
园艺学报 ›› 2025, Vol. 52 ›› Issue (6): 1488-1504.doi: 10.16420/j.issn.0513-353x.2024-0373
陈敏氡, 王彬, 白昌辉, 裘波音, 刘建汀, 温庆放, 朱海生*( ), 李永平*(
), 李永平*( )
)
收稿日期:2025-02-06
修回日期:2025-03-14
出版日期:2025-06-20
发布日期:2025-06-20
通讯作者:
基金资助:
CHEN Mindong, WANG Bin, BAI Changhui, QIU Boyin, LIU Jianting, WEN Qingfang, ZHU Haisheng*( ), and LI Yongping*(
), and LI Yongping*( )
)
Received:2025-02-06
Revised:2025-03-14
Published:2025-06-20
Online:2025-06-20
摘要:
为揭示普通丝瓜(Luffa cylindrica)扩展蛋白(Expansin)家族基因的组成、结构和进化过程以及在果实膨大中的作用,利用生物信息技术从普通丝瓜基因组中鉴定出50个Expansin基因,分属于EXPA(18个)、EXPB(3个)、EXLA(27个)和EXLB(2个)4个亚家族,其中EXLA亚家族发生4对基因串联复制和6对基因片段复制,导致该亚家族基因数量明显扩增。系统发育树显示,4个亚家族成员各自聚集成群,且与同科的黄瓜亲缘关系较近。基因结构和保守基序分析显示,4个亚家族的外显子—内含子结构和保守基序种类及分布在进化上趋于保守,可以作为亚家族分型的重要依据。启动子元件预测分析发现,Expansin基因启动子中含有大量的光诱导、厌氧诱导、茉莉酸甲酯和脱落酸元件,而数量最多的光诱导元件只在EXLA/EXLB亚家族预测到。基于RNA-seq数据筛选出8个与果实横纵径发育极显著正相关的Expansin基因,其中4个已报道的基因LcEXPA8、LcEXPA18、LcEXPA14、LcEXPA1可作为丝瓜果形控制的潜在候选基因。对另外4个基因进行RT-qPCR分析,LcEXPA15在花后4 d显著高表达,LcEXPA3、LcEXPA16和LcEXPA7在花后8 ~ 15 d显著高表达。花后2 ~ 15 d是普通丝瓜的果实膨大期,这时期的果实细胞数量增加、单细胞明显膨大、伸长。由此推测,4个Expansin可能在催化细胞生长,促进果实膨大过程中发挥重要作用。通过构建8个Expansin互作网络,提示部分Expansin可能与ERF、bHLH、TCP和GATA这4类转录因子家族存在互作。以上8个Expansin基因可作为普通丝瓜果实膨大相关候选基因进行深入研究。
陈敏氡, 王彬, 白昌辉, 裘波音, 刘建汀, 温庆放, 朱海生, 李永平. 普通丝瓜Expansin家族基因的鉴定及其在果实膨大中的表达分析[J]. 园艺学报, 2025, 52(6): 1488-1504.
CHEN Mindong, WANG Bin, BAI Changhui, QIU Boyin, LIU Jianting, WEN Qingfang, ZHU Haisheng, and LI Yongping. Identification of Expansin Family Gene and Its Expression Analysis During Fruit Enlargement in Sponge Gourd[J]. Acta Horticulturae Sinica, 2025, 52(6): 1488-1504.
| 基因Gene | 引物序列Primer sequence(5′-3′) |
|---|---|
| LcEXPA16 | F:AAACGGAAATAGTAGCGG;R:TCCAATCAGCAGGGAGTA |
| LcEXPA3 | F:CCCAAGTGGTGTCTTCCTG;R:GCACGGTACTGAGCGATT |
| LcEXPA7 | F:AGAAAGGTGGAGTGAGGTTCA;R:TATCGACACGGCATGGAC |
| LcEXPA15 | F:CTCCAGTAGCCTCCCATTT;R:GCCATAACCTGCCTTGACT |
| 18sRNA | F:GCTTGGGTGCTCGACAAACT;R:TCCACAGAGCAATGTCAATGG |
表1 本研究中所用RT-qPCR引物
Table 1 RT-qPCR primers used in this study
| 基因Gene | 引物序列Primer sequence(5′-3′) |
|---|---|
| LcEXPA16 | F:AAACGGAAATAGTAGCGG;R:TCCAATCAGCAGGGAGTA |
| LcEXPA3 | F:CCCAAGTGGTGTCTTCCTG;R:GCACGGTACTGAGCGATT |
| LcEXPA7 | F:AGAAAGGTGGAGTGAGGTTCA;R:TATCGACACGGCATGGAC |
| LcEXPA15 | F:CTCCAGTAGCCTCCCATTT;R:GCCATAACCTGCCTTGACT |
| 18sRNA | F:GCTTGGGTGCTCGACAAACT;R:TCCACAGAGCAATGTCAATGG |
| 基因名称 Gene name | 基因ID Gene ID | 染色体位置 Chromosome position | 氨基酸/aa Amino acid number | 分子量/kD Molecular weight | 理论等电点 Theory isoelectric point | |||
|---|---|---|---|---|---|---|---|---|
| LcEXPA1 | Maker00039481 | Scaffold10:44274633..44276318(-) | 242 | 59.74 | 5.12 | |||
| LcEXPA2 | Maker00039350 | Scaffold10:42862024..42863052(-) | 271 | 67.93 | 5.07 | |||
| LcEXPA3 | Maker00039137 | Scaffold10:45546872..45547695(+) | 247 | 61.82 | 5.08 | |||
| LcEXPA4 | Maker00036366 | Scaffold5:48146177..48147973(+) | 242 | 59.97 | 5.14 | |||
| LcEXPA5 | Maker00033676 | Scaffold13:37888029..37890899(-) | 258 | 63.83 | 5.10 | |||
| LcEXPA6 | Maker00030282 | Scaffold6:14070765..14072443(+) | 261 | 64.67 | 5.11 | |||
| LcEXPA7 | Maker00028836 | Scaffold7:11466255..11470101(-) | 252 | 64.36 | 5.05 | |||
| LcEXPA8 | Maker00026031 | Scaffold7:2531229..2532764(-) | 266 | 66.01 | 5.10 | |||
| LcEXPA9 | Maker00018301 | Scaffold3:15686323..15688378(-) | 264 | 65.19 | 5.10 | |||
| LcEXPA10 | Maker00016100 | Scaffold6:42665910..42667187(+) | 279 | 69.54 | 5.08 | |||
| LcEXPA11 | Maker00013380 | Scaffold1:12581253..12583334(-) | 289 | 71.29 | 5.07 | |||
| LcEXPA12 | Maker00012399 | Scaffold1:13900544..13901668(+) | 260 | 65.89 | 5.05 | |||
| LcEXPA13 | Maker00011031 | Scaffold4:27627243..27635585(-) | 510 | 129.04 | 4.94 | |||
| LcEXPA14 | Maker00009740 | Scaffold7:8940229..8942250(-) | 264 | 66.11 | 5.06 | |||
| LcEXPA15 | Maker00008644 | Scaffold10:33380501..33385747(-) | 338 | 84.60 | 5.05 | |||
| LcEXPA16 | Maker00006224 | Scaffold4:5275140..5276227(-) | 252 | 61.61 | 5.16 | |||
| LcEXPA17 | Maker00003094 | Scaffold12:37764073..37766325(-) | 287 | 71.50 | 5.08 | |||
| LcEXPA18 | Maker00000768 | Scaffold8:44424275..44425647(+) | 259 | 64.59 | 5.08 | |||
| LcEXPB1 | Maker00000264 | Scaffold8:41943956..41945654(-) | 353 | 87.82 | 5.03 | |||
| LcEXPB2 | Maker00031725 | Scaffold11:4460008..:44601243(+) | 274 | 68.75 | 5.07 | |||
| LcEXPB3 | Maker00008065 | Scaffold4:7569358..7572344(-) | 276 | 68.08 | 5.10 | |||
| LcEXLA1 | Maker00037537 | Scaffold11:32691879..32693098(+) | 259 | 63.88 | 5.17 | |||
| LcEXLA2 | Maker00037507 | Scaffold11:32677443..32678122(+) | 137 | 33.94 | 5.28 | |||
| LcEXLA3 | Maker00022629 | Scaffold2:14547662..14548896(-) | 264 | 65.08 | 5.18 | |||
| LcEXLA4 | Maker00022507 | Scaffold2:14624877..14626093(+) | 264 | 64.94 | 5.17 | |||
| LcEXLA5 | Maker00022429 | Scaffold2:14529590..14530833(-) | 259 | 63.86 | 5.18 | |||
| LcEXLA6 | Maker00022378 | Scaffold2:14666085..14667284(+) | 265 | 65.59 | 5.16 | |||
| LcEXLA7 | Maker00020500 | Scaffold1:36285431..36288306(+) | 474 | 119.30 | 4.99 | |||
| LcEXLA8 | Maker00015327 | Scaffold3:29504343..29505828(-) | 305 | 75.57 | 5.13 | |||
| LcEXLA9 | Maker00015140 | Scaffold3:30071104..30076372(-) | 264 | 64.55 | 5.13 | |||
| LcEXLA10 | Maker00015111 | Scaffold3:29507534..29508742(-) | 265 | 65.23 | 5.18 | |||
| LcEXLA11 | Maker00015029 | Scaffold3:29059681..29062050(-) | 356 | 87.97 | 5.07 | |||
| LcEXLA12 | Maker00015000 | Scaffold3:29055967..29057177(-) | 266 | 65.49 | 5.17 | |||
| LcEXLA13 | Maker00014988 | Scaffold3:29376358..29377434(+) | 263 | 65.19 | 5.17 | |||
| LcEXLA14 | Maker00014757 | Scaffold3:29145720..29146827(-) | 263 | 64.64 | 5.18 | |||
| LcEXLA15 | Maker00014734 | Scaffold3:28994980..28996188(-) | 265 | 64.84 | 5.18 | |||
| LcEXLA16 | Maker00014637 | Scaffold3:28901855..28903092(-) | 267 | 65.25 | 5.14 | |||
| LcEXLA17 | Maker00014364 | Scaffold3:29435506..29437459(-) | 266 | 65.41 | 5.16 | |||
| LcEXLA18 | Maker00014148 | Scaffold3:30094458..30096603(-) | 286 | 71.17 | 5.14 | |||
| LcEXLA19 | Maker00013924 | Scaffold3:28963173..28964068(-) | 235 | 57.10 | 5.21 | |||
| LcEXLA20 | Maker00013850 | Scaffold3:29069173..29075111(-) | 414 | 104.23 | 5.05 | |||
| LcEXLA21 | Maker00013746 | Scaffold3:29194590..29195803(-) | 247 | 60.62 | 5.20 | |||
| LcEXLA22 | Maker00013718 | Scaffold3:29455193..29456551(-) | 263 | 64.71 | 5.17 | |||
| LcEXLA23 | Maker00013618 | Scaffold3:29098873..29100084(-) | 269 | 66.51 | 5.15 | |||
| LcEXLA24 | Maker00005227 | Scaffold11:33830538..33832189(+) | 264 | 65.00 | 5.17 | |||
| LcEXLA25 | Maker00005193 | Scaffold11:33797689..33799277(-) | 298 | 74.46 | 5.13 | |||
| LcEXLA26 | Maker00005141 | Scaffold11:33820779..33822086(-) | 264 | 64.93 | 5.17 | |||
| LcEXLA27 | Maker00003985 | Scaffold4:1360141..1361912(+) | 198 | 49.36 | 5.22 | |||
| LcEXLB1 | Maker00038210 | Scaffold12:39908175..39910085(-) | 253 | 62.53 | 5.14 | |||
| LcEXLB2 | Maker00038088 | Scaffold12:40370339..40373048(+) | 256 | 62.82 | 5.15 | |||
| 基因名称 Gene name | 基因ID Gene ID | 蛋白不稳定指数 Protein instability index | 脂溶指数 Lipid solubility index | 疏水性 Total average hydrophobicity | 亚细胞定位 Subcellular localization | |||
| LcEXPA1 | Maker00039481 | 50.82 | 22.18 | 0.72 | 细胞外Extracellular | |||
| LcEXPA2 | Maker00039350 | 51.01 | 22.26 | 0.79 | 细胞外Extracellular | |||
| LcEXPA3 | Maker00039137 | 57.17 | 21.32 | 0.84 | 细胞外Extracellular | |||
| LcEXPA4 | Maker00036366 | 41.14 | 21.35 | 0.62 | 内质网Endoplasmic | |||
| LcEXPA5 | Maker00033676 | 48.80 | 24.42 | 0.80 | 细胞外Extracellular | |||
| LcEXPA6 | Maker00030282 | 45.04 | 21.71 | 0.66 | 细胞外Extracellular | |||
| LcEXPA7 | Maker00028836 | 61.26 | 19.97 | 0.92 | 叶绿体Chloroplast | |||
| LcEXPA8 | Maker00026031 | 56.01 | 23.06 | 0.74 | 内质网Endoplasmic | |||
| LcEXPA9 | Maker00018301 | 52.41 | 23.74 | 0.76 | 叶绿体Chloroplast | |||
| LcEXPA10 | Maker00016100 | 53.80 | 24.25 | 0.77 | 叶绿体Chloroplast | |||
| LcEXPA11 | Maker00013380 | 41.43 | 23.76 | 0.82 | 叶绿体Chloroplast | |||
| LcEXPA12 | Maker00012399 | 56.19 | 18.85 | 0.85 | 细胞外Extracellular | |||
| LcEXPA13 | Maker00011031 | 48.98 | 21.83 | 0.83 | 液泡Vacuole | |||
| LcEXPA14 | Maker00009740 | 52.76 | 21.34 | 0.86 | 细胞外Extracellular | |||
| LcEXPA15 | Maker00008644 | 48.99 | 21.79 | 0.71 | 叶绿体Chloroplast | |||
| LcEXPA16 | Maker00006224 | 43.52 | 27.25 | 0.65 | 内质网Endoplasmic | |||
| LcEXPA17 | Maker00003094 | 48.54 | 24.62 | 0.80 | 细胞质Cytoplasm | |||
| LcEXPA18 | Maker00000768 | 50.04 | 21.24 | 0.81 | 细胞外Extracellular | |||
| LcEXPB1 | Maker00000264 | 48.95 | 23.23 | 0.76 | 叶绿体Chloroplast | |||
| LcEXPB2 | Maker00031725 | 50.10 | 20.07 | 0.74 | 细胞外Extracellular | |||
| LcEXPB3 | Maker00008065 | 43.53 | 22.34 | 0.68 | 液泡Vacuole | |||
| LcEXLA1 | Maker00037537 | 42.07 | 30.76 | 0.70 | 叶绿体Chloroplast | |||
| LcEXLA2 | Maker00037507 | 42.46 | 27.01 | 0.61 | 叶绿体Chloroplast | |||
| LcEXLA3 | Maker00022629 | 41.60 | 32.20 | 0.69 | 叶绿体Chloroplast | |||
| LcEXLA4 | Maker00022507 | 40.72 | 32.45 | 0.72 | 细胞外Extracellular | |||
| LcEXLA5 | Maker00022429 | 38.76 | 32.43 | 0.68 | 叶绿体Chloroplast | |||
| LcEXLA6 | Maker00022378 | 36.96 | 32.20 | 0.72 | 叶绿体Chloroplast | |||
| LcEXLA7 | Maker00020500 | 51.72 | 24.40 | 0.74 | 叶绿体Chloroplast | |||
| LcEXLA8 | Maker00015327 | 41.21 | 30.27 | 0.71 | 叶绿体Chloroplast | |||
| LcEXLA9 | Maker00015140 | 37.36 | 26.39 | 0.71 | 细胞外Extracellular | |||
| LcEXLA10 | Maker00015111 | 38.05 | 29.94 | 0.64 | 细胞外Extracellular | |||
| LcEXLA11 | Maker00015029 | 47.43 | 27.15 | 0.71 | 细胞质Cytoplasm | |||
| LcEXLA12 | Maker00015000 | 43.60 | 30.08 | 0.66 | 细胞外Extracellular | |||
| LcEXLA13 | Maker00014988 | 38.12 | 27.63 | 0.60 | 细胞外Extracellular | |||
| LcEXLA14 | Maker00014757 | 39.28 | 29.66 | 0.61 | 叶绿体Chloroplast | |||
| LcEXLA15 | Maker00014734 | 37.80 | 30.06 | 0.63 | 叶绿体Chloroplast | |||
| LcEXLA16 | Maker00014637 | 39.87 | 25.97 | 0.66 | 细胞外Extracellular | |||
| LcEXLA17 | Maker00014364 | 42.27 | 31.70 | 0.72 | 细胞外Extracellular | |||
| LcEXLA18 | Maker00014148 | 39.96 | 29.49 | 0.70 | 细胞外Extracellular | |||
| LcEXLA19 | Maker00013924 | 33.66 | 30.64 | 0.62 | 叶绿体Chloroplast | |||
| LcEXLA20 | Maker00013850 | 41.44 | 26.17 | 0.67 | 内质网Endoplasmic | |||
| LcEXLA21 | Maker00013746 | 37.90 | 30.90 | 0.62 | 叶绿体Chloroplast | |||
| LcEXLA22 | Maker00013718 | 42.64 | 30.42 | 0.68 | 细胞外Extracellular | |||
| LcEXLA23 | Maker00013618 | 42.52 | 30.61 | 0.70 | 叶绿体Chloroplast | |||
| LcEXLA24 | Maker00005227 | 39.20 | 30.18 | 0.67 | 细胞外Extracellular | |||
| LcEXLA25 | Maker00005193 | 47.09 | 31.10 | 0.72 | 叶绿体Chloroplast | |||
| LcEXLA26 | Maker00005141 | 34.91 | 30.43 | 0.67 | 细胞外Extracellular | |||
| LcEXLA27 | Maker00003985 | 43.48 | 29.12 | 0.64 | 液泡Vacuole | |||
| LcEXLB1 | Maker00038210 | 41.61 | 26.48 | 0.68 | 细胞外Extracellular | |||
| LcEXLB2 | Maker00038088 | 37.14 | 26.69 | 0.66 | 细胞外Extracellular | |||
表2 普通丝瓜Expansin家族基因及其理化性质
Table 2 Expansin family genes and their physicochemical properties of sponge gourd
| 基因名称 Gene name | 基因ID Gene ID | 染色体位置 Chromosome position | 氨基酸/aa Amino acid number | 分子量/kD Molecular weight | 理论等电点 Theory isoelectric point | |||
|---|---|---|---|---|---|---|---|---|
| LcEXPA1 | Maker00039481 | Scaffold10:44274633..44276318(-) | 242 | 59.74 | 5.12 | |||
| LcEXPA2 | Maker00039350 | Scaffold10:42862024..42863052(-) | 271 | 67.93 | 5.07 | |||
| LcEXPA3 | Maker00039137 | Scaffold10:45546872..45547695(+) | 247 | 61.82 | 5.08 | |||
| LcEXPA4 | Maker00036366 | Scaffold5:48146177..48147973(+) | 242 | 59.97 | 5.14 | |||
| LcEXPA5 | Maker00033676 | Scaffold13:37888029..37890899(-) | 258 | 63.83 | 5.10 | |||
| LcEXPA6 | Maker00030282 | Scaffold6:14070765..14072443(+) | 261 | 64.67 | 5.11 | |||
| LcEXPA7 | Maker00028836 | Scaffold7:11466255..11470101(-) | 252 | 64.36 | 5.05 | |||
| LcEXPA8 | Maker00026031 | Scaffold7:2531229..2532764(-) | 266 | 66.01 | 5.10 | |||
| LcEXPA9 | Maker00018301 | Scaffold3:15686323..15688378(-) | 264 | 65.19 | 5.10 | |||
| LcEXPA10 | Maker00016100 | Scaffold6:42665910..42667187(+) | 279 | 69.54 | 5.08 | |||
| LcEXPA11 | Maker00013380 | Scaffold1:12581253..12583334(-) | 289 | 71.29 | 5.07 | |||
| LcEXPA12 | Maker00012399 | Scaffold1:13900544..13901668(+) | 260 | 65.89 | 5.05 | |||
| LcEXPA13 | Maker00011031 | Scaffold4:27627243..27635585(-) | 510 | 129.04 | 4.94 | |||
| LcEXPA14 | Maker00009740 | Scaffold7:8940229..8942250(-) | 264 | 66.11 | 5.06 | |||
| LcEXPA15 | Maker00008644 | Scaffold10:33380501..33385747(-) | 338 | 84.60 | 5.05 | |||
| LcEXPA16 | Maker00006224 | Scaffold4:5275140..5276227(-) | 252 | 61.61 | 5.16 | |||
| LcEXPA17 | Maker00003094 | Scaffold12:37764073..37766325(-) | 287 | 71.50 | 5.08 | |||
| LcEXPA18 | Maker00000768 | Scaffold8:44424275..44425647(+) | 259 | 64.59 | 5.08 | |||
| LcEXPB1 | Maker00000264 | Scaffold8:41943956..41945654(-) | 353 | 87.82 | 5.03 | |||
| LcEXPB2 | Maker00031725 | Scaffold11:4460008..:44601243(+) | 274 | 68.75 | 5.07 | |||
| LcEXPB3 | Maker00008065 | Scaffold4:7569358..7572344(-) | 276 | 68.08 | 5.10 | |||
| LcEXLA1 | Maker00037537 | Scaffold11:32691879..32693098(+) | 259 | 63.88 | 5.17 | |||
| LcEXLA2 | Maker00037507 | Scaffold11:32677443..32678122(+) | 137 | 33.94 | 5.28 | |||
| LcEXLA3 | Maker00022629 | Scaffold2:14547662..14548896(-) | 264 | 65.08 | 5.18 | |||
| LcEXLA4 | Maker00022507 | Scaffold2:14624877..14626093(+) | 264 | 64.94 | 5.17 | |||
| LcEXLA5 | Maker00022429 | Scaffold2:14529590..14530833(-) | 259 | 63.86 | 5.18 | |||
| LcEXLA6 | Maker00022378 | Scaffold2:14666085..14667284(+) | 265 | 65.59 | 5.16 | |||
| LcEXLA7 | Maker00020500 | Scaffold1:36285431..36288306(+) | 474 | 119.30 | 4.99 | |||
| LcEXLA8 | Maker00015327 | Scaffold3:29504343..29505828(-) | 305 | 75.57 | 5.13 | |||
| LcEXLA9 | Maker00015140 | Scaffold3:30071104..30076372(-) | 264 | 64.55 | 5.13 | |||
| LcEXLA10 | Maker00015111 | Scaffold3:29507534..29508742(-) | 265 | 65.23 | 5.18 | |||
| LcEXLA11 | Maker00015029 | Scaffold3:29059681..29062050(-) | 356 | 87.97 | 5.07 | |||
| LcEXLA12 | Maker00015000 | Scaffold3:29055967..29057177(-) | 266 | 65.49 | 5.17 | |||
| LcEXLA13 | Maker00014988 | Scaffold3:29376358..29377434(+) | 263 | 65.19 | 5.17 | |||
| LcEXLA14 | Maker00014757 | Scaffold3:29145720..29146827(-) | 263 | 64.64 | 5.18 | |||
| LcEXLA15 | Maker00014734 | Scaffold3:28994980..28996188(-) | 265 | 64.84 | 5.18 | |||
| LcEXLA16 | Maker00014637 | Scaffold3:28901855..28903092(-) | 267 | 65.25 | 5.14 | |||
| LcEXLA17 | Maker00014364 | Scaffold3:29435506..29437459(-) | 266 | 65.41 | 5.16 | |||
| LcEXLA18 | Maker00014148 | Scaffold3:30094458..30096603(-) | 286 | 71.17 | 5.14 | |||
| LcEXLA19 | Maker00013924 | Scaffold3:28963173..28964068(-) | 235 | 57.10 | 5.21 | |||
| LcEXLA20 | Maker00013850 | Scaffold3:29069173..29075111(-) | 414 | 104.23 | 5.05 | |||
| LcEXLA21 | Maker00013746 | Scaffold3:29194590..29195803(-) | 247 | 60.62 | 5.20 | |||
| LcEXLA22 | Maker00013718 | Scaffold3:29455193..29456551(-) | 263 | 64.71 | 5.17 | |||
| LcEXLA23 | Maker00013618 | Scaffold3:29098873..29100084(-) | 269 | 66.51 | 5.15 | |||
| LcEXLA24 | Maker00005227 | Scaffold11:33830538..33832189(+) | 264 | 65.00 | 5.17 | |||
| LcEXLA25 | Maker00005193 | Scaffold11:33797689..33799277(-) | 298 | 74.46 | 5.13 | |||
| LcEXLA26 | Maker00005141 | Scaffold11:33820779..33822086(-) | 264 | 64.93 | 5.17 | |||
| LcEXLA27 | Maker00003985 | Scaffold4:1360141..1361912(+) | 198 | 49.36 | 5.22 | |||
| LcEXLB1 | Maker00038210 | Scaffold12:39908175..39910085(-) | 253 | 62.53 | 5.14 | |||
| LcEXLB2 | Maker00038088 | Scaffold12:40370339..40373048(+) | 256 | 62.82 | 5.15 | |||
| 基因名称 Gene name | 基因ID Gene ID | 蛋白不稳定指数 Protein instability index | 脂溶指数 Lipid solubility index | 疏水性 Total average hydrophobicity | 亚细胞定位 Subcellular localization | |||
| LcEXPA1 | Maker00039481 | 50.82 | 22.18 | 0.72 | 细胞外Extracellular | |||
| LcEXPA2 | Maker00039350 | 51.01 | 22.26 | 0.79 | 细胞外Extracellular | |||
| LcEXPA3 | Maker00039137 | 57.17 | 21.32 | 0.84 | 细胞外Extracellular | |||
| LcEXPA4 | Maker00036366 | 41.14 | 21.35 | 0.62 | 内质网Endoplasmic | |||
| LcEXPA5 | Maker00033676 | 48.80 | 24.42 | 0.80 | 细胞外Extracellular | |||
| LcEXPA6 | Maker00030282 | 45.04 | 21.71 | 0.66 | 细胞外Extracellular | |||
| LcEXPA7 | Maker00028836 | 61.26 | 19.97 | 0.92 | 叶绿体Chloroplast | |||
| LcEXPA8 | Maker00026031 | 56.01 | 23.06 | 0.74 | 内质网Endoplasmic | |||
| LcEXPA9 | Maker00018301 | 52.41 | 23.74 | 0.76 | 叶绿体Chloroplast | |||
| LcEXPA10 | Maker00016100 | 53.80 | 24.25 | 0.77 | 叶绿体Chloroplast | |||
| LcEXPA11 | Maker00013380 | 41.43 | 23.76 | 0.82 | 叶绿体Chloroplast | |||
| LcEXPA12 | Maker00012399 | 56.19 | 18.85 | 0.85 | 细胞外Extracellular | |||
| LcEXPA13 | Maker00011031 | 48.98 | 21.83 | 0.83 | 液泡Vacuole | |||
| LcEXPA14 | Maker00009740 | 52.76 | 21.34 | 0.86 | 细胞外Extracellular | |||
| LcEXPA15 | Maker00008644 | 48.99 | 21.79 | 0.71 | 叶绿体Chloroplast | |||
| LcEXPA16 | Maker00006224 | 43.52 | 27.25 | 0.65 | 内质网Endoplasmic | |||
| LcEXPA17 | Maker00003094 | 48.54 | 24.62 | 0.80 | 细胞质Cytoplasm | |||
| LcEXPA18 | Maker00000768 | 50.04 | 21.24 | 0.81 | 细胞外Extracellular | |||
| LcEXPB1 | Maker00000264 | 48.95 | 23.23 | 0.76 | 叶绿体Chloroplast | |||
| LcEXPB2 | Maker00031725 | 50.10 | 20.07 | 0.74 | 细胞外Extracellular | |||
| LcEXPB3 | Maker00008065 | 43.53 | 22.34 | 0.68 | 液泡Vacuole | |||
| LcEXLA1 | Maker00037537 | 42.07 | 30.76 | 0.70 | 叶绿体Chloroplast | |||
| LcEXLA2 | Maker00037507 | 42.46 | 27.01 | 0.61 | 叶绿体Chloroplast | |||
| LcEXLA3 | Maker00022629 | 41.60 | 32.20 | 0.69 | 叶绿体Chloroplast | |||
| LcEXLA4 | Maker00022507 | 40.72 | 32.45 | 0.72 | 细胞外Extracellular | |||
| LcEXLA5 | Maker00022429 | 38.76 | 32.43 | 0.68 | 叶绿体Chloroplast | |||
| LcEXLA6 | Maker00022378 | 36.96 | 32.20 | 0.72 | 叶绿体Chloroplast | |||
| LcEXLA7 | Maker00020500 | 51.72 | 24.40 | 0.74 | 叶绿体Chloroplast | |||
| LcEXLA8 | Maker00015327 | 41.21 | 30.27 | 0.71 | 叶绿体Chloroplast | |||
| LcEXLA9 | Maker00015140 | 37.36 | 26.39 | 0.71 | 细胞外Extracellular | |||
| LcEXLA10 | Maker00015111 | 38.05 | 29.94 | 0.64 | 细胞外Extracellular | |||
| LcEXLA11 | Maker00015029 | 47.43 | 27.15 | 0.71 | 细胞质Cytoplasm | |||
| LcEXLA12 | Maker00015000 | 43.60 | 30.08 | 0.66 | 细胞外Extracellular | |||
| LcEXLA13 | Maker00014988 | 38.12 | 27.63 | 0.60 | 细胞外Extracellular | |||
| LcEXLA14 | Maker00014757 | 39.28 | 29.66 | 0.61 | 叶绿体Chloroplast | |||
| LcEXLA15 | Maker00014734 | 37.80 | 30.06 | 0.63 | 叶绿体Chloroplast | |||
| LcEXLA16 | Maker00014637 | 39.87 | 25.97 | 0.66 | 细胞外Extracellular | |||
| LcEXLA17 | Maker00014364 | 42.27 | 31.70 | 0.72 | 细胞外Extracellular | |||
| LcEXLA18 | Maker00014148 | 39.96 | 29.49 | 0.70 | 细胞外Extracellular | |||
| LcEXLA19 | Maker00013924 | 33.66 | 30.64 | 0.62 | 叶绿体Chloroplast | |||
| LcEXLA20 | Maker00013850 | 41.44 | 26.17 | 0.67 | 内质网Endoplasmic | |||
| LcEXLA21 | Maker00013746 | 37.90 | 30.90 | 0.62 | 叶绿体Chloroplast | |||
| LcEXLA22 | Maker00013718 | 42.64 | 30.42 | 0.68 | 细胞外Extracellular | |||
| LcEXLA23 | Maker00013618 | 42.52 | 30.61 | 0.70 | 叶绿体Chloroplast | |||
| LcEXLA24 | Maker00005227 | 39.20 | 30.18 | 0.67 | 细胞外Extracellular | |||
| LcEXLA25 | Maker00005193 | 47.09 | 31.10 | 0.72 | 叶绿体Chloroplast | |||
| LcEXLA26 | Maker00005141 | 34.91 | 30.43 | 0.67 | 细胞外Extracellular | |||
| LcEXLA27 | Maker00003985 | 43.48 | 29.12 | 0.64 | 液泡Vacuole | |||
| LcEXLB1 | Maker00038210 | 41.61 | 26.48 | 0.68 | 细胞外Extracellular | |||
| LcEXLB2 | Maker00038088 | 37.14 | 26.69 | 0.66 | 细胞外Extracellular | |||
| 物种Species | EXPA亚家族 EXPA subfamily | EXPB亚家族 EXPB subfamily | EXLA亚家族 EXLA subfamily | EXLB亚家族 EXLB subfamily |
|---|---|---|---|---|
| 普通丝瓜Sponge gourd | 18 | 3 | 27 | 2 |
| 拟南芥Arabidopsis | 25 | 6 | 3 | 1 |
| 葡萄Grape | 20 | 4 | 1 | 4 |
| 陆地棉Upland cotton | 46 | 8 | 6 | 12 |
| 苹果Apple | 34 | 1 | 2 | 4 |
| 甜瓜Melon | 21 | 3 | 8 | 3 |
| 黄瓜Cucumber | 21 | 3 | 9 | 2 |
表3 不同作物Expansin亚家族基因数目
Table 3 Gene number of Expansin subfamilies in different crops
| 物种Species | EXPA亚家族 EXPA subfamily | EXPB亚家族 EXPB subfamily | EXLA亚家族 EXLA subfamily | EXLB亚家族 EXLB subfamily |
|---|---|---|---|---|
| 普通丝瓜Sponge gourd | 18 | 3 | 27 | 2 |
| 拟南芥Arabidopsis | 25 | 6 | 3 | 1 |
| 葡萄Grape | 20 | 4 | 1 | 4 |
| 陆地棉Upland cotton | 46 | 8 | 6 | 12 |
| 苹果Apple | 34 | 1 | 2 | 4 |
| 甜瓜Melon | 21 | 3 | 8 | 3 |
| 黄瓜Cucumber | 21 | 3 | 9 | 2 |
| 复制事件 Replication event | 基因对 Gene pair | 同义替换率/Ks Synonymous substitution rate | 非同义替换率/Ka Non synonymous substitution rate | Ka/Ks | 进化时间(×106年) Evolution time |
|---|---|---|---|---|---|
| 串联复制 Tandem replication | LcEXLA1/LcEXLA2 | 0.0130 | 0.0425 | 0.3052 | 3.27 |
| LcEXLA24/LcEXLA26 | 0.0634 | 0.1533 | 0.4136 | 11.79 | |
| LcEXLA8/LcEXLA22 | 0.1165 | 0.3072 | 0.3791 | 23.63 | |
| LcEXLA3/LcEXLA5 | 0.0202 | 0.2132 | 0.0948 | 16.40 | |
| 片段复制 Fragment replication | LcEXLA25/LcEXLA12 | 0.2793 | 0.8348 | 0.3345 | 64.22 |
| LcEXLA12/LcEXLA6 | 0.2716 | 0.6760 | 0.4017 | 52.00 | |
| LcEXLA8/LcEXLA6 | 0.3015 | 1.2057 | 0.2501 | 92.74 | |
| LcEXLA14/LcEXLA22 | 0.2783 | 0.8367 | 0.3326 | 64.36 | |
| LcEXLA7/LcEXLA14 | 0.0964 | 0.7387 | 0.1305 | 56.82 | |
| LcEXLA7/LcEXLA22 | 0.2854 | 1.0210 | 0.2795 | 78.53 |
表4 普通丝瓜Expansin基因的复制事件分析
Table 4 Analysis of replication events of Expansin gene in sponge gourd
| 复制事件 Replication event | 基因对 Gene pair | 同义替换率/Ks Synonymous substitution rate | 非同义替换率/Ka Non synonymous substitution rate | Ka/Ks | 进化时间(×106年) Evolution time |
|---|---|---|---|---|---|
| 串联复制 Tandem replication | LcEXLA1/LcEXLA2 | 0.0130 | 0.0425 | 0.3052 | 3.27 |
| LcEXLA24/LcEXLA26 | 0.0634 | 0.1533 | 0.4136 | 11.79 | |
| LcEXLA8/LcEXLA22 | 0.1165 | 0.3072 | 0.3791 | 23.63 | |
| LcEXLA3/LcEXLA5 | 0.0202 | 0.2132 | 0.0948 | 16.40 | |
| 片段复制 Fragment replication | LcEXLA25/LcEXLA12 | 0.2793 | 0.8348 | 0.3345 | 64.22 |
| LcEXLA12/LcEXLA6 | 0.2716 | 0.6760 | 0.4017 | 52.00 | |
| LcEXLA8/LcEXLA6 | 0.3015 | 1.2057 | 0.2501 | 92.74 | |
| LcEXLA14/LcEXLA22 | 0.2783 | 0.8367 | 0.3326 | 64.36 | |
| LcEXLA7/LcEXLA14 | 0.0964 | 0.7387 | 0.1305 | 56.82 | |
| LcEXLA7/LcEXLA22 | 0.2854 | 1.0210 | 0.2795 | 78.53 |
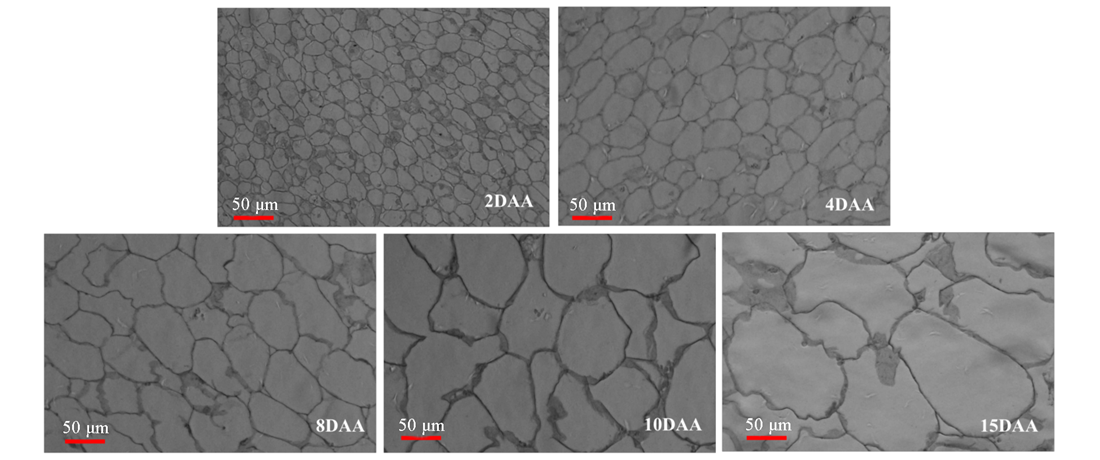
图5 普通丝瓜果实膨大期细胞形态差异 2、4、8、10和15 DAA:花后2、4、8、10和15 d
Fig. 5 The cell morphology of fruit enlargement stage in sponge gourd 2,4,8,10 and 15 DAA:2,4,8,10,15 days after anthesis
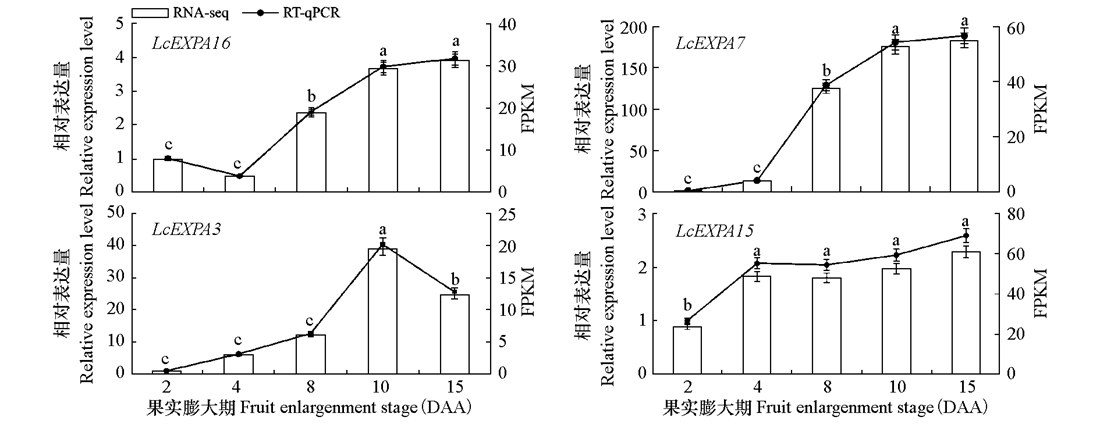
图6 4个Expansin的RT-qRCR和RNA-seq结果 不同字母表示差异显著(P < 0.05)
Fig. 6 RT-qRCR and RNA-seq results of 4 Expansin genes Different letters indicate significant differences at 0.05 level

图7 普通丝瓜中8个Expansin基因的互作网络 图中连线颜色代表Expansin基因与关联基因的相关性类型。红色代表正相关,蓝色代表负相关;线条粗细代表相关性数值大小。关联的转录因子用红色标注
Fig. 7 Interaction network of 8 Expansin genes in sponge gourd The color of the line in the figure represents the type of correlation between Expansin genes and associated genes. Red represents positive correlation and blue represents negative correlation;The thickness of the line represents the magnitude of the correlation value. The associated transcription factors are marked in red
| [1] |
|
| [2] |
|
| [3] |
|
|
蔡兆琴, 龙婷晞, 肖冬, 王爱勤, 黄咏梅, 李慧峰, 詹洁, 何龙飞. 2023. 甘薯扩展蛋白IbEXPA2基因克隆和表达分析. 分子植物育种, 21 (5):1401-1407.
|
|
| [4] |
|
|
蔡兆琴, 吴强, 陈慧, 肖冬, 王爱勤, 黄咏梅, 李慧峰, 詹洁, 何龙飞. 2021. 甘薯扩展蛋白基因IbEXPA4的克隆和表达分析. 农业生物技术学报, 29 (10):1914-1925.
|
|
| [5] |
doi: 10.3864/j.issn.0578-1752.2023.22.012 |
|
陈敏氡, 王彬, 刘建汀, 李永平, 白昌辉, 叶新如, 裘波音, 温庆放, 朱海生. 2023. 基于转录组和WGCNA筛选丝瓜果长和果径发育调控相关基因. 中国农业科学, 56 (22):4506-4522.
doi: 10.3864/j.issn.0578-1752.2023.22.012 |
|
| [6] |
|
| [7] |
|
| [8] |
|
| [9] |
|
|
|
|
| [10] |
|
| [11] |
|
|
韩阳阳, 王天晓, 王玮. 2015. 扩展蛋白与激素调控的生长发育. 生命的化学,(6):778-784.
|
|
| [12] |
|
| [13] |
|
| [14] |
doi: 10.3732/ajb.1400273 pmid: 25587144 |
| [15] |
|
| [16] |
|
|
李必聪, 李慧英, 肖遥, 罗莎, 周庆红, 黄英金, 朱强龙. 2023. 芋扩展蛋白基因家族的全基因组鉴定及其在球茎膨大中的表达分析. 浙江农业学报, 35 (7):1604-1616.
doi: 10.3969/j.issn.1004-1524.20220960 |
|
| [17] |
doi: 10.7668/hbnxb.2017.05.007 |
|
理向阳, 代丹丹, 杨铁钢, 郝西. 2017. 马铃薯扩展蛋白基因家族的鉴定与表达分析. 华北农学报, 32 (5):37-44.
doi: 10.7668/hbnxb.2017.05.007 |
|
| [18] |
|
|
刘昌. 2018. 甜瓜扩展蛋白基因家族成员的鉴定及CmEXPA8和CmEXPA12基因在果实发育中功能的初步分析[博士论文]. 内蒙古: 内蒙古大学.
|
|
| [19] |
|
|
刘春香, 张卫华, 曹齐卫, 孙小镭. 2011. 黄瓜扩张蛋白CsEXP10基因5'序列的克隆和表达的初步分析. 华北农学报, 26 (1):4.
|
|
| [20] |
|
|
刘维妙. 2021. 芸薹属基本种扩展蛋白基因家族的进化分析及花粉发育相关扩展蛋白基因的功能分化研究[博士论文]. 浙江: 浙江大学.
|
|
| [21] |
doi: 10.1006/meth.2001.1262 pmid: 11846609 |
| [22] |
|
|
鲁广宇, 任竹. 2019. 丝瓜常见生理性病害的发生与防治. 园艺与种苗, 39 (7):6-8.
|
|
| [23] |
|
|
罗萍, 王晓萍, 张昊楠, 范春节, 王玉娇, 徐建民. 2023. 巨桉扩展蛋白EgrEXPA8和EgrEXPA10基因的克隆和表达特性分析. 热带亚热带植物学报, 31 (6):827-834.
|
|
| [24] |
|
|
罗少波, 罗剑宁, 郑晓明. 2006. 我国丝瓜育种研究进展与展望. 广东农业科学,1:15-17.
|
|
| [25] |
doi: 10.7525/j.issn.1673-5102.2022.01.004 |
|
马霜, 王博雅, 曹颖, 胡尚连, 高志民. 2022. 毛竹扩展蛋白基因韵鉴定及其表达分析. 植物研究, 42 (1):29-38.
doi: 10.7525/j.issn.1673-5102.2022.01.004 |
|
| [26] |
|
|
毛彦芝. 2013. 番茄和马铃薯扩展蛋白研究进展. 黑龙江八一农垦大学学报, 25 (5):5-8,47. (in Chinese)
|
|
| [27] |
doi: 10.7525/j.issn.1673-5102.2021.01.014 |
|
任梦轩, 张洋, 王爽, 王瑞琪, 刘聪, 魏志刚. 2021. 毛果杨GATA基因家族全基因组水平鉴定及表达分析. 植物研究, 41 (1):107-118.
doi: 10.7525/j.issn.1673-5102.2021.01.014 |
|
| [28] |
doi: 10.1104/pp.123.4.1583 pmid: 10938374 |
| [29] |
|
| [30] |
|
|
施杨, 徐筱, 李昊阳, 徐倩, 徐吉臣. 2014. 水稻扩展蛋白家族的生物信息学分析. 遗传, 36 (8):809-820.
|
|
| [31] |
|
|
王兰伟. 2011. 异源多倍体物种的形成和演化:来自菊科蓍草属的证据[博士论文]. 开封: 河南大学.
|
|
| [32] |
|
|
王玮, 赵新西, 马千全, 郭启芳, 邹琦. 2004. 扩展蛋白生理与分子生物学. 植物生理学通讯, 40 (1):1-6.
|
|
| [33] |
|
| [34] |
doi: 10.16420/j.issn.0513-353x.2020-0903 |
|
辛海青, 周军永, 孙耀星, 穆文磊, 杨健, 马福利, 孙俊, 薛峥嵘, 陆丽娟, 孙其宝. 2021. 枣易裂与抗裂品种灌水后果皮结构和扩张蛋白基因表达差异研究. 园艺学报, 48 (9):1785-1793.
doi: 10.16420/j.issn.0513-353x.2020-0903 |
|
| [35] |
|
| [36] |
|
|
许世达, 耿兴敏, 王露露. 2021. 植物乙烯响应因子(ERF)的结构、功能及表达调控研究进展. 浙江农林大学学报, 38 (3):624-633.
|
|
| [37] |
|
|
徐筱, 徐倩, 张钅習, 徐吉臣. 2010. 植物扩展蛋白基因的研究进展. 北京林业大学学报, 32 (5):154-162.
|
|
| [38] |
|
|
张建鹏, 尚霄丽. 2024. 葡萄扩展蛋白家族全基因组分析及VvEXPA27功能研究. 农业生物技术学报, 32 (4):762-774.
|
|
| [39] |
|
|
张奇艳, 雷忠萍, 宋银, 海江波, 贺道华. 2019. 陆地棉扩展蛋白基因的鉴定与特征分析. 中国农业科学, 52 (21):3713-3732.
doi: 10.3864/j.issn.0578-1752.2019.21.001 |
|
| [40] |
|
| [41] |
|
|
张伟. 2014. 玉米EXPANSIN家族胚乳特异表达基因鉴定及ZmEXPB13功能分析[博士论文]. 安徽: 安徽农业大学.
|
|
| [42] |
|
|
赵美荣. 2014. 植物扩展蛋白基因及其表达调控的研究进展. 赤峰学院学报(自然科学版), 30 (14):1-5.
|
| [1] | 孙廷珍, 疏琴, 马玮, 史玉滋, 张蒙, 向成钢, 薄凯亮, 段颖, 王长林. 印度南瓜GAox家族基因鉴定及其在不同蔓长种质中的表达[J]. 园艺学报, 2025, 52(3): 603-622. |
| [2] | 王珊珊, 郭瑞, 何棱, 吴春红, 陈禅友, 万何平, 赵慧霞. 长豇豆Lhc家族基因鉴定及其在盐胁迫下的表达分析[J]. 园艺学报, 2025, 52(1): 111-122. |
| [3] | 赵佳莹, 曾周婷, 岑欣颖, 施姣淇, 李效贤, 沈晓霞, 俞振明. 铁皮石斛CCO基因家族鉴定及其在花发育中的表达分析[J]. 园艺学报, 2024, 51(9): 2075-2088. |
| [4] | 王艳红, 席克勇, 田野, 刘德麒, 周克贵, 尹军良, 刘奕清, 朱永兴. 姜GST基因家族成员鉴定与表达分析[J]. 园艺学报, 2024, 51(8): 1803-1822. |
| [5] | 陈佳悦, 段英明, 周雁, 肖扬, 边银丙, 龚钰华. 香菇ALDH家族基因鉴定、表达及功能分析[J]. 园艺学报, 2024, 51(5): 1033-1046. |
| [6] | 刘晋红, 王峥, 于昊, 辛依睿, 亓果宁, 柳参奎, 任慧敏. 毛竹SLAC家族基因鉴定及PheSLAC1功能分析[J]. 园艺学报, 2024, 51(3): 545-559. |
| [7] | 罗新锐, 张晓旭, 王玉萍, 王智, 马媛媛, 周丙月. 普通菜豆Trihelix基因家族鉴定及表达分析[J]. 园艺学报, 2024, 51(12): 2775-2790. |
| [8] | 黄治皓, 刘婷婷, 董旭杰, 严明理, 刘志祥, 曾超珍. 芥菜HMA家族基因鉴定及其在镉胁迫下的表达分析[J]. 园艺学报, 2023, 50(6): 1230-1242. |
| [9] | 于婷婷, 李欢, 宁源生, 宋建飞, 彭璐琳, 贾竣淇, 张玮玮, 杨洪强. 苹果GRAS全基因组鉴定及其对生长素的响应分析[J]. 园艺学报, 2023, 50(2): 397-409. |
| [10] | 王炫榛, 陈敏氡, 刘建汀, 曾美娟, 叶新如, 李永平, 温庆放, 朱海生. 普通丝瓜幼苗响应低温弱光核心基因共表达网络的WGCNA鉴定[J]. 园艺学报, 2023, 50(12): 2601-2618. |
| [11] | 申玉晓, 邹金玉, 罗平, 尚文倩, 李永华, 何松林, 王政, 石力匀. ‘月月粉’月季PP2C家族基因鉴定及非生物胁迫响应分析[J]. 园艺学报, 2023, 50(10): 2139-2156. |
| [12] | 翟含含, 翟宇杰, 田义, 张叶, 杨丽, 温陟良, 陈海江. 桃SAUR家族基因分析及PpSAUR5功能鉴定[J]. 园艺学报, 2023, 50(1): 1-14. |
| [13] | 高彦龙, 吴玉霞, 张仲兴, 王双成, 张瑞, 张德, 王延秀. 苹果ELO家族基因鉴定及其在低温胁迫下的表达分析[J]. 园艺学报, 2022, 49(8): 1621-1636. |
| [14] | 刘金明, 郭彩华, 袁星, 亢超, 全绍文, 牛建新. 梨Dof家族基因鉴定及其在宿存与脱落萼片中的表达分析[J]. 园艺学报, 2022, 49(8): 1637-1649. |
| [15] | 马维峰, 李艳梅, 马宗桓, 陈佰鸿, 毛娟. 苹果POD家族基因的鉴定与MdPOD15的功能分析[J]. 园艺学报, 2022, 49(6): 1181-1199. |
| 阅读次数 | ||||||
|
全文 |
|
|||||
|
摘要 |
|
|||||
版权所有 © 2012 《园艺学报》编辑部 京ICP备10030308号-2 国际联网备案号 11010802023439
编辑部地址: 北京市海淀区中关村南大街12号中国农业科学院蔬菜花卉研究所 邮编: 100081
电话: 010-82109523 E-Mail: yuanyixuebao@126.com
技术支持:北京玛格泰克科技发展有限公司