
园艺学报 ›› 2021, Vol. 48 ›› Issue (7): 1371-1385.doi: 10.16420/j.issn.0513-353x.2021-0205
贾会霞1, 李锡香1, 宋江萍1, 林毓娥2, 张晓辉1, 邱杨1, 阳文龙1, 娄群峰3, 王海平1,*( )
)
收稿日期:2021-02-22
修回日期:2021-04-06
出版日期:2021-07-25
发布日期:2021-08-10
通讯作者:
王海平
E-mail:wanghaiping@caas.cn
基金资助:
JIA Huixia1, LI Xixiang1, SONG Jiangping1, LIN Yu’e2, ZHANG Xiaohui1, QIU Yang1, YANG Wenlong1, LOU Qunfeng3, WANG Haiping1,*( )
)
Received:2021-02-22
Revised:2021-04-06
Online:2021-07-25
Published:2021-08-10
Contact:
WANG Haiping
E-mail:wanghaiping@caas.cn
摘要:
以231份来自世界各地的黄瓜核心种质为材料,于2017和2018年在北京和广州进行白粉病抗性鉴定,利用基因组重测序检测的950 382个SNPs进行全基因组关联分析。结果表明,黄瓜白粉病在广州地区的自然发病率显著高于北京地区,不同种质在两年病情指数(DI)之间存在较高的相关性;基于混合线性模型,共检测到12个显著关联信号,分布于1号、5号和6号染色体上,其中,pm1.1在北京的两年中被重复检测到,pm5.7在广州的两年中被重复检测到;显著关联信号中共有68个超过阈值[-log10(P value) = 6.0]的SNP,包含1个外显子区非同义突变SNP(Chr5:16 676 542),位于编码异嗜性和多嗜性逆转录病毒受体蛋白基因XPR(Csa5G471600)上;在与白粉病抗性相关联SNP的LD区段内共检测到63个候选基因,其中7个候选基因的表达量在高抗材料接种白粉病前后出现显著差异。
中图分类号:
贾会霞, 李锡香, 宋江萍, 林毓娥, 张晓辉, 邱杨, 阳文龙, 娄群峰, 王海平. 黄瓜核心种质白粉病抗性的全基因组关联分析[J]. 园艺学报, 2021, 48(7): 1371-1385.
JIA Huixia, LI Xixiang, SONG Jiangping, LIN Yu’e, ZHANG Xiaohui, QIU Yang, YANG Wenlong, LOU Qunfeng, WANG Haiping. Genome-Wide Association Study of Powdery Mildew Resistance of Cucumber Core Germplasms[J]. Acta Horticulturae Sinica, 2021, 48(7): 1371-1385.
| 国家 Country | 省 Province | 样本数 Number of samples | 国家 Country | 样本数 Number of samples |
|---|---|---|---|---|
| 中国China | 广东Guangdong | 39 | 美国America | 10 |
| 北京Beijing | 14 | 日本Japan | 7 | |
| 山东Shandong | 14 | 印度India | 6 | |
| 辽宁Liaoning | 12 | 德国Germany | 5 | |
| 云南Yunnan | 12 | 俄罗斯Russia | 4 | |
| 吉林Jilin | 11 | 土耳其Turkey | 3 | |
| 河北Hebei | 9 | 匈牙利Hungary | 3 | |
| 四川Sichuan | 7 | 伊朗Iran | 3 | |
| 天津Tianjin | 7 | 阿富汗Afghanistan | 2 | |
| 河南Henan | 5 | 荷兰Holland | 2 | |
| 湖北Hubei | 5 | 挪威Norway | 2 | |
| 湖南Hunan | 5 | 西班牙Spain | 2 | |
| 福建Fujian | 3 | 叙利亚Syria | 2 | |
| 黑龙江Heilongjiang | 3 | 以色列Israel | 2 | |
| 内蒙Inner Mongolia | 3 | 埃及Egypt | 1 | |
| 山西Shanxi | 3 | 巴基斯坦Pakistan | 1 | |
| 安徽Anhui | 2 | 波多黎各Puerto Rico | 1 | |
| 贵州Guizhou | 2 | 丹麦Denmark | 1 | |
| 新疆Xinjiang | 2 | 哈萨克斯坦Kazakhstan | 1 | |
| 重庆Chongqing | 2 | 津巴布韦Zimbabwe | 1 | |
| 广西Guangxi | 1 | 克罗地亚Croatia | 1 | |
| 江苏Jiangsu | 1 | 肯尼亚Kenya | 1 | |
| 江西Jiangxi | 1 | 毛里求斯Mauritius | 1 | |
| 昆明Kunming | 1 | 尼泊尔Nepal | 1 | |
| 宁夏Ningxia | 1 | 塞尔维亚Serbia | 1 | |
| 泰国Thailand | 1 | |||
| 希腊Greece | 1 |
表1 黄瓜抗白粉病鉴定材料
Table 1 Cucumber materials for resistance identification of powdery mildew
| 国家 Country | 省 Province | 样本数 Number of samples | 国家 Country | 样本数 Number of samples |
|---|---|---|---|---|
| 中国China | 广东Guangdong | 39 | 美国America | 10 |
| 北京Beijing | 14 | 日本Japan | 7 | |
| 山东Shandong | 14 | 印度India | 6 | |
| 辽宁Liaoning | 12 | 德国Germany | 5 | |
| 云南Yunnan | 12 | 俄罗斯Russia | 4 | |
| 吉林Jilin | 11 | 土耳其Turkey | 3 | |
| 河北Hebei | 9 | 匈牙利Hungary | 3 | |
| 四川Sichuan | 7 | 伊朗Iran | 3 | |
| 天津Tianjin | 7 | 阿富汗Afghanistan | 2 | |
| 河南Henan | 5 | 荷兰Holland | 2 | |
| 湖北Hubei | 5 | 挪威Norway | 2 | |
| 湖南Hunan | 5 | 西班牙Spain | 2 | |
| 福建Fujian | 3 | 叙利亚Syria | 2 | |
| 黑龙江Heilongjiang | 3 | 以色列Israel | 2 | |
| 内蒙Inner Mongolia | 3 | 埃及Egypt | 1 | |
| 山西Shanxi | 3 | 巴基斯坦Pakistan | 1 | |
| 安徽Anhui | 2 | 波多黎各Puerto Rico | 1 | |
| 贵州Guizhou | 2 | 丹麦Denmark | 1 | |
| 新疆Xinjiang | 2 | 哈萨克斯坦Kazakhstan | 1 | |
| 重庆Chongqing | 2 | 津巴布韦Zimbabwe | 1 | |
| 广西Guangxi | 1 | 克罗地亚Croatia | 1 | |
| 江苏Jiangsu | 1 | 肯尼亚Kenya | 1 | |
| 江西Jiangxi | 1 | 毛里求斯Mauritius | 1 | |
| 昆明Kunming | 1 | 尼泊尔Nepal | 1 | |
| 宁夏Ningxia | 1 | 塞尔维亚Serbia | 1 | |
| 泰国Thailand | 1 | |||
| 希腊Greece | 1 |
| 染色体 Chromosome | 显著关联 信号 Significantly associated signals | SNP位置/bp SNP Position | 基因型 Genotype | SNP注释 SNP annotation | P值 P value | |||
|---|---|---|---|---|---|---|---|---|
| 2017- Beijing | 2018- Beijing | 2017- Guangzhou | 2018- Guangzhou | |||||
| Chr1 | pm1.1 | 20 946 043 | A/C | 基因间区 Intergenic | 7.34E-07 | |||
| 20 946 325 | C/T | 基因间区 Intergenic | 1.72E-08 | |||||
| 20 946 384 | C/T | 基因间区 Intergenic | 6.04E-08 | |||||
| 20 946 425 | A/G | 基因间区 Intergenic | 2.44E-07 | |||||
| 20 946 565 | C/T | 基因间区 Intergenic | 1.67E-08 | |||||
| 20 947 445 | T/C | 基因间区 Intergenic | 2.13E-08 | |||||
| 20 947 746 | G/T | 基因间区 Intergenic | 4.18E-08 | |||||
| 20 947 997 | G/A | 基因间区 Intergenic | 1.53E-08 | |||||
| 20 948 176 | T/A | 基因间区 Intergenic | 4.91E-09 | |||||
| 20 948 443 | C/T | 基因间区 Intergenic | 3.54E-08 | |||||
| 20 949 791 | A/G | 上游区 Upstream | 2.41E-08 | |||||
| 20 950 222 | T/C | 内含子区 Intronic | 1.17E-08 | |||||
| 20 950 313 | T/C | 内含子区 Intronic | 6.97E-07 | 1.17E-08 | ||||
| 20 951 178 | T/C | 内含子区 Intronic | 9.50E-08 | |||||
| 20 951 719 | G/A | 内含子区 Intronic | 2.86E-08 | |||||
| 20 951 938 | G/A | 内含子区 Intronic | 2.45E-08 | |||||
| 20 952 168 | T/C | 内含子区 Intronic | 1.43E-08 | |||||
| 20 952 590 | A/G | 内含子区 Intronic | 3.64E-08 | 1.59E-09 | ||||
| 20 952 617 | G/A | 内含子区 Intronic | 8.71E-08 | 1.24E-09 | ||||
| 20 953 029 | T/C | 内含子区 Intronic | 7.98E-09 | |||||
| 20 953 187 | T/C | 内含子区 Intronic | 9.32E-09 | |||||
| 20 953 655 | A/C | 内含子区 Intronic | 5.23E-09 | |||||
| 20 954 157 | T/C | 内含子区 Intronic | 1.21E-07 | 3.17E-09 | ||||
| 20 955 127 | T/C | 内含子区 Intronic | 8.56E-08 | |||||
| 20 955 284 | T/A | 内含子区 Intronic | 6.32E-10 | |||||
| 20 955 289 | G/A | 内含子区 Intronic | 3.57E-07 | 1.25E-09 | ||||
| 20 955 466 | A/C | 内含子区 Intronic | 3.12E-07 | 8.79E-09 | ||||
| 20 955 556 | G/A | 内含子区 Intronic | 1.78E-07 | 4.36E-09 | ||||
| 20 956 182 | A/C | 外显子区(同义突变)Exonic(synonymous) | 1.43E-08 | |||||
| 20 956 822 | T/A | 内含子区 Intronic | 1.34E-08 | |||||
| 20 959 137 | G/A | 下游区 Downstream | 7.29E-07 | |||||
| 20 960 732 | T/C | 内含子区 Intronic | 7.54E-08 | |||||
| 20 960 788 | C/T | 内含子区 Intronic | 2.25E-08 | |||||
| 20 961 611 | A/C | 内含子区 Intronic | 1.23E-08 | |||||
| 20 963 321 | A/G | 上游区/下游区 Upstream/Downstream | 2.31E-07 | |||||
| 20 963 487 | A/G | 上游区/下游区 Upstream/Downstream | 5.23E-07 | 3.40E-09 | ||||
| 20 963 500 | T/C | 上游区/下游区 Upstream/Downstream | 6.56E-08 | |||||
| 20 964 042 | A/G | 内含子区 Intronic | 9.32E-07 | |||||
| 20 964 068 | T/C | 内含子区 Intronic | 9.32E-07 | |||||
| 20 964 073 | T/G | 内含子区 Intronic | 1.79E-07 | |||||
| 20 964 207 | A/G | 内含子区 Intronic | 1.51E-07 | |||||
| 20 965 920 | G/C | 内含子区 Intronic | 1.30E-07 | |||||
| 20 966 914 | T/A | 上游区 Upstream | 3.98E-07 | |||||
| 20 967 021 | C/T | 上游区 Upstream | 8.88E-07 | |||||
| pm1.2 | 21 133 564 | C/T | 基因间区 Intergenic | 4.74E-07 | ||||
| pm1.3 | 21 453 973 | T/C | 内含子区 Intronic | 6.68E-07 | ||||
| 21 454 062 | A/G | 内含子区 Intronic | 5.04E-07 | |||||
| 21 459 154 | T/C | 基因间区Intergenic | 7.54E-07 | |||||
| 21 459 157 | G/A | 基因间区Intergenic | 2.46E-07 | |||||
| 21 459 889 | A/T | 上游区 Upstream | 1.46E-07 | |||||
| 21 459 973 | C/T | 上游区 Upstream | 1.29E-07 | |||||
| 21 461 960 | A/G | 内含子区 Intronic | 1.74E-07 | |||||
| 染色体 Chromosome | 显著关联 信号 Significantly associated signals | SNP位置/bp SNP Position | 基因型 Genotype | SNP注释 SNP annotation | P值 P value | |||
| 2017- Beijing | 2018- Beijing | 2017- Guangzhou | 2018- Guangzhou | |||||
| Chr5 | pm5.1 | 1 858 575 | C/T | 上游区 Upstream | 4.14E-08 | |||
| pm5.2 | 7 910 694 | C/T | 基因间区 Intergenic | 3.20E-07 | ||||
| pm5.3 | 15 769 974 | G/C | 基因间区 Intergenic | 9.23E-07 | ||||
| pm5.4 | 16 149 984 | G/A | 内含子区 Intronic | 5.83E-07 | ||||
| pm5.5 | 16 316 760 | T/A | 内含子区 Intronic | 1.13E-07 | ||||
| 16 327 596 | G/C | 内含子区 Intronic | 6.55E-07 | |||||
| pm5.6 | 16 357 208 | T/C | 内含子区 Intronic | 4.66E-07 | ||||
| 16 380 085 | A/T | 内含子区 Intronic | 1.38E-08 | |||||
| 16 390 016 | G/A | 上游区 Upstream | 2.96E-07 | |||||
| pm5.7 | 16 666 839 | C/G | 基因间区 Intergenic | 4.64E-07 | 5.95E-08 | |||
| 16 676 542 | G/C | 外显子区(非同义突变Exonic(Nonsynonymous) | 2.44E-10 | 2.70E-07 | ||||
| 16 683 577 | C/A | 基因间区 Intergenic | 2.24E-08 | 4.22E-11 | ||||
| pm5.8 | 22 992 521 | A/G | 基因间区 Intergenic | 3.44E-08 | ||||
| 22 992 567 | A/T | 基因间区 Intergenic | 3.34E-07 | |||||
| 22 993 739 | T/A | 基因间区 Intergenic | 8.32E-07 | |||||
| Chr6 | pm6.1 | 15 844 016 | T/G | 基因间区 Intergenic | 6.97E-07 | |||
表2 与白粉病抗性显著关联的SNP位点
Table 2 SNPs associated with powdery mildew resistance
| 染色体 Chromosome | 显著关联 信号 Significantly associated signals | SNP位置/bp SNP Position | 基因型 Genotype | SNP注释 SNP annotation | P值 P value | |||
|---|---|---|---|---|---|---|---|---|
| 2017- Beijing | 2018- Beijing | 2017- Guangzhou | 2018- Guangzhou | |||||
| Chr1 | pm1.1 | 20 946 043 | A/C | 基因间区 Intergenic | 7.34E-07 | |||
| 20 946 325 | C/T | 基因间区 Intergenic | 1.72E-08 | |||||
| 20 946 384 | C/T | 基因间区 Intergenic | 6.04E-08 | |||||
| 20 946 425 | A/G | 基因间区 Intergenic | 2.44E-07 | |||||
| 20 946 565 | C/T | 基因间区 Intergenic | 1.67E-08 | |||||
| 20 947 445 | T/C | 基因间区 Intergenic | 2.13E-08 | |||||
| 20 947 746 | G/T | 基因间区 Intergenic | 4.18E-08 | |||||
| 20 947 997 | G/A | 基因间区 Intergenic | 1.53E-08 | |||||
| 20 948 176 | T/A | 基因间区 Intergenic | 4.91E-09 | |||||
| 20 948 443 | C/T | 基因间区 Intergenic | 3.54E-08 | |||||
| 20 949 791 | A/G | 上游区 Upstream | 2.41E-08 | |||||
| 20 950 222 | T/C | 内含子区 Intronic | 1.17E-08 | |||||
| 20 950 313 | T/C | 内含子区 Intronic | 6.97E-07 | 1.17E-08 | ||||
| 20 951 178 | T/C | 内含子区 Intronic | 9.50E-08 | |||||
| 20 951 719 | G/A | 内含子区 Intronic | 2.86E-08 | |||||
| 20 951 938 | G/A | 内含子区 Intronic | 2.45E-08 | |||||
| 20 952 168 | T/C | 内含子区 Intronic | 1.43E-08 | |||||
| 20 952 590 | A/G | 内含子区 Intronic | 3.64E-08 | 1.59E-09 | ||||
| 20 952 617 | G/A | 内含子区 Intronic | 8.71E-08 | 1.24E-09 | ||||
| 20 953 029 | T/C | 内含子区 Intronic | 7.98E-09 | |||||
| 20 953 187 | T/C | 内含子区 Intronic | 9.32E-09 | |||||
| 20 953 655 | A/C | 内含子区 Intronic | 5.23E-09 | |||||
| 20 954 157 | T/C | 内含子区 Intronic | 1.21E-07 | 3.17E-09 | ||||
| 20 955 127 | T/C | 内含子区 Intronic | 8.56E-08 | |||||
| 20 955 284 | T/A | 内含子区 Intronic | 6.32E-10 | |||||
| 20 955 289 | G/A | 内含子区 Intronic | 3.57E-07 | 1.25E-09 | ||||
| 20 955 466 | A/C | 内含子区 Intronic | 3.12E-07 | 8.79E-09 | ||||
| 20 955 556 | G/A | 内含子区 Intronic | 1.78E-07 | 4.36E-09 | ||||
| 20 956 182 | A/C | 外显子区(同义突变)Exonic(synonymous) | 1.43E-08 | |||||
| 20 956 822 | T/A | 内含子区 Intronic | 1.34E-08 | |||||
| 20 959 137 | G/A | 下游区 Downstream | 7.29E-07 | |||||
| 20 960 732 | T/C | 内含子区 Intronic | 7.54E-08 | |||||
| 20 960 788 | C/T | 内含子区 Intronic | 2.25E-08 | |||||
| 20 961 611 | A/C | 内含子区 Intronic | 1.23E-08 | |||||
| 20 963 321 | A/G | 上游区/下游区 Upstream/Downstream | 2.31E-07 | |||||
| 20 963 487 | A/G | 上游区/下游区 Upstream/Downstream | 5.23E-07 | 3.40E-09 | ||||
| 20 963 500 | T/C | 上游区/下游区 Upstream/Downstream | 6.56E-08 | |||||
| 20 964 042 | A/G | 内含子区 Intronic | 9.32E-07 | |||||
| 20 964 068 | T/C | 内含子区 Intronic | 9.32E-07 | |||||
| 20 964 073 | T/G | 内含子区 Intronic | 1.79E-07 | |||||
| 20 964 207 | A/G | 内含子区 Intronic | 1.51E-07 | |||||
| 20 965 920 | G/C | 内含子区 Intronic | 1.30E-07 | |||||
| 20 966 914 | T/A | 上游区 Upstream | 3.98E-07 | |||||
| 20 967 021 | C/T | 上游区 Upstream | 8.88E-07 | |||||
| pm1.2 | 21 133 564 | C/T | 基因间区 Intergenic | 4.74E-07 | ||||
| pm1.3 | 21 453 973 | T/C | 内含子区 Intronic | 6.68E-07 | ||||
| 21 454 062 | A/G | 内含子区 Intronic | 5.04E-07 | |||||
| 21 459 154 | T/C | 基因间区Intergenic | 7.54E-07 | |||||
| 21 459 157 | G/A | 基因间区Intergenic | 2.46E-07 | |||||
| 21 459 889 | A/T | 上游区 Upstream | 1.46E-07 | |||||
| 21 459 973 | C/T | 上游区 Upstream | 1.29E-07 | |||||
| 21 461 960 | A/G | 内含子区 Intronic | 1.74E-07 | |||||
| 染色体 Chromosome | 显著关联 信号 Significantly associated signals | SNP位置/bp SNP Position | 基因型 Genotype | SNP注释 SNP annotation | P值 P value | |||
| 2017- Beijing | 2018- Beijing | 2017- Guangzhou | 2018- Guangzhou | |||||
| Chr5 | pm5.1 | 1 858 575 | C/T | 上游区 Upstream | 4.14E-08 | |||
| pm5.2 | 7 910 694 | C/T | 基因间区 Intergenic | 3.20E-07 | ||||
| pm5.3 | 15 769 974 | G/C | 基因间区 Intergenic | 9.23E-07 | ||||
| pm5.4 | 16 149 984 | G/A | 内含子区 Intronic | 5.83E-07 | ||||
| pm5.5 | 16 316 760 | T/A | 内含子区 Intronic | 1.13E-07 | ||||
| 16 327 596 | G/C | 内含子区 Intronic | 6.55E-07 | |||||
| pm5.6 | 16 357 208 | T/C | 内含子区 Intronic | 4.66E-07 | ||||
| 16 380 085 | A/T | 内含子区 Intronic | 1.38E-08 | |||||
| 16 390 016 | G/A | 上游区 Upstream | 2.96E-07 | |||||
| pm5.7 | 16 666 839 | C/G | 基因间区 Intergenic | 4.64E-07 | 5.95E-08 | |||
| 16 676 542 | G/C | 外显子区(非同义突变Exonic(Nonsynonymous) | 2.44E-10 | 2.70E-07 | ||||
| 16 683 577 | C/A | 基因间区 Intergenic | 2.24E-08 | 4.22E-11 | ||||
| pm5.8 | 22 992 521 | A/G | 基因间区 Intergenic | 3.44E-08 | ||||
| 22 992 567 | A/T | 基因间区 Intergenic | 3.34E-07 | |||||
| 22 993 739 | T/A | 基因间区 Intergenic | 8.32E-07 | |||||
| Chr6 | pm6.1 | 15 844 016 | T/G | 基因间区 Intergenic | 6.97E-07 | |||
| 显著关联信号 Significantly associated signals | LD区段/bp LD block | 候选基因ID Candidate genes ID | 功能注释 Functional annotation |
|---|---|---|---|
| pm1.1 | Chr1:20 913 000 ~ | Csa1G569320 | 泛素蛋白连接酶RFI2 E3 ubiquitin-protein ligase RFI2 |
| 21 000 000 | Csa1G569330 | 丝氨酸/精氨酸富集剪接因子SR30 Serine/arginine-rich splicing factor SR30 | |
| Csa1G569340 | 嘌呤渗透酶11 Probable purine permease 11 | ||
| Csa1G569350 | 磷酸葡萄糖胺变位酶 Phosphoglucosamine mutase | ||
| Csa1G569360 | 缺氧诱导蛋白 Hypoxia induced protein | ||
| Csa1G569370 | SOSS复合体亚基B同系物 SOSS complex subunit B homolog | ||
| Csa1G569380 | 60S核糖体蛋白L22-2 60S Ribosomal protein L22-2 | ||
| Csa1G569390 | 内质网膜蛋白复合体亚基6 ER membrane protein complex subunit 6 | ||
| Csa1G569400 | 休眠相关蛋白3 Dormancy-associated protein 3 | ||
| Csa1G569410 | Tafazzin蛋白 Tafazzin protein | ||
| Csa1G569420 | 未知蛋白Unknown protein | ||
| Csa1G569430 | 橡胶延长因子Rubber elongation factor | ||
| Csa1G569440 | 重金属相关异戊二烯化植物蛋白 Heavy metal-associated isoprenylated plant protein | ||
| Csa1G569450 | 呼吸爆发氧化酶同源蛋白B Respiratory burst oxidase homolog protein B | ||
| Csa1G569460 | Formin蛋白11 Formin protein 11 | ||
| pm1.2 | Chr1:21 130 000 ~ | Csa1G570200 | 短链脱氢酶/还原酶 Short-chain dehydrogenase/reductase |
| 21 137 000 | Csa1G570210 | 核糖体生物发生蛋白类BRX1蛋白 Ribosome biogenesis protein BRX1-like protein | |
| pm1.3 | Chr1:21 452 000 ~ | Csa1G573670 | 不依赖sec移位酶TATA Sec-independent translocase protein TATA |
| 21 468 000 | Csa1G573680 | 转录因子 bHLH transcription factor bHLH(UNE12) | |
| pm5.1 | Chr5:1 800 000 ~ | Csa5G055570 | 未知蛋白 Unknown protein |
| 1 886 000 | Csa5G056070 | 未知蛋白 Unknown protein | |
| Csa5G056080 | 富含羟脯氨酸糖蛋白Hydroxyproline-rich glycoprotein-like protein | ||
| Csa5G056090 | 富含羟脯氨酸糖蛋白Hydroxyproline-rich glycoprotein-like protein | ||
| Csa5G056100 | 邻氨基苯甲酸盐合酶β亚单元Anthranilate synthase beta subunit 2 | ||
| Csa5G056110 | 未知蛋白 Unknown protein | ||
| Csa5G056610 | 未知蛋白 Unknown protein | ||
| Csa5G056620 | 3-(3-羟苯基)丙酸盐/3-羟基桂皮酸羟化酶3-(3-hydroxy-phenyl)propionate/3-hydroxycinnamic acid hydroxylase | ||
| Csa5G056630 | 香豆素酰基辅酶a:花青素3-O-葡萄糖-6"-O-香豆素酰基转移酶 2 Coumaroyl-CoA:anthocyanidin 3-O-glucoside-6''-O-coumaroyltransferase 2 | ||
| pm5.2 | Chr5:7 910 000 ~ | Csa5G180870 | RNA结合蛋白 RNA binding protein |
| 7 921 000 | Csa5G180880 | 三角状五肽重复蛋白Pentatricopeptide repeat-containing protein | |
| pm5.3 | Chr5:15 700 000 ~ | Csa5G431540 | 花期控制蛋白 Flowering time control protein FY |
| 15 847 000 | Csa5G432540 | 泛素羧基末端水解酶 Ubiquitin carboxyl-terminal hydrolase | |
| Csa5G432550 | 泛素羧基末端水解酶 Ubiquitin carboxyl-terminal hydrolase | ||
| Csa5G434550 | 双因子响应调节蛋白 Two-component response regulator ORR10 | ||
| Csa5G435050 | DNA定向RNA聚合酶V亚单位1 DNA-directed RNA polymerase V subunit 1 | ||
| Csa5G435060 | 未知蛋白 Unknown protein | ||
| Csa5G435070 | 磷脂酰肌醇4-磷酸5-激酶6 Phosphatidylinositol 4-phosphate 5-kinase 6 | ||
| Csa5G435080 | 醌氧化还原酶NAD(P)H:quinone oxidoreductase | ||
| pm5.4 | Chr5:16 130 000 ~ | Csa5G456730 | 尿苷5'-磷酸合酶 Uridine 5'-monophosphate synthase |
| 16 170 000 | Csa5G457230 | ADP核糖基化因子GTP激活蛋白AGD11 同类型X1蛋白 ADP-ribosylation factor GTPase-activating protein AGD11 isoform X1 | |
| Csa5G457240 | 对红光的敏感性降低蛋白 Sensitivity to red light reduced protein | ||
| pm5.5 | Chr5:16 312 000 ~ | Csa5G466330 | rRNA-加工蛋白质UTP23 rRNA-processing protein UTP23 |
| 16 354 000 | Csa5G466340 | 未知蛋白Unknown protein | |
| Csa5G466350 | 富含脯氨酸受体蛋白激酶PERK10 Proline-rich receptor-like protein kinase PERK10 | ||
| Csa5G466360 | L型凝集素结构域的受体激酶L-type lectin-domain containing receptor kinase | ||
| pm5.6 | Chr5:16 355 000 ~ 16 382 000 | Csa5G466370 | 组氨酸生物合成双功能蛋白hisIE Histidine biosynthesis bifunctional protein hisIE |
| Csa5G466870 | 40S核糖体蛋白S27 40S ribosomal protein S27 | ||
| 显著关联信号 Significantly associated signals | LD区段/bp LD block | 候选基因ID Candidate genes ID | 功能注释 Functional annotation |
| pm5.7 | Chr5:16 665 000 ~ 16 706 000 | Csa5G471600 | 异嗜性和多嗜性逆转录病毒受体蛋白 Xenotropic and polytropic retrovirus receptor protein |
| Csa5G472100 | 肽基脯氨酸顺反式异构酶NIMA相互作用蛋白4 Peptidyl-prolyl cis-trans isomerase NIMA-interacting 4 | ||
| pm5.8 | Chr5:22 994 000 ~ | Csa5G606690 | 三角状五肽重复蛋白 Pentatricopeptide repeat-containing protein |
| 23 053 000 | Csa5G606700 | 未知蛋白 Unknown protein | |
| Csa5G606710 | 未知蛋白 Unknown protein | ||
| Csa5G606720 | 未知蛋白 Unknown protein | ||
| Csa5G606730 | Ninja家族蛋白 Ninja-family protein AFP3 | ||
| Csa5G606740 | 天冬酰胺合成酶蛋白 Asparagine synthetase domain-containing protein 1 | ||
| Csa5G606750 | 细胞分裂控制蛋白48同源物B Cell division control protein 48 homolog B | ||
| Csa5G606760 | 核糖体成熟因子rimP Ribosome maturation factor rimP | ||
| Csa5G606770 | 6-磷酸葡萄糖酸内酯酶1 6-phosphogluconolactonase 1 | ||
| Csa5G606780 | CRABS CLAW蛋白CRABS CLAW protein | ||
| Csa5G606790 | 类大谷粒蛋白EBIG GRAIN 1-like E | ||
| pm6.1 | Chr6:15 801 000 ~ | Csa6G351870 | 细胞壁连接的类受体激酶3 Wall-associated receptor kinase 3 |
| 15 847 000 | Csa6G352870 | 细胞壁连接的类受体激酶2 Wall-associated receptor kinase 2 | |
| Csa6G354370 | 细胞壁连接的类受体激酶5 Wall-associated receptor kinase 5-like |
表3 与白粉病抗性显著关联的候选基因
Table 3 Candidate genes associated with powdery mildew resistantance
| 显著关联信号 Significantly associated signals | LD区段/bp LD block | 候选基因ID Candidate genes ID | 功能注释 Functional annotation |
|---|---|---|---|
| pm1.1 | Chr1:20 913 000 ~ | Csa1G569320 | 泛素蛋白连接酶RFI2 E3 ubiquitin-protein ligase RFI2 |
| 21 000 000 | Csa1G569330 | 丝氨酸/精氨酸富集剪接因子SR30 Serine/arginine-rich splicing factor SR30 | |
| Csa1G569340 | 嘌呤渗透酶11 Probable purine permease 11 | ||
| Csa1G569350 | 磷酸葡萄糖胺变位酶 Phosphoglucosamine mutase | ||
| Csa1G569360 | 缺氧诱导蛋白 Hypoxia induced protein | ||
| Csa1G569370 | SOSS复合体亚基B同系物 SOSS complex subunit B homolog | ||
| Csa1G569380 | 60S核糖体蛋白L22-2 60S Ribosomal protein L22-2 | ||
| Csa1G569390 | 内质网膜蛋白复合体亚基6 ER membrane protein complex subunit 6 | ||
| Csa1G569400 | 休眠相关蛋白3 Dormancy-associated protein 3 | ||
| Csa1G569410 | Tafazzin蛋白 Tafazzin protein | ||
| Csa1G569420 | 未知蛋白Unknown protein | ||
| Csa1G569430 | 橡胶延长因子Rubber elongation factor | ||
| Csa1G569440 | 重金属相关异戊二烯化植物蛋白 Heavy metal-associated isoprenylated plant protein | ||
| Csa1G569450 | 呼吸爆发氧化酶同源蛋白B Respiratory burst oxidase homolog protein B | ||
| Csa1G569460 | Formin蛋白11 Formin protein 11 | ||
| pm1.2 | Chr1:21 130 000 ~ | Csa1G570200 | 短链脱氢酶/还原酶 Short-chain dehydrogenase/reductase |
| 21 137 000 | Csa1G570210 | 核糖体生物发生蛋白类BRX1蛋白 Ribosome biogenesis protein BRX1-like protein | |
| pm1.3 | Chr1:21 452 000 ~ | Csa1G573670 | 不依赖sec移位酶TATA Sec-independent translocase protein TATA |
| 21 468 000 | Csa1G573680 | 转录因子 bHLH transcription factor bHLH(UNE12) | |
| pm5.1 | Chr5:1 800 000 ~ | Csa5G055570 | 未知蛋白 Unknown protein |
| 1 886 000 | Csa5G056070 | 未知蛋白 Unknown protein | |
| Csa5G056080 | 富含羟脯氨酸糖蛋白Hydroxyproline-rich glycoprotein-like protein | ||
| Csa5G056090 | 富含羟脯氨酸糖蛋白Hydroxyproline-rich glycoprotein-like protein | ||
| Csa5G056100 | 邻氨基苯甲酸盐合酶β亚单元Anthranilate synthase beta subunit 2 | ||
| Csa5G056110 | 未知蛋白 Unknown protein | ||
| Csa5G056610 | 未知蛋白 Unknown protein | ||
| Csa5G056620 | 3-(3-羟苯基)丙酸盐/3-羟基桂皮酸羟化酶3-(3-hydroxy-phenyl)propionate/3-hydroxycinnamic acid hydroxylase | ||
| Csa5G056630 | 香豆素酰基辅酶a:花青素3-O-葡萄糖-6"-O-香豆素酰基转移酶 2 Coumaroyl-CoA:anthocyanidin 3-O-glucoside-6''-O-coumaroyltransferase 2 | ||
| pm5.2 | Chr5:7 910 000 ~ | Csa5G180870 | RNA结合蛋白 RNA binding protein |
| 7 921 000 | Csa5G180880 | 三角状五肽重复蛋白Pentatricopeptide repeat-containing protein | |
| pm5.3 | Chr5:15 700 000 ~ | Csa5G431540 | 花期控制蛋白 Flowering time control protein FY |
| 15 847 000 | Csa5G432540 | 泛素羧基末端水解酶 Ubiquitin carboxyl-terminal hydrolase | |
| Csa5G432550 | 泛素羧基末端水解酶 Ubiquitin carboxyl-terminal hydrolase | ||
| Csa5G434550 | 双因子响应调节蛋白 Two-component response regulator ORR10 | ||
| Csa5G435050 | DNA定向RNA聚合酶V亚单位1 DNA-directed RNA polymerase V subunit 1 | ||
| Csa5G435060 | 未知蛋白 Unknown protein | ||
| Csa5G435070 | 磷脂酰肌醇4-磷酸5-激酶6 Phosphatidylinositol 4-phosphate 5-kinase 6 | ||
| Csa5G435080 | 醌氧化还原酶NAD(P)H:quinone oxidoreductase | ||
| pm5.4 | Chr5:16 130 000 ~ | Csa5G456730 | 尿苷5'-磷酸合酶 Uridine 5'-monophosphate synthase |
| 16 170 000 | Csa5G457230 | ADP核糖基化因子GTP激活蛋白AGD11 同类型X1蛋白 ADP-ribosylation factor GTPase-activating protein AGD11 isoform X1 | |
| Csa5G457240 | 对红光的敏感性降低蛋白 Sensitivity to red light reduced protein | ||
| pm5.5 | Chr5:16 312 000 ~ | Csa5G466330 | rRNA-加工蛋白质UTP23 rRNA-processing protein UTP23 |
| 16 354 000 | Csa5G466340 | 未知蛋白Unknown protein | |
| Csa5G466350 | 富含脯氨酸受体蛋白激酶PERK10 Proline-rich receptor-like protein kinase PERK10 | ||
| Csa5G466360 | L型凝集素结构域的受体激酶L-type lectin-domain containing receptor kinase | ||
| pm5.6 | Chr5:16 355 000 ~ 16 382 000 | Csa5G466370 | 组氨酸生物合成双功能蛋白hisIE Histidine biosynthesis bifunctional protein hisIE |
| Csa5G466870 | 40S核糖体蛋白S27 40S ribosomal protein S27 | ||
| 显著关联信号 Significantly associated signals | LD区段/bp LD block | 候选基因ID Candidate genes ID | 功能注释 Functional annotation |
| pm5.7 | Chr5:16 665 000 ~ 16 706 000 | Csa5G471600 | 异嗜性和多嗜性逆转录病毒受体蛋白 Xenotropic and polytropic retrovirus receptor protein |
| Csa5G472100 | 肽基脯氨酸顺反式异构酶NIMA相互作用蛋白4 Peptidyl-prolyl cis-trans isomerase NIMA-interacting 4 | ||
| pm5.8 | Chr5:22 994 000 ~ | Csa5G606690 | 三角状五肽重复蛋白 Pentatricopeptide repeat-containing protein |
| 23 053 000 | Csa5G606700 | 未知蛋白 Unknown protein | |
| Csa5G606710 | 未知蛋白 Unknown protein | ||
| Csa5G606720 | 未知蛋白 Unknown protein | ||
| Csa5G606730 | Ninja家族蛋白 Ninja-family protein AFP3 | ||
| Csa5G606740 | 天冬酰胺合成酶蛋白 Asparagine synthetase domain-containing protein 1 | ||
| Csa5G606750 | 细胞分裂控制蛋白48同源物B Cell division control protein 48 homolog B | ||
| Csa5G606760 | 核糖体成熟因子rimP Ribosome maturation factor rimP | ||
| Csa5G606770 | 6-磷酸葡萄糖酸内酯酶1 6-phosphogluconolactonase 1 | ||
| Csa5G606780 | CRABS CLAW蛋白CRABS CLAW protein | ||
| Csa5G606790 | 类大谷粒蛋白EBIG GRAIN 1-like E | ||
| pm6.1 | Chr6:15 801 000 ~ | Csa6G351870 | 细胞壁连接的类受体激酶3 Wall-associated receptor kinase 3 |
| 15 847 000 | Csa6G352870 | 细胞壁连接的类受体激酶2 Wall-associated receptor kinase 2 | |
| Csa6G354370 | 细胞壁连接的类受体激酶5 Wall-associated receptor kinase 5-like |
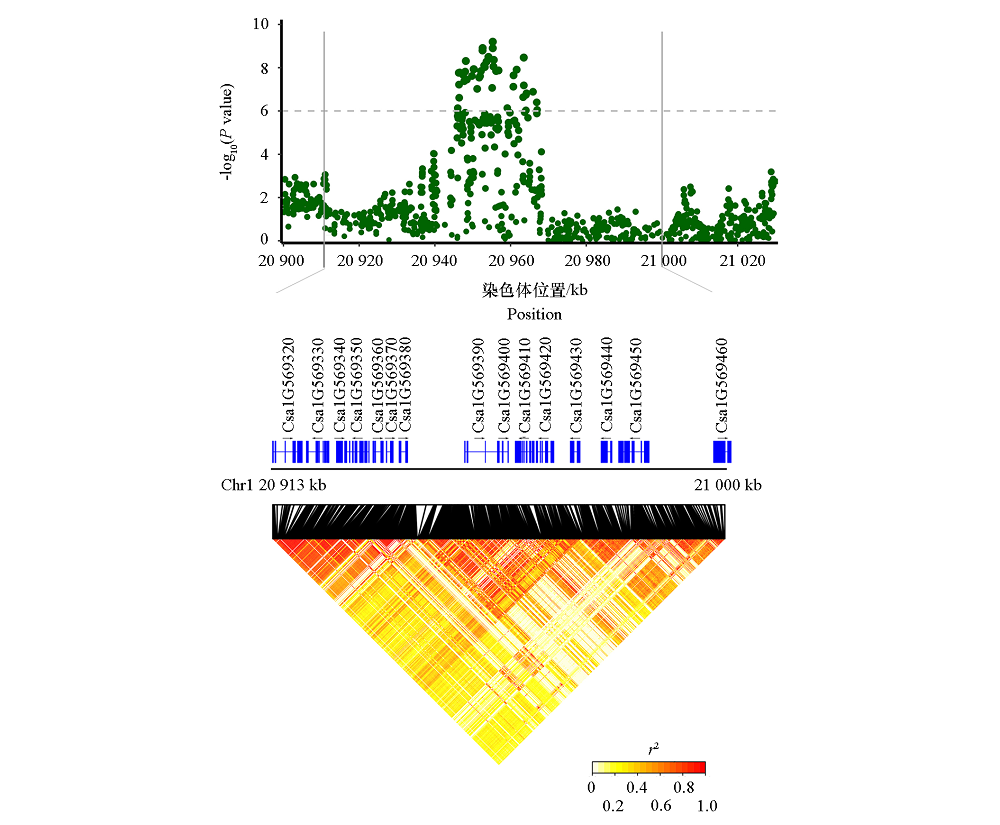
图4 北京两年均检出显著关联信号pm1.1的曼哈顿图和候选基因预测
Fig. 4 Manhattan diagram and candidate gene prediction of significant associated signal pm1.1 detected in two-year in Beijing
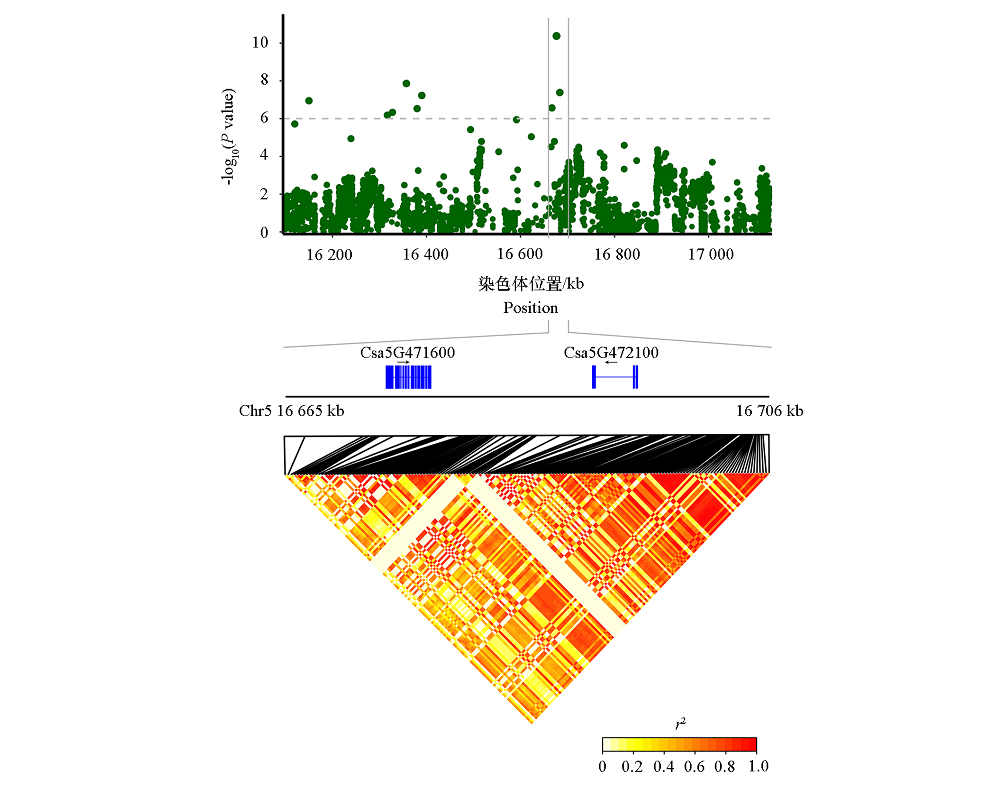
图5 广州两年均检出显著关联信号pm5.7的曼哈顿图和候选基因预测
Fig. 5 Manhattan diagram and candidate gene prediction of significant associated signal pm5.7 detected in two-year in Guangzhou
| [1] | Abd Hamid N A, Zainal Z, Ismai I. 2018. Two members of unassigned type of short-chain dehydrogenase/reductase superfamily(SDR)isolated from Persicaria minor show response towards ABA and drought stress. Journal of Plant Biochemistry & Biotechnology, 27:260-271. |
| [2] |
Barrett J C, Fry B, Maller J, Daly M J. 2005. Haploview:analysis and visualization of LD and haplotype maps. Bioinformatics, 21(5):263-265.
doi: 10.1093/bioinformatics/bth457 URL |
| [3] |
Bo K, Wei S, Wang W, Miao H, Dong S, Zhang S, Gu X. 2019. QTL mapping and genome-wide association study reveal two novel loci associated with green flesh color in cucumber. BMC Plant Biology, 19(1):243.
doi: 10.1186/s12870-019-1835-6 URL |
| [4] |
Cheng C, Xu X, Gao M, Li J, Guo C, Song J, Wang X. 2013. Genome-wide analysis of respiratory burst oxidase homologs in grape(Vitis vinifera L.). International Journal of Molecular Sciences, 14(12):24169-24186.
doi: 10.3390/ijms141224169 URL |
| [5] | Cheng Jia-qi, Shen Di, Li Xi-xiang, Song Jiang-ping, Wang Hai-ping, Qiu Yang. 2011. Field resistance evaluation of cucumber core collection to powdery mildew. China Vegetables, (20):15-19. (in Chinese) |
| 程嘉琪, 沈镝, 李锡香, 宋江萍, 王海平, 邱杨. 2011. 黄瓜核心种质对白粉病的田间抗性评价. 中国蔬菜, (20):15-19. | |
| [6] |
Du X, Huang G, He S, Yang Z, Sun G, Ma X, Li N, Zhang X, Sun J, Liu M. 2018. Resequencing of 243 diploid cotton accessions based on an updated A genome identifies the genetic basis of key agronomic traits. Nature Genetics, 50:796-802.
doi: 10.1038/s41588-018-0116-x URL |
| [7] |
Fukino N, Yoshioka Y, Sugiyama M, Sakata Y, Matsumoto S. 2013. Identification and validation of powdery mildew(Podosphaera xanthii)-resistant loci in recombinant inbred lines of cucumber(Cucumis sativus L.). Molecular Breeding, 32:267-277.
doi: 10.1007/s11032-013-9867-3 URL |
| [8] | Hao Jun-jie, Li Lei, Wang Bo, Qin Yu-hong, Cui Jian, Wang Ying, Wang Pei-sheng, Jiang Zhi-xun, Sun Ji-lu, Wang Zhen-qing, Yue Huan, Zhang Shou-cai. 2018. Fine mapping and analysis candidate gene to powdery mildew in cucumber(Cucumis sativus L.). Scientia Agricultura Sinica, 51(17):3427-3434. (in Chinese) |
| 郝俊杰, 李磊, 王波, 秦玉红, 崔健, 王瑛, 王佩圣, 江志训, 孙吉禄, 王珍青, 岳欢, 张守才. 2018. 黄瓜白粉病抗性基因定位及候选基因分析. 中国农业科学, 51(17):3427-3434. | |
| [9] | He X, Li Y, Pandey S, Yandell B S, Pathak M, Weng Y. 2013. QTL mapping of powdery mildew resistance in WI 2757 cucumber(Cucumis sativus L.). Theoretical & Applied Genetics, 126(8):2149-2161. |
| [10] |
Javierre B M, Burren O S, Wilder S P, Kreuzhuber R, Hill S M, Sewitz S, Cairns J, Wingett S W, Várnai C, Thiecke M J, Burden F, Farrow S, Cutler A J, Rehnström K, Downes K, Grassi L, Kostadima M, Freire-Pritchett P, Wang F, Consortium B, Stunnenberg H G, Todd J A, Zerbino D R, Stegle O, Ouwehand W H, Frontini M, Wallace C, Spivakov M, Fraser P. 2016. Lineage-specific genome architecture links enhancers and non-coding disease variants to target gene promoters. Cell, 167(5):1369-1384.
doi: S0092-8674(16)31322-8 pmid: 27863249 |
| [11] | Kallberg Y, Oppermann U, Jörnvall H, Persson B. 2008. Medium-and short-chain dehydrogenase/reductase gene and protein families. Cellular & Molecular Life Sciences, 65(24):3950. |
| [12] |
Khurana E, Fu Y, Chakravarty D, Demichelis F, Rubin M A, Gerstein M. 2016. Role of non-coding sequence variants in cancer. Nature Reviews Genetics, 17(2):93-108.
doi: 10.1038/nrg.2015.17 pmid: 26781813 |
| [13] |
Kikuchi M, Hara N, Hasegawa M, Miyashita A, Kuwano R, Ikeuchi T, Nakaya A. 2019. Enhancer variants associated with Alzheimer’s disease affect gene expression via chromatin looping. BMC Medical Genomics, 12:128.
doi: 10.1186/s12920-019-0574-8 URL |
| [14] |
Kristina B, Anne N, Scott J N, Chris K, Jennifer A C, David A, Tobias L, Giulia G, Shiana M, Crystal N D, Jessalyn B, Nayana L, Elodie P, Simon C W, Colúm C, Rebecca A S, REGISTRY I E H D N, Sarah J T, Ola H, Douglas R L, Michael R H, Wyeth W W, Blair R L. 2015. A SNP in the HTT promoter alters NF-κB binding and is a bidirectional genetic modifier of Huntington disease. Nature Neuroscience, 18:807-816.
doi: 10.1038/nn.4014 URL |
| [15] | Li Xi-xiang, Zhu De-wei. 2005. Description specification and data standard of cucumber germplasm resources. Beijing:China Agriculture Press. (in Chinese) |
| 李锡香, 朱德蔚. 2005. 黄瓜种质资源描述规范和数据标准, 北京:中国农业出版社. | |
| [16] |
Li Y, Li Q, Guo G, He T, Gao R, Muhammad F, Huang J, Lu R, Liu C. 2018. Transient overexpression of HvSERK2 improves barley resistance to powdery mildew. International Journal of Molecular Sciences, 19(4):1226.
doi: 10.3390/ijms19041226 URL |
| [17] | Liu Dong, Xin Ming, Zhou Xiu-yan, Wang Chun-hua, Qin Zhi-wei. 2017. Resistance evaluation of main cucumber germplasms. Journal of Northeast Agricultural University, 48(3):10-16. (in Chinese) |
| 刘东, 辛明, 周秀艳, 王春华, 秦智伟. 2017. 主要黄瓜种质资源抗病性评价. 东北农业大学学报, 48(3):10-16. | |
| [18] | Liu Long-zhou, Cai Rui, Yuan Xiao-jun, He Huan-le, Pan Jun-song. 2008. QTL mapping of powdery mildew resistance in cucumber. Chinese Science, 9(6):851-856. (in Chinese) |
| 刘龙洲, 蔡润, 袁晓君, 何欢乐, 潘俊松. 2008. 黄瓜抗白粉病QTL分子标记定位. 中国科学, 9(6):851-856. | |
| [19] |
Liu X, Lu H, Liu P, Miao H, Bai Y, Gu X, Zhang S. 2020. Identification of novel loci and candidate genes for cucumber downy mildew resistance using GWAS. Plants, 9:1659.
doi: 10.3390/plants9121659 URL |
| [20] | Lou H Q, Fan W, Jin J F, Xu J M, Zheng S J. 2020. A NAC‐type transcription factor confers aluminium resistance by regulating cell wall‐associated receptor kinase 1 and cell wall pectin. Plant, Cell & Environment, 43(2):463-478. |
| [21] |
Lu P, Guo L, Wang Z, Li B, Liu Z. 2020. A rare gain of function mutation in a wheat tandem kinase confers resistance to powdery mildew. Nature Communications, 11(1):680.
doi: 10.1038/s41467-020-14294-0 URL |
| [22] | Luo Jing-jing, Qi Xiao-hua, Chen Xue-hao. 2010. Advances on genetic mechanisms of powdery mildew resistance in cucurbit crops. Molecular Plant Breeding, 8(3):556-562. (in Chinese) |
| 罗晶晶, 齐晓花, 陈学好. 2010. 瓜类作物白粉病抗性遗传机制. 分子植物育种, 8(3):556-562. | |
| [23] |
Lü J, Qi J, Shi Q, Shen D, Zhang S, Shao G, Li H, Sun Z, Weng Y, Shang Y, Gu X, Li X, Zhu X, Zhang J, Treuren R, Dooijeweert W, Zhang Z, Huang S. 2012. Genetic diversity and population structure of cucumber(Cucumis sativus L.). PLoS ONE, 7(10):e46919.
doi: 10.1371/journal.pone.0046919 URL |
| [24] | Mailer R K, Allende M, Heestermans M, Schweizer M, Renné T. 2020. Xenotropic and polytropic retrovirus receptor 1 regulates procoagulant platelet polyphosphate. Blood, 9(15):4617. |
| [25] |
Maurano M T, Humbert R, Rynes E, Thurman R E, Haugen E, Wang H, Reynolds A P, Sandstrom R, Qu H, Brody J. 2012. Systematic localization of common disease-associated variation in regulatory DNA. Science, 337(6099):1190-1195.
doi: 10.1126/science.1222794 URL |
| [26] |
Mukhtar M S, Carvunis A R, Dreze M, Epple P, Steinbrenner J, Moore J, Tasan M, Galli M, Hao T, Nishimura M T, Pevzner S J, Donovan S E, Ghamsari L, Santhanam B, Romero V, Poulin M M, Gebreab F, Gutierrez B J, Tam S, Monachello D, Boxem M, Harbort C J, McDonald N, Gai L T, Chen H M, He Y J, European Union Effectoromics Consortium, Vandenhaute J, Roth F P, Hill D E, Ecker J R, Vidal M, Beynon J, Braun P, Dangl J L. 2011. Independently evolved virulence effectors converge onto hubs in a plant immune system network. Science, 333(6042):596-601.
doi: 10.1126/science.1203659 URL pmid: 21798943 |
| [27] |
Nie J, He H, Peng J, Yang X, Bie B, Zhao J, Wang Y, Si L, Pan J S, Cai R. 2015. Identification and fine mapping of pm5.1:a recessive gene for powdery mildew resistance in cucumber(Cucumis sativus L.). Molecular Breeding, 35(1):7.
doi: 10.1007/s11032-015-0206-8 URL |
| [28] |
Nyquist W E, Baker R J. 1991. Estimation of heritability and prediction of selection response in plant population. Critical Reviews in Plant Sciences, 10(3):235-322.
doi: 10.1080/07352689109382313 URL |
| [29] |
Tanabe N, Yoshimura K, Kimura A, Yabuta Y, Shigeoka S. 2007. Differential expression of alternatively spliced mRNAs of Arabidopsis SR protein homologs,atSR30 and atSR45a,in response to environmental stress. Plant Cell Physiology, 48(12):1826.
doi: 10.1093/pcp/pcm156 URL |
| [30] |
Pathak S, Meng W J, Zhang H, Gnosa S, Nandy S K, Adell G, Holmlund B, Sun X F. 2014. Tafazzin protein expression is associated with tumorigenesis and radiation response in rectal cancer:a study of swedish clinical trial on preoperative radiotherapy. PLoS ONE, 9(5):e98317.
doi: 10.1371/journal.pone.0098317 URL |
| [31] |
Peng Y, Xiong D, Zhao L, Ouyang W, Wang S, Sun J, Zhang Q, Guan P, Xie L, Li W, Li G, Yan J, Li X. 2019. Chromatin interaction maps reveal genetic regulation for quantitative traits in maize. Nature Communications, 10(1):2632.
doi: 10.1038/s41467-019-10602-5 URL |
| [32] |
Pinski A, Betekhtin A, Sala K, Godel-Jedrychowska K, Kurczynska E, Hasterok R. 2019. Hydroxyproline-rich glycoproteins as markers of temperature stress in the leaves of Brachypodium distachyon. International Journal of Molecular Sciences, 20(10):2571.
doi: 10.3390/ijms20102571 URL |
| [33] | Sakata Y, Kubo N, Morishita M, Kitadani E, Hirai M. 2006. QTL analysis of powdery mildew resistance in cucumber(Cucumis sativus L.). Theoretical & Applied Genetics, 112(2):243-250. |
| [34] |
Tian F, Bradbury P J, Brown P J, Hung H, Sun Q, Flint-Garcia S, Rocheford T R, Mcmullen M D, Holland J B, Buckler E S. 2011. Genome-wide association study of leaf architecture in the maize nested association mapping population. Nature Genetics, 43(2):159-162.
doi: 10.1038/ng.746 URL |
| [35] |
Ueda Y, Frimpong F, Qi Y, Matthus E, Wu L, Hller S, Kraska T, Frei M. 2015. Genetic dissection of ozone tolerance in rice(Oryza sativa L.) by a genome-wide association study. Journal of Experimental Botany, 66(1):293-306.
doi: 10.1093/jxb/eru419 URL |
| [36] |
Wang X, Bao K, Reddy U K, Bai Y, Hammar S A, Jiao C, Wehner T C, Ramírez-Madera A O, Weng Y, Grumet R, Fei Z. 2018. The USDA cucumber(Cucumis sativus L.)collection:genetic diversity,population structure,genome-wide association studies,and core collection development. Horticulture Research, 5:64.
doi: 10.1038/s41438-018-0080-8 pmid: 30302260 |
| [37] | Wei Shuang, Zhang Song, Bo Kai-liang, Wang Wei-ping, Miao Han, Dong Shao-yun, Gu Xing-fang, Zhang Sheng-ping. 2019. Evaluation and genome-wide association study(GWAS)of seedling thermotolerance in cucumber core germplasm. Journal of Plant Genetic Resources, 20(5):1223-1231. (in Chinese) |
| 魏爽, 张松, 薄凯亮, 王伟平, 苗晗, 董邵云, 顾兴芳, 张圣平. 2019. 黄瓜核心种质幼苗耐热性评价及 GWAS 分析. 植物遗传资源学报, 20(5):1223-1231. | |
| [38] | Wu Feng-ying, Liu Meng-qi, Wang Yue-jin. 2020. Research on control of disease resistance by stilbene synthase genes derived from Chinese wild Vitis quinquangularis. Acta Horticulturae Sinica, 47(2):205-219. (in Chinese) |
| 吴凤颖, 刘梦琦, 王跃进. 2020. 中国野生毛葡萄芪合酶基因抗白粉病功能分析. 园艺学报, 47(2):205-219. | |
| [39] | Xu X, Yu, T, Xu R, Shi Y, Lin X, Xu Q, Qi X, Weng Y, Chen X. 2016. Fine mapping of a dominantly inherited powdery mildew resistance major-effect QTL,Pm1.1,in cucumber identifies a 41.1kb region containing two tandemly arrayed cysteine-rich receptor-like protein kinase genes. Theoretical & Applied Genetics, 129(3):507-516. |
| [40] |
Yang J, Lee S H, Goddard M E, Visscher P M. 2011. GCTA:a tool for genome-wide complex trait analysis. American Journal of Human Genetics, 88(1):76-82.
doi: 10.1016/j.ajhg.2010.11.011 URL |
| [41] |
Yang Z, Li G, Tieman D, Zhu G. 2019. Genomics approaches to domestication studies of horticultural crops. Horticultural Plant Journal, 5(6):240-246.
doi: 10.1016/j.hpj.2019.11.001 URL |
| [42] |
Zhang H, Zhang X, Liu J, Niu Y, Chen Y, Hao Y, Zhao J, Sun L, Wang H, Xiao J, Wang X. 2020. Characterization of the Heavy-Metal-Associated Isoprenylated Plant Protein(HIPP)gene family from Triticeae species. International Journal of Molecular Sciences, 21(17):6191.
doi: 10.3390/ijms21176191 URL |
| [43] | Zhang K, Wang X, Zhu W, Qin X, Xu J, Cheng C, Lou Q, Li J, Chen J. 2018. Complete resistance to powdery mildew and partial resistance to downy mildew in a Cucumis hystrix introgression line of cucumber were controlled by a co-localized locus. Theoretical & Applied Genetics, 131:2229-2243. |
| [44] | Zhang Sheng-ping, Liu Miao-miao, Miao Han, Zhang Su-qin, Yang Yu-hong, Xie Bing-yan, Gu Xing-fang. 2011. QTL mapping of resistance genes to powdery mildew in cucumber(Cucumis sativus L.). Scientia Agricultura Sinica, 44(17):3584-3593. (in Chinese) |
| 张圣平, 刘苗苗, 苗晗, 张素勤, 杨宇红, 谢丙炎, 顾兴芳. 2011. 黄瓜白粉病抗性基因的QTL定位. 中国农业科学, 44(17):3584-3593. | |
| [45] |
Zhang Y, Bai Y, Wu G, Zou S, Tang D. 2017. Simultaneous modification of three homoeologs of TaEDR1 by genome editing enhances powdery mildew resistance in wheat. Plant Journal, 91(4):714-724.
doi: 10.1111/tpj.2017.91.issue-4 URL |
| [46] | Zhao Kai-qian, Ding Qian, Wang Yue-jin. 2020. Function analysis of the stilbene synthase genes VqSTS11 and VqSTS23 of the resistance to powdery mildew in Vitis quinquangularis. Acta Horticulturae Sinica, 47(7):1264-1276. (in Chinese) |
| 赵凯茜, 丁茜, 王跃进. 2020. 中国野生毛葡萄芪合酶基因VqSTS11和VqSTS23调控抗白粉病的研究. 园艺学报, 47(7):1264-1276. | |
| [47] | Zheng L, Zhang M, Zhuo Z, Wang Y, Gao X, Li Y, Liu W, Zhang W. 2020. Transcriptome profiling analysis reveals distinct resistance response of cucumber leaves infected with powdery mildew. Plant Biology, 10:13213. |
| [48] |
Zhou X, Stephens M. 2012. Genome-wide efficient mixed model analysis for association studies. Nature Genetics, 44(7):821-824.
doi: 10.1038/ng.2310 URL |
| [49] |
Zhu G, Wang S, Huang Z, Zhang S, Liao Q, Zhang C, Lin T, Qin M, Peng M, Yang C. 2018. Rewiring of the fruit metabolome in tomato breeding. Cell, 172(1-2):249-261.
doi: 10.1016/j.cell.2017.12.019 URL |
| [50] |
Zou S, Wang H, Li Y, Kong Z, Tang D. 2017. The NB‐LRR gene Pm60 confers powdery mildew resistance in wheat. New Phytologist, 218(1):298-309.
doi: 10.1111/nph.14964 URL |
| [1] | 罗天宽, 吴海涛, 张圣美, 黄宗安, 孙 继, 水德聚, 陈先知. 黄瓜新品种‘瓯翠1号’[J]. 园艺学报, 2022, 49(S2): 125-126. |
| [2] | 王鹤冰, 向华丰, 陈新中, 张 生, 张洪成. 华南型黄瓜新品种‘新燕095’[J]. 园艺学报, 2022, 49(S1): 79-80. |
| [3] | 许春梅, 张作标, 柳景兰, 王 昕, 杨 龙, 赵 丹, 刘思宇, 贾云鹤, 孟雪娇, 崔嵩岑. 黄瓜新品种‘绿春2号’[J]. 园艺学报, 2022, 49(S1): 81-82. |
| [4] | 张利东, 黄洪宇, 孔维良, 李加旺, 李愚鹤, . 华北型黄瓜新品种‘津优355’[J]. 园艺学报, 2022, 49(S1): 83-84. |
| [5] | 王惠哲, 杨瑞环, 邓 强, 曹明明, 李淑菊, . 抗黑星病黄瓜新品种‘津冬369’[J]. 园艺学报, 2022, 49(S1): 85-86. |
| [6] | 聂鑫淼, 栾恒, 冯改利, 王超, 李岩, 魏珉. 硅营养和嫁接砧木对黄瓜幼苗耐冷性的影响[J]. 园艺学报, 2022, 49(8): 1795-1804. |
| [7] | 魏晓羽, 王跃进. 中国野生葡萄果皮解剖结构与白粉病抗性的相关性研究[J]. 园艺学报, 2022, 49(6): 1200-1212. |
| [8] | 韩鲁杰, 冯一清, 杨秀华, 张宁, 毕焕改, 艾希珍. 有机肥化肥配施对大棚黄瓜根区土壤与根系特征的影响[J]. 园艺学报, 2022, 49(5): 1047-1059. |
| [9] | 权建华, 段誉, 罗天, 袁强, 齐鑫, 王勤礼. 黄瓜新品种‘裕研9号’[J]. 园艺学报, 2022, 49(3): 703-704. |
| [10] | 卢甜甜, 刘志远, 徐兆生, 张合龙, 李国亮, 折红兵, 钱伟. 菜豆花色全基因组关联分析[J]. 园艺学报, 2022, 49(2): 332-340. |
| [11] | 宋蒙飞, 查高辉, 陈劲枫, 娄群峰. 黄瓜株型性状分子基础研究进展[J]. 园艺学报, 2022, 49(12): 2683-2702. |
| [12] | 王荣花, 王树彬, 刘栓桃, 李巧云, 张志刚, 王立华, 赵智中. 大白菜花茎蜡粉近等基因系转录组分析[J]. 园艺学报, 2022, 49(1): 62-72. |
| [13] | 王团团, 缑晨星, 夏磊, 朱拼玉, 李季, 陈劲枫. 黄瓜10个基因型材料外植体内源激素水平及比例对其离体再生的影响[J]. 园艺学报, 2021, 48(9): 1731-1742. |
| [14] | 赵昌博, 郑世伟, 边婷, 王爽, 张小兰, 富宏丹, 孙周平, 李天来. 日光温室黄瓜不同连作茬次土壤的中量与微量元素含量变化[J]. 园艺学报, 2021, 48(9): 1805-1814. |
| [15] | 甘彩霞, 崔磊, 庞文星, 汪爱华, 於校青, 邓晓辉, 宋莉萍, 朴钟云. 基于萝卜高密度遗传图谱的抽薹和开花性状的QTL定位[J]. 园艺学报, 2021, 48(7): 1273-1281. |
| 阅读次数 | ||||||
|
全文 |
|
|||||
|
摘要 |
|
|||||
版权所有 © 2012 《园艺学报》编辑部 京ICP备10030308号-2 国际联网备案号 11010802023439
编辑部地址: 北京市海淀区中关村南大街12号中国农业科学院蔬菜花卉研究所 邮编: 100081
电话: 010-82109523 E-Mail: yuanyixuebao@126.com
技术支持:北京玛格泰克科技发展有限公司