjSEP3 | AGCTCGGGTACCCGGGATGCTCAAAACACTTGAGA | TCTAGAGGATCTGGTAACCATCCTGGCAT | |
| ZjSEP4 | AGCTCGGGTACCCGGGATGGGGAGGGGTAGAGTTGA | TCTAGAGGATCAAGCATCCATCCAGGAA |
表1 本研究中所用引物名称及序列
Table 1 Primer names and sequences used in this study
| 用途 Usage | 基因名称 Gene name | 正向序列(5′-3′) Forward primer sequence | 反向序列(5′-3′) Reverse primer sequence |
|---|---|---|---|
| q-RT-PCR Quantitative real-time PCR | ZjSEP1/2 | GGGGAGAGGTAAGGTGGAG | GGTTGGAGAAGATGATAAGAGC |
| ZjSEP3 | GGGGAAGACCTTGGACCTTT | AGCAAGTGTTCCTTGCGTTG | |
| ZjSEP4 | GGGGTTCTTCCAGGCCTTAG | ACTTGCTGGGCATGACTTGT | |
| 内参 Primers for internal control | ZjACT | AGCCTTCCTGCCAACGAGT | TTGCTTCTCACCCTTGATGC |
| 酵母双杂交 Yeast two- Hybrid(Y2H) | ZjBDZjSEP1/2 | ATGGCCATGGAGGCCGATGGGGAGAGGTAAGGTGGA | GGATCCCCGGGAATTTCAAAGCATCCACCCAGGG |
| ZjBDZjSEP3 | GGATCCCCGGGAATTTCAAAGCATCCACCCAGGG | GGATCCCCGGGAATTTCATGGTAACCATCCTGGCA | |
| ZjBDZjSEP4 | ATGGCCATGGAGGCCGATGGGGAGGGGTAGAGTTGA | GGATCCCCGGGAATTTCAAAGCATCCATCCAGGAA | |
| ZjADZjSEP1/2 | CCATGGAGGCCAGTGATGGGGAGAGGTAAGGTGGA | CACCCGGGTGGAATTTCAAAGCATCCACCCAGGG | |
| ZjADZjSEP3 | CCATGGAGGCCAGTGATGGGAAGAGGTAGAGTGGA | CACCCGGGTGGAATTTCATGGTAACCATCCTGGCA | |
| ZjADZjSEP4 | CCATGGAGGCCAGTGATGGGGAGGGGTAGAGTTGA | CACCCGGGTGGAATTTCAAAGCATCCATCCAGGAA | |
| ZjADZjMADS46 | CCATGGAGGCCAGTGATGGAGTTCCCAAATCAAGCA | CACCCGGGTGGAATTTCAAACAAGTTGGAGAGCTG | |
| ZjADMADS24 | CCATGGAGGCCAGTGATGGGGAGGGGAAGGGTTC | CACCCGGGTGGAATTGCATCCAAGCGGGCAGGAG | |
| 亚细胞定位 Subcellular localization | ZjSEP1/2 | AGCTCGGGTACCCGGGATGGGGAGAGGTAAGGTGGA | TCTAGAGGATCAAGCATCCACCCAGGG |
| ZjSEP3 | AGCTCGGGTACCCGGGATGCTCAAAACACTTGAGA | TCTAGAGGATCTGGTAACCATCCTGGCAT | |
| ZjSEP4 | AGCTCGGGTACCCGGGATGGGGAGGGGTAGAGTTGA | TCTAGAGGATCAAGCATCCATCCAGGAA |
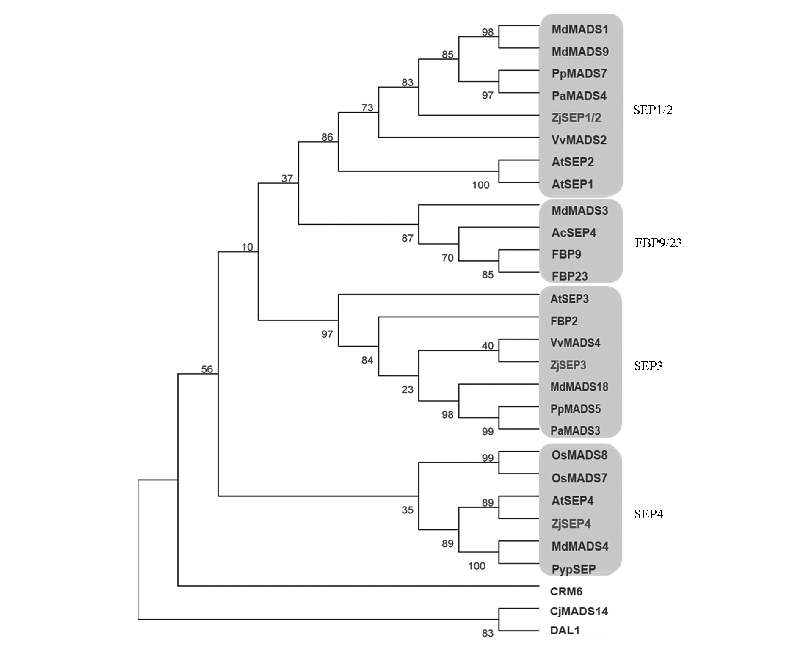
图1 枣和其他物种的E类蛋白系统进化树 At:拟南芥;Md:苹果;Pp:桃;Pa:樱桃;Ac:猕猴桃;Vv:葡萄;Pyp:沙梨;Os:水稻;Cj:日本柳杉;DAL1:欧洲云杉;CRM6:水蕨;FBP:矮牵牛。
Fig. 1 Phylogenetic tree of E-type proteins from Chinese jujube and other species At:Arabidopsis thaliana;Md:Malus × domestica;Pp:Prunus persica;Pa:Prunus avium;Ac:Actinidia chinensis;Vv:Vitis vinifera;Pyp:Pyrus pyrifolia;Os:Oryza sativa;Cj:Cryptomeria japonica;DAL1:Picea abies;CRM6:Ceratopteris pteridoides;FBP:Petunia hybrida.

图2 枣E类基因在不同发育时期花(A)、果实(B)和四轮花器官(C)中的表达模式 根据邓肯氏多组样本差异显著性分析,不同小写字母表示每个基因的表达在花、果实不同发育阶段和不同花器官中差异显著(P < 0.05)。
Fig. 2 Expression pattern of jujube E-type genes in flowers(A),fruit(B)and four-whorl floral organs(C) According to the Duncan’s multiple range test,the different letters above the column indicate the significant differences in flower,fruit at different development stages and four-wheel flower organs for each gene(P < 0.05).
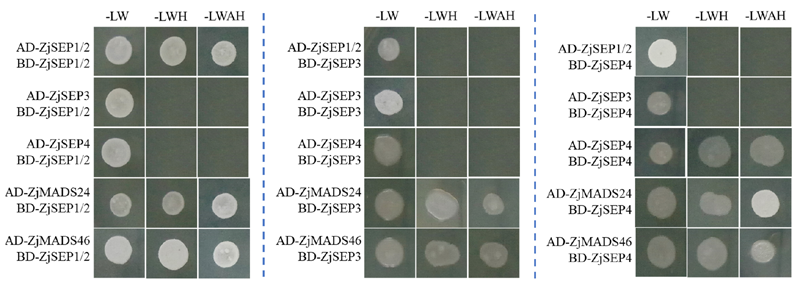
图6 枣E类MADS蛋白的酵母双杂交筛选
Fig. 6 Yeast two-hybrid results of E-type proteins in Chinese jujube -LW:SD/-Trp/-Leu;-LWH:SD/-Trp/-Leu/-His;-LWAH:SD/-Trp/-Leu/-His/-Ade.
| [1] |
Becker A, Theissen G. 2003. The major clades of MADS-box genes and their role in the development and evolution of flowering plants. Molecular Phylogenetics and Evolution, 29 (3):464-489.
doi: 10.1016/S1055-7903(03)00207-0 URL |
| [2] |
Coen E S, Meyerowitz E M. 1991. The war of the whorls:genetic interactions controlling flower development. Nature, 353 (6339):31-37.
doi: 10.1038/353031a0 URL |
| [3] |
Ditta G, Pinyopich A, Robles P, Pelaz S, Yanofsky M F. 2004. The SEP 4 gene of Arabidopsis thaliana functions in floral organ and meristem identity. Current Biology, 14 (21):1935-1940.
pmid: 15530395 |
| [4] |
Günter Theißen. 2001. Development of floral organ identity:stories from the MADS house. Current Opinion in Plant Biology, 4 (1):75-85.
doi: 10.1016/S1369-5266(00)00139-4 URL |
| [5] |
Honma T, Goto K. 2001. Complexes of MADS-box proteins are sufficient to convert leaves into floral organs. Nature, 409 (6819):525-529.
doi: 10.1038/35054083 URL |
| [6] | Huang Fang. 2007. Cloning and functional analysis of flower development related genes in soybean(Glycine max L.)[Ph. D. Dissertation]. Nanjing: Nanjing Agricultural University. (in Chinese) |
| 黄方. 2007. 大豆花发育相关基因的克隆与功能研究[博士论文]. 南京: 南京农业大学. | |
| [7] |
Huang J, Zhang C, Zhao X, Fei Z, Wan K, Zhang Z, Pang X, Yin X, Bai Y, Sun X, Gao L, Li R, Zhang J, Li X. 2016. The jujube genome provides insights into genome evolution and the domestication of sweetness/acidity taste in fruit trees. PLoS Genetics, 12 (12):e1006433.
doi: 10.1371/journal.pgen.1006433 URL |
| [8] |
Immink R G, Tonaco I A, de Folter S, Shchennikova A, van Dijk A D, Busscher-Lange J, Borst J W, Angenent G C. 2009. SEPALLATA3:the‘glue’for MADS box transcription factor complex formation. Genome Biology, 10 (2):R24.
doi: 10.1186/gb-2009-10-2-r24 URL |
| [9] |
Ireland H S, Yao J L, Tomes S, Sutherland P W, Nieuwenhuizen N, Gunaseelan K, Winz R A, David K M, Schaffer R J. 2013. Apple SEPALLATA1/2-like genes control fruit flesh development and ripening. Plant Journal, 73 (6):1044-1056.
doi: 10.1111/tpj.12094 URL |
| [10] |
Kobayashi K, Yasuno N, Sato Y, Yoda M, Yamazaki R, Kimizu M, Yoshida H, Nagamura Y, Kyozuka J. 2012. Inflorescence meristem identity in rice is specified by overlapping functions of three AP 1 /FUL-like MADS box genes and PAP2,a SEPALLATA MADS box gene. Plant Cell, 24 (5):1848-1859.
doi: 10.1105/tpc.112.097105 URL |
| [11] |
Krizek B A, Meyerowitz E M. 1996. The Arabidopsis homeotic genes APETALA3 and PISTILLATA are sufficient to provide the B class organ identity function. Development, 122 (1):11-22.
pmid: 8565821 |
| [12] | Li Chengru, Dong Na, Li Xiaoping, Wu Shasha, Liu Zhongjian, Zhai Junwen. 2020. A review of MADS-box genes,the molecular regulatory genes for floral organ development in Orchidaceae. Acta Horticulturae Sinica, 47 (10):2047-2062. (in Chinese) |
| 李成儒, 董钠, 李笑平, 吴沙沙, 刘仲健, 翟俊文. 2020. 兰科植物花发育调控MADS-box基因家族研究进展. 园艺学报, 47 (10):2047-2062. | |
| [13] |
Liao Y T, Lin S S, Lin S J, Shen B N, Cheng H P, Lin C P. 2019. Structural insights into the interaction between phytoplasmal effector causing phyllody 1(PHYL1)and MADS transcription factor. Plant Journal, 100 (4):706-719.
doi: 10.1111/tpj.14463 URL |
| [14] |
Lin X, Wu F, Du X, Shi X, Liu Y, Liu S, Hu Y, Theissen G, Meng Z. 2014. The pleiotropic SEPALLATA-like gene OsMADS34 reveals that the‘empty glumes’of rice(Oryza sativa)spikelets are in fact rudimentary lemmas. New Phytologist, 202 (2):689-702.
doi: 10.1111/nph.12657 URL |
| [15] | Liu M J, Zhao J, Cai Q L, Liu G C, Wang J R, Zhao Z H, Liu P, Dai L, Yan G, Wang W J, Li X S, Chen Y, Sun Y D, Liu Z G, Lin M J, Xiao J, Chen Y Y, Li X F, Wu B, Ma Y, Jian J B, Yang W, Yuan Z, Sun X C, Wei Y L, Yu L L, Zhang C, Liao S G, He R J, Guang X M, Wang Z, Zhang Y Y, Luo L H. 2014. The complex jujube genome provides insights into fruit tree biology. Nature Communications, 28 (5):5315. |
| [16] | Liu Shinan, Qi Tiantian, Ma Jingjing, Ma Lyuyi, Lin Xinchun. 2016. Cloning and functional analysis of SEP-like gene from phyllostachys violascens. Journal of Nuclear Agricultural Sciences, 30 (8):1453-1459. (in Chinese) |
| 刘世男, 戚田田, 马晶晶, 马履一, 林新春. 2016. 雷竹SEP-like基因的克隆及功能分析. 核农学报, 30 (8):1453-1459. | |
| [17] |
Ma H, Yanofsky M F, Meyerowitz E M. 1991. AGL1AGL6,an Arabidopsis gene family with similarity to floral hometic and transcription factor genes. Genes and Development, 5 (3):484-495.
pmid: 1672119 |
| [18] | MacLean A M, Sugio A, Makarova O V, Findlay K C, Grieve V M. 2011. Phytoplasma effector SAP54 induces indeterminate leaf-like flower development in Arabidopsis plants. Plant Physiology 157:831-841. |
| [19] |
Maejima K, Iwai R, Himeno M, Komatsu K, Kitazawa Y, Fujita N, Ishikawa K, Fukuoka M, Minato N, Yamaji Y, Oshima K, Namba S. 2014. Recognition of floral homeotic MADS domain transcription factors by a phytoplasmal effector,phyllogen,induces phyllody. Plant Journal, 78 (4):541-554.
doi: 10.1111/tpj.12495 URL |
| [20] |
Maejima K, Kitazawa Y, Tomomitsu T, Yusa A, Neriya Y, Himeno M, Yamaji Y, Oshima K, Namba S. 2015. Degradation of class E MADs-domain transcription factors in Arabidopsis by a phytoplasmal effector,phyllogen. Plant Signaling and Behavior, 10 (8):e1042635.
doi: 10.1080/15592324.2015.1042635 URL |
| [21] |
Melzer R, Verelst W, Theissen G. 2009. The class E floral homeotic protein SEPALLATA 3 is sufficient to loop DNA in‘floral quartet’-like complexes in vitro. Nucleic Acids Research, 37 (1):144-157.
doi: 10.1093/nar/gkn900 URL |
| [22] |
Meng X W, Li Y, Yuan Y, Zhang Y, Li H T, Zhao J, Liu M J. 2020. The regulatory pathways of distinct flowering characteristics in Chinese jujube. Horticulture Research, 7 (1):13-19.
doi: 10.1038/s41438-019-0236-1 URL |
| [23] |
Ng M, Yanofsky M F. 2001. Function and evolution of the plant MADS box gene family. Nature Reviews Genetics, 2:186-195.
pmid: 11256070 |
| [24] |
Pelaz S, Ditta G S, Baumann E, Wisman E, Yanofsky M F. 2000. B and C floral organ identity functions require SEPALLATA MADS box genes. Nature, 405 (6783):200-203.
doi: 10.1038/35012103 URL |
| [25] |
Pelaz S, Tapia-López R, Alvarez-Buylla E R, Yanofsky M F. 2001. Conversion of leaves into petals in Arabidopsis. Current Biology, 11 (3):182-184.
pmid: 11231153 |
| [26] |
Seymour G B., Ryder C D, Cevik V, Hammond J P, Popovich A., King G J, Vrebalov J, Giovannoni J J, Manning K. 2011. A SEPALLATA gene is involved in the development and ripening of strawberry(Fragaria × ananassa Duch.)fruit,a non-climacteric tissue. Journal of Experimental Botany, 62 (3):1179-1188.
doi: 10.1093/jxb/erq360 pmid: 21115665 |
| [27] | Tamura K, Stecher G, Peterson D, Filipski A, Kumar S. 2013. MEGA6:molecular evolutionary genetics analysis Version 6.0. Molecular Biology & Evolution, 30 (12):2725-2729. |
| [28] | Teo Z W N, Zhou W, Shen L. 2019. Dissecting the function of MADS-box transcription factors in orchid reproductive development. Frontiers in Plant Science, 15 (10):1474. |
| [29] |
Xu K, Huang X, Wu M, Wang Y, Chang Y, Liu K, Zhang J, Zhang Y, Zhang F, Yi L, Li T, Wang R, Tan G, Li C. 2014a. A rapid,highly efficient and economical method of Agrobacterium-mediated in planta transient transformation in living onion epidermis. PLoS ONE, 9 (1):e83556.
doi: 10.1371/journal.pone.0083556 URL |
| [30] | Xu Y, Zhang L, Xie H, Zhang Y Q, Oliveira M M, Ma R C. 2008. Expression analysis and genetic mapping of three SEPALLATA -like genes from peach(Prunus persica(L.)Batsch). Tree Genetics & Genomes, 4 (4):693-703. |
| [31] |
Xu Z, Zhang Q, Sun L, Du D, Cheng T, Pan H, Yang W, Wang J. 2014b. Genome-wide identification,characterisation and expression analysis of the MADS-box gene family in Prunus mume. Molecular Genetics and Genomics, 289 (5):903-920.
doi: 10.1007/s00438-014-0863-z URL |
| [32] |
Zhang L M, Zhao J, Feng C F, Liu M J, Wang J R, Hu Y F. 2017. Genome-wide identification,characterization of the MADS-box gene family in Chinese jujube and their involvement in flower development. Scientific Reports, 7 (1):1025.
doi: 10.1038/s41598-017-01159-8 URL |
| [33] |
Zhou Y, Xu Z, Yong X, Ahmad S, Yang W, Cheng T, Wang J, Zhang Q. 2017. SEP-class genes in Prunus mume and their likely role in floral organ development. BMC Plant Biology, 17 (1):10.
doi: 10.1186/s12870-016-0954-6 URL |
| [34] | Zong Chengwen, Fang Jinggui, Tao Jianmin, Zhang Zhen, Qian Yaming. 2007. Cloning and sequence analysis of MADS-box family protein gene fragments from grape. Journal of Fruit Science, 25 (1):27-32. (in Chinese) |
| 宗成文, 房经贵, 陶建敏, 章镇, 钱亚明. 2007. 葡萄MADS-box家族基因保守片段的克隆与序列分析. 果树学报, 25 (1):27-32. |
| [1] | 杨植, 张川疆, 杨芯芳, 董梦怡, 王振磊, 闫芬芬, 吴翠云, 王玖瑞, 刘孟军, 林敏娟. 枣与酸枣杂交后代果实遗传倾向及混合遗传分析[J]. 园艺学报, 2023, 50(1): 36-52. |
| [2] | 赵雪艳, 王琪, 王莉, 王方圆, 王庆, 李艳. 基于比较转录组的延胡索组织差异性表达分析[J]. 园艺学报, 2023, 50(1): 177-187. |
| [3] | 王建新, 马廷军. 枣新品种‘佳油1号’[J]. 园艺学报, 2022, 49(S2): 55-56. |
| [4] | 张玉平, 武 阳, 路东晔, 姚砚武, 潘青华. 枣新品种‘平安葫芦枣’[J]. 园艺学报, 2022, 49(S2): 57-58. |
| [5] | 王晓玲, 仇晓靖, 刘淑怡, 李智慧, 李旭茂, 毛永民, 申连英. 仁用酸枣新品种‘丽园珍珠4号’[J]. 园艺学报, 2022, 49(S2): 59-60. |
| [6] | 王晓玲, 仇晓靖, 李旭茂, 刘淑怡, 李智慧, 毛永民, 申连英. 酸枣新品种‘丽园珍珠8号’[J]. 园艺学报, 2022, 49(S2): 61-62. |
| [7] | 王晓玲, 仇晓靖, 李智慧, 刘淑怡, 李旭茂, 毛永民, 申连英. 酸枣新品种‘丽园珍珠10号’[J]. 园艺学报, 2022, 49(S2): 63-64. |
| [8] | 王晓玲, 仇晓靖, 李智慧, 刘淑怡, 李旭茂, 毛永民, 申连英. 酸枣新品种‘丽园珍珠14号’[J]. 园艺学报, 2022, 49(S2): 65-66. |
| [9] | 帕提古丽 • 买买提吐尔逊, 罗青红, 古丽尼沙 • 卡斯木, 盛 伟, 蒋 腾. 新疆大果沙枣新品种‘红铃’[J]. 园艺学报, 2022, 49(S2): 67-68. |
| [10] | 帕提古丽 • 买买提吐尔逊, 古丽尼沙 • 卡斯木, 罗青红, 刘丽燕, 刘巧玲, 热依汗 • 阿吾提塔什. 新疆大果沙枣新品种‘雅丰’[J]. 园艺学报, 2022, 49(S2): 69-70. |
| [11] | 周军永, 陆丽娟, 孙 俊, 马福利, 刘 茂, 朱淑芳, 孙其宝, . 中晚熟鲜食枣新品种‘高王枣’[J]. 园艺学报, 2022, 49(S1): 35-36. |
| [12] | 高彦龙, 吴玉霞, 张仲兴, 王双成, 张瑞, 张德, 王延秀. 苹果ELO家族基因鉴定及其在低温胁迫下的表达分析[J]. 园艺学报, 2022, 49(8): 1621-1636. |
| [13] | 邱子文, 刘林敏, 林永盛, 林晓洁, 李永裕, 吴少华, 杨超. 千层金MbEGS基因的克隆与功能分析[J]. 园艺学报, 2022, 49(8): 1747-1760. |
| [14] | 许海峰, 王中堂, 陈新, 刘志国, 王利虎, 刘平, 刘孟军, 张琼. 冬枣果皮着色相关类黄酮靶向代谢组学及潜在MYB转录因子分析[J]. 园艺学报, 2022, 49(8): 1761-1771. |
| [15] | 刘平, 王玖瑞, 刘志国, 王立新, 赵璇, 李建兵, 宋韬亮, 冯建华, 刘孟军. 鲜食枣新品种‘雨虹’[J]. 园艺学报, 2022, 49(8): 1833-1834. |
| 阅读次数 | ||||||
|
全文 |
|
|||||
|
摘要 |
|
|||||
版权所有 © 2012 《园艺学报》编辑部 京ICP备10030308号-2 国际联网备案号 11010802023439
编辑部地址: 北京市海淀区中关村南大街12号中国农业科学院蔬菜花卉研究所 邮编: 100081
电话: 010-82109523 E-Mail: yuanyixuebao@126.com
技术支持:北京玛格泰克科技发展有限公司