
Acta Horticulturae Sinica ›› 2025, Vol. 52 ›› Issue (7): 1780-1788.doi: 10.16420/j.issn.0513-353x.2024-0767
• Genetic & Breeding·Germplasm Resources·Molecular Biology • Previous Articles Next Articles
LI Xuesong1,2, HUA Rong1,2, SUN Dafeng1,2, YUE Wansong1, ZHANG Junbo1, LI Jianying1, and LIU Shaoxiong1,2,*( )
)
Received:2025-02-02
Revised:2025-05-14
Online:2025-07-23
Published:2025-07-23
Contact:
and LIU Shaoxiong
LI Xuesong, HUA Rong, SUN Dafeng, YUE Wansong, ZHANG Junbo, LI Jianying, and LIU Shaoxiong. Development of SSR Markers Based on Transcriptiome of Naematelia aurantialba[J]. Acta Horticulturae Sinica, 2025, 52(7): 1780-1788.
Add to citation manager EndNote|Ris|BibTeX
URL: https://www.ahs.ac.cn/EN/10.16420/j.issn.0513-353x.2024-0767
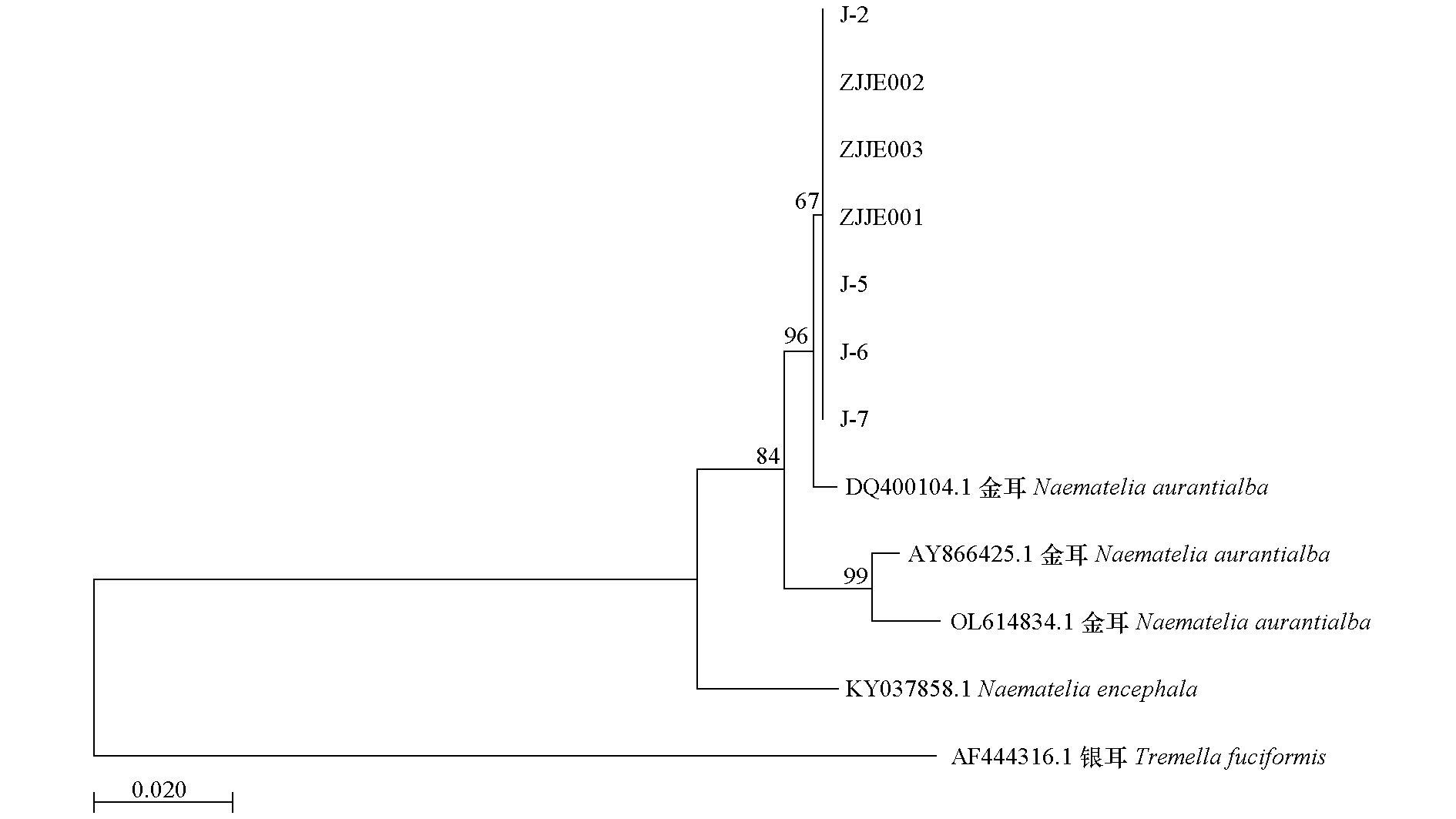
Fig. 1 phylogenetic tree using neighbor-joining method for Naematelia aurantialba The numbers in the figure represent the support rates for each branch node,with a maximum value of 100. Higher values indicate greater confidence in the node,while unmarked nodes are assumed to have a support rate of 100
| 标记名称 Markers | 上游引物(5′-3′) Forward primer | 下游引物(5′-3′) Reverse primer | SSR 类型 SSR type | SSR 基元 SSR motif |
|---|---|---|---|---|
| JESSR003 | CATCCCTATGCAATGATTCCACG | TAGAACAAGACTAGAACCGGGAC | P2 | (TC)8 |
| JESSR010 | GAAGGACGTACAAGGCCGTG | GTGACTAGATTTGATGCTGGCAG | P6 | (CCCAGC)4 |
| JESSR032 | GCTCTGTTTACGAGGGGATAGG | CCGAATCTGACACCATCATCTCT | P3 | (TTG)6 |
| JESSR075 | GAGTCTTGAGCGGAGGAGTAC | GACGTACCATGCTGATGCTGAT | P6 | (GATGCC)4 |
| JESSR090 | TAAAGCACTTCAATTGCGTCTCG | CTAGAGGAGGCCAAGAGGAAGAA | P6 | (CATGTG)4 |
| JESSR098 | CCTATACCTTAGCCTGAGACAGC | GAGGATCACCATGTCCGTCTC | P3 | (CGT)6 |
| JESSR100 | CTTGGGGAGGTGGAGAGTATAGG | ACTGTTCTTCGCTTGACCAAATT | P6 | (CTGGCA)6 |
Table 1 information statistics of 7 SSR primers
| 标记名称 Markers | 上游引物(5′-3′) Forward primer | 下游引物(5′-3′) Reverse primer | SSR 类型 SSR type | SSR 基元 SSR motif |
|---|---|---|---|---|
| JESSR003 | CATCCCTATGCAATGATTCCACG | TAGAACAAGACTAGAACCGGGAC | P2 | (TC)8 |
| JESSR010 | GAAGGACGTACAAGGCCGTG | GTGACTAGATTTGATGCTGGCAG | P6 | (CCCAGC)4 |
| JESSR032 | GCTCTGTTTACGAGGGGATAGG | CCGAATCTGACACCATCATCTCT | P3 | (TTG)6 |
| JESSR075 | GAGTCTTGAGCGGAGGAGTAC | GACGTACCATGCTGATGCTGAT | P6 | (GATGCC)4 |
| JESSR090 | TAAAGCACTTCAATTGCGTCTCG | CTAGAGGAGGCCAAGAGGAAGAA | P6 | (CATGTG)4 |
| JESSR098 | CCTATACCTTAGCCTGAGACAGC | GAGGATCACCATGTCCGTCTC | P3 | (CGT)6 |
| JESSR100 | CTTGGGGAGGTGGAGAGTATAGG | ACTGTTCTTCGCTTGACCAAATT | P6 | (CTGGCA)6 |
| 标记名称Markers | 荧光标记Fluorescence labeling | 扩增片段数Fragment number | 片段大小/bp Fragment size |
|---|---|---|---|
| JESSR003 | FAM | 3 | 172,174,182 |
| JESSR010 | FAM | 4 | 166,172,178,184 |
| JESSR032 | HEX | 3 | 161,164,167 |
| JESSR075 | ROX | 3 | 159.161,179 |
| JESSR090 | TAMRA | 3 | 162,168,174 |
| JESSR098 | TAMRA | 3 | 129,131,135 |
| JESSR100 | TAMRA | 3 | 136,138,142 |
Table 2 Statistics of amplification information of 7 SSR primers
| 标记名称Markers | 荧光标记Fluorescence labeling | 扩增片段数Fragment number | 片段大小/bp Fragment size |
|---|---|---|---|
| JESSR003 | FAM | 3 | 172,174,182 |
| JESSR010 | FAM | 4 | 166,172,178,184 |
| JESSR032 | HEX | 3 | 161,164,167 |
| JESSR075 | ROX | 3 | 159.161,179 |
| JESSR090 | TAMRA | 3 | 162,168,174 |
| JESSR098 | TAMRA | 3 | 129,131,135 |
| JESSR100 | TAMRA | 3 | 136,138,142 |
| 引物Primer | 赋值Assignment | ||||
|---|---|---|---|---|---|
| 0 | 1 | 2 | 3 | 4 | |
| JESSR003 | - | 173 | 174 | 182 | - |
| JESSR010 | - | 166 | 172 | 178 | 184 |
| JESSR032 | - | 161 | 164 | 167 | - |
| JESSR075 | - | 159 | 161 | 179 | - |
| JESSR090 | - | 162 | 168 | 174 | - |
| JESSR098 | - | 129 | 131 | 135 | - |
| JESSR100 | - | 136 | 138 | 142 | - |
Table 3 Amplified fragments and assignment standard
| 引物Primer | 赋值Assignment | ||||
|---|---|---|---|---|---|
| 0 | 1 | 2 | 3 | 4 | |
| JESSR003 | - | 173 | 174 | 182 | - |
| JESSR010 | - | 166 | 172 | 178 | 184 |
| JESSR032 | - | 161 | 164 | 167 | - |
| JESSR075 | - | 159 | 161 | 179 | - |
| JESSR090 | - | 162 | 168 | 174 | - |
| JESSR098 | - | 129 | 131 | 135 | - |
| JESSR100 | - | 136 | 138 | 142 | - |
| 编号 Code | 引物 Primer | 指纹编号 Fingerprint code | ||||||
|---|---|---|---|---|---|---|---|---|
| JESSR003 | JESSR010 | JESSR032 | JESSR075 | JESSR090 | JESSR098 | JESSR100 | ||
| ZJJE001 | 02 | 24 | 01 | 03 | 02 | 13 | 13 | 02240103021313 |
| ZJJE002 | 01 | 01 | 02 | 02 | 03 | 01 | 02 | 01010202030102 |
| ZJJE003 | 03 | 01 | 02 | 01 | 03 | 02 | 02 | 03010201030202 |
| J-2 | 03 | 03 | 03 | 02 | 03 | 01 | 01 | 03030302030101 |
| J-5 | 03 | 01 | 02 | 02 | 03 | 01 | 01 | 03010202030101 |
| J-6 | 03 | 13 | 02 | 02 | 03 | 02 | 01 | 03130202030201 |
| J-7 | 03 | 01 | 02 | 02 | 13 | 02 | 01 | 03010202130201 |
Table 4 Fingerprint database code of Naematelia aurantialba
| 编号 Code | 引物 Primer | 指纹编号 Fingerprint code | ||||||
|---|---|---|---|---|---|---|---|---|
| JESSR003 | JESSR010 | JESSR032 | JESSR075 | JESSR090 | JESSR098 | JESSR100 | ||
| ZJJE001 | 02 | 24 | 01 | 03 | 02 | 13 | 13 | 02240103021313 |
| ZJJE002 | 01 | 01 | 02 | 02 | 03 | 01 | 02 | 01010202030102 |
| ZJJE003 | 03 | 01 | 02 | 01 | 03 | 02 | 02 | 03010201030202 |
| J-2 | 03 | 03 | 03 | 02 | 03 | 01 | 01 | 03030302030101 |
| J-5 | 03 | 01 | 02 | 02 | 03 | 01 | 01 | 03010202030101 |
| J-6 | 03 | 13 | 02 | 02 | 03 | 02 | 01 | 03130202030201 |
| J-7 | 03 | 01 | 02 | 02 | 13 | 02 | 01 | 03010202130201 |
| [1] |
|
| [2] |
doi: 10.3864/j.issn.0578-1752.2011.10.013 |
|
陈昌文, 曹珂, 王力荣, 朱更瑞, 方伟超. 2011. 中国桃主要品种资源及其野生近缘种的分子身份证构建. 中国农业科学, 44 (10):2081-2093.
doi: 10.3864/j.issn.0578-1752.2011.10.013 |
|
| [3] |
|
|
陈世军, 张明泽, 姚玉仙, 谢维斌. 2017. 基于SSR标记的黔南茶树种质资源DNA 指纹图谱构建. 植物遗传资源学报, 18 (1):106-111.
doi: 10.13430/j.cnki.jpgr.2017.01.013 |
|
| [4] |
|
|
杜秀菊, 张劲松, 刘艳芳, 唐庆九, 贾薇, 杨焱, 潘迎捷. 2010. 金耳子实体不同提取物的抗氧化性和对PC12细胞氧化损伤的保护作用. 上海农业学报, 26 (2):49-52.
|
|
| [5] |
|
|
高锋, 何明霞, 张春霞, 刘静, 许欣景, 曹旸. 2021. 暗褐网柄牛肝菌高多态性SSR标记的开发. 热带农业科技, 44 (1):24-28.
|
|
| [6] |
|
|
桂明英, 马绍斌, 郭相. 2016. 西南大型真菌(Ⅰ). 上海: 上海科学技术文献出版社.
|
|
| [7] |
|
| [8] |
|
|
李春牛, 苏群, 李先民, 黄展文, 孙明艳, 卢家仕, 王虹妍, 卜朝阳. 2024. 茉莉花全基因组SSR 标记开发及其亲缘关系鉴定. 园艺学报, 51 (10):2343-2357.
|
|
| [9] |
|
|
李雪松, 华蓉, 孙达锋, 岳万松, 尚陆娥, 刘绍雄. 2023. 基于花脸香蘑基因组的SSR分子标记开发. 北方园艺,(2):112-120.
|
|
| [10] |
doi: 10.3969/j.issn.1004-1524.20221149 |
|
李雪松, 孙达锋, 刘绍雄, 张俊波, 岳万松, 李建英, 华蓉. 2024. 基于大球盖菇基因组的SSR分子标记开发及指纹数据库应用. 浙江农业学报, 36 (1):84-93.
doi: 10.3969/j.issn.1004-1524.20221149 |
|
| [11] |
|
|
刘春滟, 李南羿, 张玉琼. 2010. 香菇EST-SSR标记的开发及应用. 食用菌学报, 17 (2):1-6.
|
|
| [12] |
|
| [13] |
|
|
马杰, 屈雯, 陈春艳, 王磊, 马维, 刘针杉, 马俊, 杨珊, 丁丽, 高强, 孙勃. 2020. 基于转录组序列的羊肚菌EST-SSR标记开发与遗传多样性分析. 江苏农业学报, 36 (5):1282-1290.
|
|
| [14] |
|
|
彭寅斌. 1986. 金耳学名问题. 食用菌,(3):3-5.
|
|
| [15] |
doi: 10.16420/j.issn.0513-353x.2023-0444 |
|
秦宇, 郭荣琨, 农惠兰, 王燕, 崔凯, 董宁光. 2024. 利用SSR 荧光标记构建山楂种质分子身份证. 园艺学报, 51 (6):1227-1240.
|
|
| [16] |
|
| [17] |
doi: 10.1016/j.tibtech.2007.07.013 pmid: 17945369 |
| [18] |
|
| [19] |
|
|
魏涛, 张静, 高兆兰. 2012. 金耳菌丝体多糖对高血糖小鼠血糖作用的研究. 安徽农业科学, 40 (12):7034-7035,7052.
|
|
| [20] |
|
|
王艺琴, 张素勤, 郭豪, 田怀志, 熊兴伟, 刘进平, 耿广东. 2024. 脆木耳转录组EST-SSR标记特征及多态性分析. 分子植物育种, 22 (8):2611-2619.
|
|
| [21] |
|
|
谢红, 刘春卉, 苏槟楠, 陈丽梅, 秦峰松, 荣福雄. 2000. 金耳8254的营养价值和药理研究. 中国食用菌,(6):39-41.
|
|
| [22] |
|
|
杨林雷, 李荣春, 曹瑶, 李梦杰, 罗祥英, 杨晓君, 闻绍峰, 沈真辉, 陆青青, 子灵山. 2020. 金耳的学名及分类地位考证. 食药用菌, 28 (4):252-255,276.
|
|
| [23] |
|
|
岳万松, 李雪松, 刘绍雄, 华蓉, 孙达锋, 刘春丽, 尚陆娥, 罗孝坤. 2022. 金耳栽培常见问题及对策探讨. 中国食菌, 41 (8):80-85.
|
|
| [24] |
|
|
张丹, 宋春艳, 章炉军, 巫萍, 鲍大鹏, 尚晓冬, 谭琦. 2014. 基于全基因组序列的香菇商业菌种SSR遗传多样性分析及多位点指纹图谱构建的研究. 食用菌学报, 21 (2):1-13.
|
|
| [25] |
|
|
张俊波, 刘绍雄, 华蓉, 刘祈猛, 李建英, 刘春丽, 罗熙, 孙达锋. 2023. 金耳新品种‘中菌金耳4号’选育. 食用菌学报, 30 (6):33-44.
|
|
| [26] |
|
|
张雪梅, 姚强, 宫志远, 高兴喜, 李瑾, 刘晓, 韩建东, 任鹏飞, 万鲁长, 任海霞. 2013. 粗皮侧耳栽培种EST-SSR分子标记的开发应用及初级核心种质库的构建. 菌物学报, 32 (1):64-80.
|
| [1] | LI Zuo, XIAO Wenfang, CHEN Heming, and LÜ Fubing. Genetic Diversity and Core Collection Construction of Phalaenopsis Germplasm Resources [J]. Acta Horticulturae Sinica, 2025, 52(6): 1519-1529. |
| [2] | SUN Pei, ZHANG Hong, YANG Yuan, WANG Hua, LI Maofu, KANG Yanhui, SUN Xiangyi, JIN Wanmei. Genetic Diversity Analysis and Fingerprint Construction of Rosa Germplasm Resources Based on SSR Marker [J]. Acta Horticulturae Sinica, 2025, 52(6): 1539-1552. |
| [3] | YUAN Juanwei, JIA Li, WANG Han, YAN Congsheng, ZHANG Qi’an, YU Feifei, GAN Defang, JIANG Haikun. Screening of Resistant Germplasm and Identifying Candidate Gene for Phytophthora capsici Resistance in Pepper Germplasm Resources [J]. Acta Horticulturae Sinica, 2025, 52(4): 857-871. |
| [4] | BI Qingrui, CUI Dongsheng, MA Xinyuan, XUE Yuran, ZHANG Shikui, FAN Guoquan, NIU Yingying. Genetic Diversity and Genetic Relationship of Local Pear Cultivars in Xinjiang [J]. Acta Horticulturae Sinica, 2025, 52(3): 561-574. |
| [5] | ZHANG Lu, ZHANG Pingping, XIE Weijia, XU Feng, PENG Lüchun, SONG Jie, DU Guanghui, LI Shifeng. Genetic Diversity Analysis and SSR Fingerprint Construction of Evergreen Rhododendron Germplasms [J]. Acta Horticulturae Sinica, 2025, 52(2): 380-394. |
| [6] | QIN Zilu, XU Zhengkang, DAI Xiaogang, CHEN Yingnan. Genetic Diversity Analysis and Core Collection Construction of Magnolia biondii Germplasm [J]. Acta Horticulturae Sinica, 2024, 51(8): 1823-1832. |
| [7] | QIN Yu, GUO Rongkun, NONG Huilan, WANG Yan, CUI Kai, DONG Ningguang. Using the Fluorescent Labeled SSR Markers to Establish Molecular Identity of Hawthorn [J]. Acta Horticulturae Sinica, 2024, 51(6): 1227-1240. |
| [8] | YANG Xi, RAO Dehua, LIU Hong, YANG Zhe, SU Zhenzhu, JIANG Yuan, YIN Jiwei, XU Zhenjiang. Association Analysis of Cattleya DUS Test Characteristics and SSR Markers [J]. Acta Horticulturae Sinica, 2024, 51(4): 787-803. |
| [9] | YUE Linqing, WU Yichao, HE Xingxing, XIANG Mingyan, WANG Jin, WU Zheng, ZHU Shiguo, DING Mengqi, ZHANG Jian, FANG Xiaomei, YI Zelin. SSR Marker Association Analysis and Fingerprint Construction of Main Traits of Plum Fruit [J]. Acta Horticulturae Sinica, 2024, 51(11): 2495-2509. |
| [10] | LI Chunniu, SU Qun, LI Xianmin, HUANG Zhanwen, SUN Mingyan, LU Jiashi, WANG Hongyan, BU Zhaoyang. SSR Molecular Markers Development and Parentage Relationship Identification Based on Whole Genomic Sequences of Jasminum sambac [J]. Acta Horticulturae Sinica, 2024, 51(10): 2343-2357. |
| [11] | NIE Xinghua, ZHANG Yu, LIU Song, YANG Jiabin, HAO Yaqiong, LIU Yang, QIN Ling, XING Yu. Study on Genetic Characteristics and Taxonomic Status of Wild Chinese Chestnut Based on Genome Re-sequencing [J]. Acta Horticulturae Sinica, 2023, 50(8): 1622-1636. |
| [12] | MEI Yulin, XU Jinjian, JIANG Mengyue, YU Junhai, ZONG Yu, CHEN Wenrong, LIAO Fanglei, GUO Weidong. Identification of F1 Hybrids and Analysis of Fruit Shape Difference Between Fingered Citron and Citron [J]. Acta Horticulturae Sinica, 2023, 50(7): 1402-1418. |
| [13] | SUN Zeshuo, JIANG Dongyue, LIU Xinhong, SHEN Xin, LI Yingang, QU Yufei, LI Yonghua. Cluster Analysis and Construction of DNA Fingerprinting of 42 Oriental Cultivars of Flowering Cherry Based on SSR Markers [J]. Acta Horticulturae Sinica, 2023, 50(3): 657-668. |
| [14] | WANG Rui, HONG Wenjuan, LUO Hua, ZHAO Lina, CHEN Ying, WANG Jun. Construction of SSR Fingerprints of Pomegranate Cultivars and Male Parent Identification of Hybrids [J]. Acta Horticulturae Sinica, 2023, 50(2): 265-278. |
| [15] | XIE Qian, ZHANG Shiyan, JIANG Lai, DING Mingyue, LIU Lingling, WU Rujian, CHEN Qingxi. Analysis of Canarium album Transcriptome SSR Information and Molecular Marker Development and Application [J]. Acta Horticulturae Sinica, 2023, 50(11): 2350-2364. |
| Viewed | ||||||
|
Full text |
|
|||||
|
Abstract |
|
|||||
Copyright © 2012 Acta Horticulturae Sinica 京ICP备10030308号-2 国际联网备案号 11010802023439
Tel: 010-82109523 E-Mail: yuanyixuebao@126.com
Support by: Beijing Magtech Co.Ltd