
Acta Horticulturae Sinica ›› 2023, Vol. 50 ›› Issue (10): 2242-2256.doi: 10.16420/j.issn.0513-353x.2022-0682
• Plant Protection • Previous Articles Next Articles
SHI Jingfang1,3, HU Chunhua1, LI Haochen1, YANG Qiaosong1,4, SHENG Ou1, BI Fangcheng1,2, DONG Tao1, LI Chunyu1, DENG Guiming1, GAO Huijun1, HE Weidi1, LIU Siwen1, YI Ganjun1,*( ), DOU Tongxin1,*(
), DOU Tongxin1,*( )
)
Received:2023-03-21
Revised:2023-08-13
Online:2023-10-25
Published:2023-10-31
SHI Jingfang, HU Chunhua, LI Haochen, YANG Qiaosong, SHENG Ou, BI Fangcheng, DONG Tao, LI Chunyu, DENG Guiming, GAO Huijun, HE Weidi, LIU Siwen, YI Ganjun, DOU Tongxin. Molecular Mechanism of MpICE1 Overexpressing Banana Resistant to Fusarium oxysporum Based on Transcriptome Analysis[J]. Acta Horticulturae Sinica, 2023, 50(10): 2242-2256.
Add to citation manager EndNote|Ris|BibTeX
URL: https://www.ahs.ac.cn/EN/10.16420/j.issn.0513-353x.2022-0682
| 基因 Gene | 引物(5′-3′) Primer |
|---|---|
| Ma06_g18840 | F:TATGGCAATGTCCCCCAACC;R:GACCAGTGTCTGAGACGCAA |
| Ma06_g16670 | F:ATCAACGTCGAGATCGGCAA;R:GGCTGAGGCTGATGGGTA |
| Ma11_g19450 | F:ATGCTCTCGGGGATCAATGG;R:TCATGACACTGTATTATCG |
| Ma04_g40060 | F:ATGGCGTCCAAGACCACAAG;R:GCTCTTTTCCTCTTGTTCAAC |
| Ma10_g15940 | F:GTCCTCACCAGCGCCTGCGG;R:CCCCGAGCCCTGCCCCCTCCG |
| Ma10_g12250 | F:TGGGTTTGCCATGGATGTTT;R:AGAACACCAGGGCCTTCCTT |
| Ma08_g05750 | F:AGCGGAGAATGTCATAGGTG;R:TGGATGCTCTGTCCATCTTG |
| Ma09_g10470 | F:ACAGTCGTCAGGAGTCGTGCTT;R:CGCTATCGATTCATGACACTGTA |
Table 1 Validation gene and primer information for qRT-PCR
| 基因 Gene | 引物(5′-3′) Primer |
|---|---|
| Ma06_g18840 | F:TATGGCAATGTCCCCCAACC;R:GACCAGTGTCTGAGACGCAA |
| Ma06_g16670 | F:ATCAACGTCGAGATCGGCAA;R:GGCTGAGGCTGATGGGTA |
| Ma11_g19450 | F:ATGCTCTCGGGGATCAATGG;R:TCATGACACTGTATTATCG |
| Ma04_g40060 | F:ATGGCGTCCAAGACCACAAG;R:GCTCTTTTCCTCTTGTTCAAC |
| Ma10_g15940 | F:GTCCTCACCAGCGCCTGCGG;R:CCCCGAGCCCTGCCCCCTCCG |
| Ma10_g12250 | F:TGGGTTTGCCATGGATGTTT;R:AGAACACCAGGGCCTTCCTT |
| Ma08_g05750 | F:AGCGGAGAATGTCATAGGTG;R:TGGATGCTCTGTCCATCTTG |
| Ma09_g10470 | F:ACAGTCGTCAGGAGTCGTGCTT;R:CGCTATCGATTCATGACACTGTA |
| 样本 Sample | 原始序列/Mb Raw reads | 过滤后序列/Mb Clean reads | Q20/% | Q30/% | 序列比值 Clean reads ratio/% | 总基因比对率 Total mapping/% |
|---|---|---|---|---|---|---|
| WT0d A | 43.82 | 42.72 | 97.26 | 93.03 | 97.49 | 87.00 |
| WT0d B | 43.82 | 42.84 | 97.22 | 92.84 | 97.76 | 86.83 |
| WT0d C | 42.61 | 41.87 | 96.76 | 91.99 | 98.26 | 86.20 |
| WT7d A | 43.82 | 43.29 | 97.12 | 92.54 | 98.80 | 88.15 |
| WT7d B | 43.82 | 43.38 | 97.03 | 92.41 | 98.99 | 87.82 |
| WT7d C | 43.82 | 42.56 | 97.15 | 92.71 | 97.12 | 87.81 |
| WT14d A | 43.82 | 42.61 | 97.19 | 92.84 | 97.23 | 87.19 |
| WT14d B | 43.82 | 43.05 | 97.22 | 92.91 | 98.25 | 87.56 |
| WT14d C | 43.82 | 42.87 | 97.38 | 93.25 | 97.84 | 87.58 |
| OE0d A | 43.82 | 42.64 | 97.17 | 92.88 | 97.32 | 87.11 |
| OE0d B | 43.82 | 43.22 | 97.12 | 92.67 | 98.62 | 87.54 |
| OE0d C | 43.82 | 43.26 | 96.99 | 92.31 | 98.72 | 86.53 |
| OE7d A | 43.82 | 43.20 | 97.19 | 92.76 | 98.59 | 87.44 |
| OE7d B | 43.82 | 42.95 | 97.31 | 93.15 | 98.02 | 87.17 |
| OE7d C | 43.82 | 42.83 | 97.02 | 92.45 | 97.74 | 87.59 |
| OE14d A | 43.82 | 42.94 | 96.97 | 92.27 | 97.99 | 87.17 |
| OE14d B | 43.82 | 42.94 | 97.00 | 92.41 | 97.98 | 87.41 |
| OE14d C | 43.82 | 43.09 | 96.99 | 92.32 | 98.33 | 86.77 |
Table 2 Summary of RNA-seq data from roots of wild(WT)and ICE1 overexpress(OE)banana before and after inoculation(0,7 and 14 d)
| 样本 Sample | 原始序列/Mb Raw reads | 过滤后序列/Mb Clean reads | Q20/% | Q30/% | 序列比值 Clean reads ratio/% | 总基因比对率 Total mapping/% |
|---|---|---|---|---|---|---|
| WT0d A | 43.82 | 42.72 | 97.26 | 93.03 | 97.49 | 87.00 |
| WT0d B | 43.82 | 42.84 | 97.22 | 92.84 | 97.76 | 86.83 |
| WT0d C | 42.61 | 41.87 | 96.76 | 91.99 | 98.26 | 86.20 |
| WT7d A | 43.82 | 43.29 | 97.12 | 92.54 | 98.80 | 88.15 |
| WT7d B | 43.82 | 43.38 | 97.03 | 92.41 | 98.99 | 87.82 |
| WT7d C | 43.82 | 42.56 | 97.15 | 92.71 | 97.12 | 87.81 |
| WT14d A | 43.82 | 42.61 | 97.19 | 92.84 | 97.23 | 87.19 |
| WT14d B | 43.82 | 43.05 | 97.22 | 92.91 | 98.25 | 87.56 |
| WT14d C | 43.82 | 42.87 | 97.38 | 93.25 | 97.84 | 87.58 |
| OE0d A | 43.82 | 42.64 | 97.17 | 92.88 | 97.32 | 87.11 |
| OE0d B | 43.82 | 43.22 | 97.12 | 92.67 | 98.62 | 87.54 |
| OE0d C | 43.82 | 43.26 | 96.99 | 92.31 | 98.72 | 86.53 |
| OE7d A | 43.82 | 43.20 | 97.19 | 92.76 | 98.59 | 87.44 |
| OE7d B | 43.82 | 42.95 | 97.31 | 93.15 | 98.02 | 87.17 |
| OE7d C | 43.82 | 42.83 | 97.02 | 92.45 | 97.74 | 87.59 |
| OE14d A | 43.82 | 42.94 | 96.97 | 92.27 | 97.99 | 87.17 |
| OE14d B | 43.82 | 42.94 | 97.00 | 92.41 | 97.98 | 87.41 |
| OE14d C | 43.82 | 43.09 | 96.99 | 92.32 | 98.33 | 86.77 |
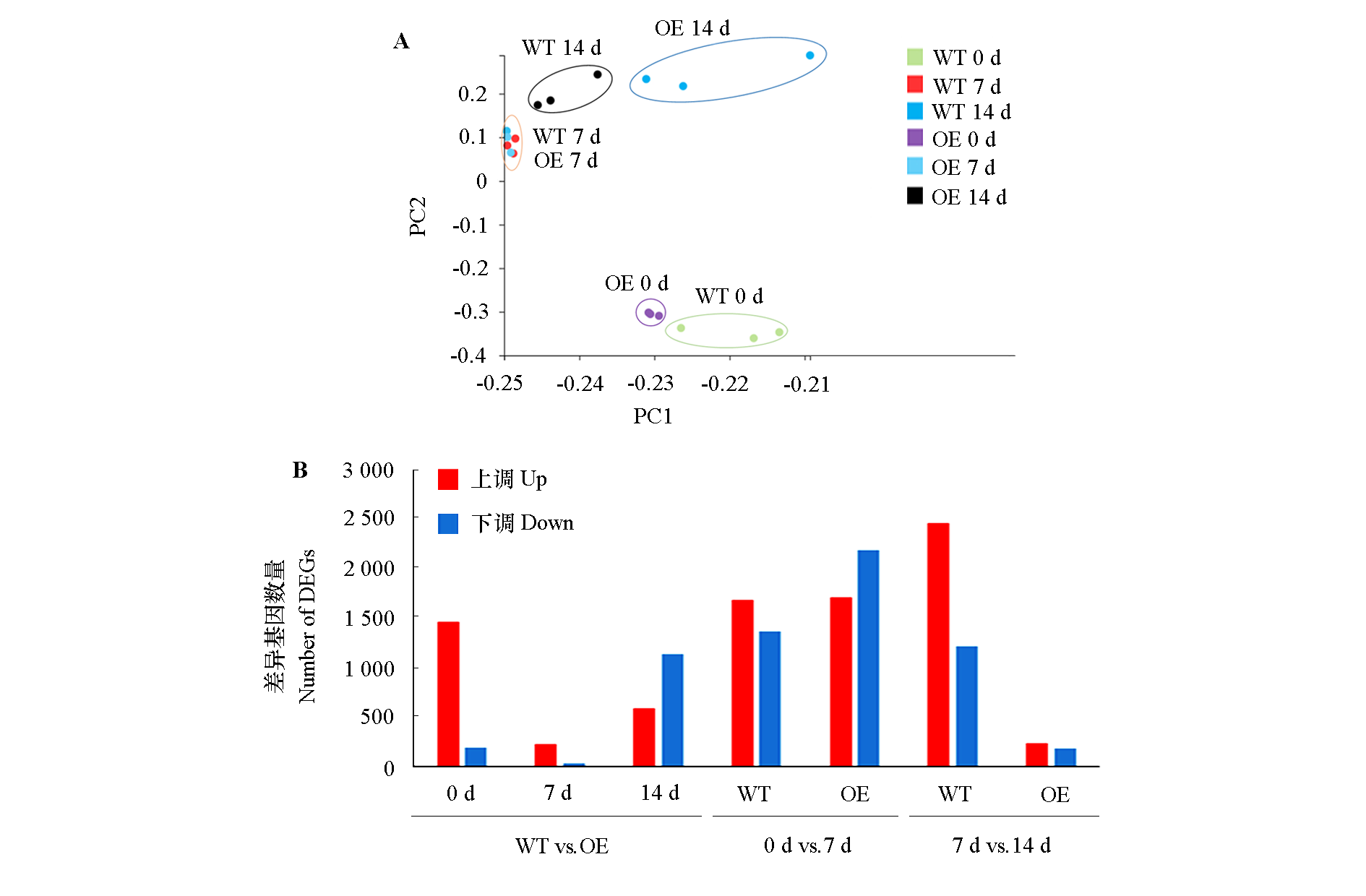
Fig. 1 PCA results(A)and statistics on number of different express genes(B)of WT and overexpress MpICE1(OE)banana root before and after inoculation(0,7 and 14 d)
| 比较组 Pair comparison | GO功能 GO term | 基因数量 Gene number | 富集率 Rich ratio | Q值 Q value |
|---|---|---|---|---|
| WT 0 d vs. OE 0 d | 细胞外区域Extracellular region | 56 | 0.19 | 6.84E-19 |
| 细胞壁组织或生物发生Cell wall organization or biogenesis | 39 | 0.16 | 6.10E-10 | |
| 细胞壁组织Cell wall organization | 34 | 0.16 | 2.23E-08 | |
| 表面封装结构组织External encapsulating structure organization | 34 | 0.15 | 3.20E-08 | |
| 细胞壁Cell wall | 27 | 0.18 | 3.66E-08 | |
| 表面封装结构External encapsulating structure | 27 | 0.18 | 3.66E-08 | |
| 糖基水解酶活性Hydrolase activity,acting on glycosyl bonds | 41 | 0.15 | 3.98E-08 | |
| O-糖基水解酶活性 Hydrolase activity,hydrolyzing O-glycosyl compounds | 39 | 0.15 | 4.60E-08 | |
| 细胞碳水化合物代谢过程Cellular carbohydrate metabolic process | 28 | 0.17 | 1.20E-07 | |
| 碳水化合物代谢过程Carbohydrate metabolic process | 59 | 0.10 | 1.44E-07 | |
| WT 7 d vs. OE 7 d | 细胞外区域Extracellular region | 18 | 0.06 | 1.68E-09 |
| 植物细胞壁组织Plant-type cell wall organization | 6 | 0.13 | 1.47E-04 | |
| 氧化还原酶活性Oxidoreductase activity | 25 | 0.02 | 2.39E-04 | |
| 植物细胞壁组织或生物发生 Plant-type cell wall organization or biogenesis | 6 | 0.10 | 2.56E-04 | |
| 细胞外周Cell periphery | 13 | 0.03 | 9.29E-03 | |
| 细胞壁Cell wall | 7 | 0.05 | 9.73E-03 | |
| 表面封装结构External encapsulating structure | 7 | 0.05 | 9.73E-03 | |
| 细胞壁组织或生物发生Cell wall organization or biogenesis | 8 | 0.03 | 9.73E-03 | |
| 质外体Apoplast | 5 | 0.06 | 2.32E-02 | |
| 细胞壁组织Cell wall organization | 7 | 0.03 | 2.32E-02 | |
| WT 14 d vs. OE 14 d | 逆转录转座子核衣壳Retrotransposon nucleocapsid | 20 | 0.40 | 7.45E-11 |
| DNA整合DNA integration | 23 | 0.29 | 5.50E-10 | |
| RNA指导的DNA聚合酶活性RNA-directed DNA polymerase activity | 10 | 0.43 | 2.49E-05 | |
| 碳水化合物代谢过程Carbohydrate metabolic process | 57 | 0.10 | 3.29E-05 | |
| 氧化还原酶活性Oxidoreductase activity | 93 | 0.08 | 2.83E-04 | |
| 细胞外区域Extracellular region | 34 | 0.11 | 5.48E-04 | |
| DNA聚合酶活性DNA polymerase activity | 10 | 0.30 | 5.48E-04 | |
| 甲基丙二酸半醛脱氢酶活性 Methylmalonate-semialdehyde dehydrogenase(acylating)activity | 5 | 0.63 | 2.20E-03 | |
| 质膜组成成分Integral component of membrane | 243 | 0.06 | 2.20E-03 | |
| 质膜固有成分Intrinsic component of membrane | 244 | 0.06 | 2.20E-03 | |
| 催化活性Catalytic activity | 353 | 0.06 | 4.09E-03 |
Table 3 GO enrichment of different express genes in wild(WT)and transgenic(OE)bananas before and after inoculation(0,7,14 d)
| 比较组 Pair comparison | GO功能 GO term | 基因数量 Gene number | 富集率 Rich ratio | Q值 Q value |
|---|---|---|---|---|
| WT 0 d vs. OE 0 d | 细胞外区域Extracellular region | 56 | 0.19 | 6.84E-19 |
| 细胞壁组织或生物发生Cell wall organization or biogenesis | 39 | 0.16 | 6.10E-10 | |
| 细胞壁组织Cell wall organization | 34 | 0.16 | 2.23E-08 | |
| 表面封装结构组织External encapsulating structure organization | 34 | 0.15 | 3.20E-08 | |
| 细胞壁Cell wall | 27 | 0.18 | 3.66E-08 | |
| 表面封装结构External encapsulating structure | 27 | 0.18 | 3.66E-08 | |
| 糖基水解酶活性Hydrolase activity,acting on glycosyl bonds | 41 | 0.15 | 3.98E-08 | |
| O-糖基水解酶活性 Hydrolase activity,hydrolyzing O-glycosyl compounds | 39 | 0.15 | 4.60E-08 | |
| 细胞碳水化合物代谢过程Cellular carbohydrate metabolic process | 28 | 0.17 | 1.20E-07 | |
| 碳水化合物代谢过程Carbohydrate metabolic process | 59 | 0.10 | 1.44E-07 | |
| WT 7 d vs. OE 7 d | 细胞外区域Extracellular region | 18 | 0.06 | 1.68E-09 |
| 植物细胞壁组织Plant-type cell wall organization | 6 | 0.13 | 1.47E-04 | |
| 氧化还原酶活性Oxidoreductase activity | 25 | 0.02 | 2.39E-04 | |
| 植物细胞壁组织或生物发生 Plant-type cell wall organization or biogenesis | 6 | 0.10 | 2.56E-04 | |
| 细胞外周Cell periphery | 13 | 0.03 | 9.29E-03 | |
| 细胞壁Cell wall | 7 | 0.05 | 9.73E-03 | |
| 表面封装结构External encapsulating structure | 7 | 0.05 | 9.73E-03 | |
| 细胞壁组织或生物发生Cell wall organization or biogenesis | 8 | 0.03 | 9.73E-03 | |
| 质外体Apoplast | 5 | 0.06 | 2.32E-02 | |
| 细胞壁组织Cell wall organization | 7 | 0.03 | 2.32E-02 | |
| WT 14 d vs. OE 14 d | 逆转录转座子核衣壳Retrotransposon nucleocapsid | 20 | 0.40 | 7.45E-11 |
| DNA整合DNA integration | 23 | 0.29 | 5.50E-10 | |
| RNA指导的DNA聚合酶活性RNA-directed DNA polymerase activity | 10 | 0.43 | 2.49E-05 | |
| 碳水化合物代谢过程Carbohydrate metabolic process | 57 | 0.10 | 3.29E-05 | |
| 氧化还原酶活性Oxidoreductase activity | 93 | 0.08 | 2.83E-04 | |
| 细胞外区域Extracellular region | 34 | 0.11 | 5.48E-04 | |
| DNA聚合酶活性DNA polymerase activity | 10 | 0.30 | 5.48E-04 | |
| 甲基丙二酸半醛脱氢酶活性 Methylmalonate-semialdehyde dehydrogenase(acylating)activity | 5 | 0.63 | 2.20E-03 | |
| 质膜组成成分Integral component of membrane | 243 | 0.06 | 2.20E-03 | |
| 质膜固有成分Intrinsic component of membrane | 244 | 0.06 | 2.20E-03 | |
| 催化活性Catalytic activity | 353 | 0.06 | 4.09E-03 |
| 比较组 Pair comparison | KEGG 通路 KEGG pathway | 基因数量 Gene number | 富集率 Rich ratio | Q值 Q value |
|---|---|---|---|---|
| WT 0 d vs. OE 0 d | 苯丙烷合成途径Phenylpropanoid biosynthesis | 80 | 0.11 | 1.43E-09 |
| 黄酮生物合成Flavonoid biosynthesis | 23 | 0.16 | 7.14E-05 | |
| 氰基氨基酸代谢Cyanoamino acid metabolism | 21 | 0.15 | 4.09E-04 | |
| ABC转运蛋白ABC transporters | 26 | 0.13 | 4.45E-04 | |
| 油菜素内酯生物合成Brassinosteroid biosynthesis | 9 | 0.27 | 6.06E-04 | |
| 单萜生物合成Monoterpenoid biosynthesis | 7 | 0.32 | 1.36E-03 | |
| 植物MAPK信号途径MAPK signaling pathway-plant | 76 | 0.08 | 1.85E-03 | |
| 戊糖和葡萄糖醛酸酯互换 Pentose and glucuronate interconversions | 34 | 0.10 | 2.16E-03 | |
| 角质、软木脂和蜡质生物合成 Cutin,suberine and wax biosynthesis | 13 | 0.15 | 4.69E-03 | |
| RNA聚合酶RNA polymerase | 40 | 0.09 | 8.29E-03 | |
| 其他聚糖降解Other glycan degradation | 17 | 0.11 | 2.11E-02 | |
| 异喹啉生物碱生物合成Isoquinoline alkaloid biosynthesis | 8 | 0.16 | 2.77E-02 | |
| WT 7 d vs. OE 7 d | 黄酮生物合成Flavonoid biosynthesis | 12 | 0.08 | 1.23E-06 |
| 类胡萝卜素生物合成Carotenoid biosynthesis | 6 | 0.06 | 6.80E-03 | |
| 苯丙烷合成途径Phenylpropanoid biosynthesis | 17 | 0.02 | 6.80E-03 | |
| 植物—病原菌互作Plant-pathogen interaction | 20 | 0.02 | 6.80E-03 | |
| ABC转运蛋白ABC transporters | 8 | 0.04 | 9.06E-03 | |
| 植物MAPK信号途径MAPK signaling pathway-plant | 20 | 0.02 | 9.06E-03 | |
| 二萜生物合成Diterpenoid biosynthesis | 5 | 0.05 | 1.45E-02 | |
| WT 14 d vs. OE 14 d | 黄酮生物合成Flavonoid biosynthesis | 23 | 0.16 | 2.03E-04 |
| 淀粉和蔗糖代谢Starch and sucrose metabolism | 56 | 0.10 | 2.09E-04 | |
| 植物MAPK信号途径MAPK signaling pathway-plant | 81 | 0.08 | 1.52E-03 | |
| ABC转运蛋白ABC transporters | 25 | 0.12 | 2.80E-03 | |
| 苯丙烷合成途径Phenylpropanoid biosynthesis | 61 | 0.09 | 3.20E-03 | |
| 植物—病原菌互作Plant-pathogen interaction | 75 | 0.08 | 3.20E-03 | |
| Alpha-亚油酸代谢alpha-Linolenic acid metabolism | 15 | 0.14 | 1.33E-02 | |
| 二萜生物合成Diterpenoid biosynthesis | 13 | 0.14 | 1.85E-02 | |
| 芪类、二苯基庚烷和姜酚生物合成 Stilbenoid,diarylheptanoid and gingerol biosynthesis | 11 | 0.15 | 2.58E-02 | |
| RNA聚合酶RNA polymerase | 40 | 0.09 | 2.58E-02 |
Table 4 KEGG enrichment of different express genes in wild(WT)and transgenic(OE)bananas before and after inoculation(0,7 and 14 d)
| 比较组 Pair comparison | KEGG 通路 KEGG pathway | 基因数量 Gene number | 富集率 Rich ratio | Q值 Q value |
|---|---|---|---|---|
| WT 0 d vs. OE 0 d | 苯丙烷合成途径Phenylpropanoid biosynthesis | 80 | 0.11 | 1.43E-09 |
| 黄酮生物合成Flavonoid biosynthesis | 23 | 0.16 | 7.14E-05 | |
| 氰基氨基酸代谢Cyanoamino acid metabolism | 21 | 0.15 | 4.09E-04 | |
| ABC转运蛋白ABC transporters | 26 | 0.13 | 4.45E-04 | |
| 油菜素内酯生物合成Brassinosteroid biosynthesis | 9 | 0.27 | 6.06E-04 | |
| 单萜生物合成Monoterpenoid biosynthesis | 7 | 0.32 | 1.36E-03 | |
| 植物MAPK信号途径MAPK signaling pathway-plant | 76 | 0.08 | 1.85E-03 | |
| 戊糖和葡萄糖醛酸酯互换 Pentose and glucuronate interconversions | 34 | 0.10 | 2.16E-03 | |
| 角质、软木脂和蜡质生物合成 Cutin,suberine and wax biosynthesis | 13 | 0.15 | 4.69E-03 | |
| RNA聚合酶RNA polymerase | 40 | 0.09 | 8.29E-03 | |
| 其他聚糖降解Other glycan degradation | 17 | 0.11 | 2.11E-02 | |
| 异喹啉生物碱生物合成Isoquinoline alkaloid biosynthesis | 8 | 0.16 | 2.77E-02 | |
| WT 7 d vs. OE 7 d | 黄酮生物合成Flavonoid biosynthesis | 12 | 0.08 | 1.23E-06 |
| 类胡萝卜素生物合成Carotenoid biosynthesis | 6 | 0.06 | 6.80E-03 | |
| 苯丙烷合成途径Phenylpropanoid biosynthesis | 17 | 0.02 | 6.80E-03 | |
| 植物—病原菌互作Plant-pathogen interaction | 20 | 0.02 | 6.80E-03 | |
| ABC转运蛋白ABC transporters | 8 | 0.04 | 9.06E-03 | |
| 植物MAPK信号途径MAPK signaling pathway-plant | 20 | 0.02 | 9.06E-03 | |
| 二萜生物合成Diterpenoid biosynthesis | 5 | 0.05 | 1.45E-02 | |
| WT 14 d vs. OE 14 d | 黄酮生物合成Flavonoid biosynthesis | 23 | 0.16 | 2.03E-04 |
| 淀粉和蔗糖代谢Starch and sucrose metabolism | 56 | 0.10 | 2.09E-04 | |
| 植物MAPK信号途径MAPK signaling pathway-plant | 81 | 0.08 | 1.52E-03 | |
| ABC转运蛋白ABC transporters | 25 | 0.12 | 2.80E-03 | |
| 苯丙烷合成途径Phenylpropanoid biosynthesis | 61 | 0.09 | 3.20E-03 | |
| 植物—病原菌互作Plant-pathogen interaction | 75 | 0.08 | 3.20E-03 | |
| Alpha-亚油酸代谢alpha-Linolenic acid metabolism | 15 | 0.14 | 1.33E-02 | |
| 二萜生物合成Diterpenoid biosynthesis | 13 | 0.14 | 1.85E-02 | |
| 芪类、二苯基庚烷和姜酚生物合成 Stilbenoid,diarylheptanoid and gingerol biosynthesis | 11 | 0.15 | 2.58E-02 | |
| RNA聚合酶RNA polymerase | 40 | 0.09 | 2.58E-02 |
| 比较组 Pair comparison | KEGG通路 KEGG pathway | 基因数量 Gene number | 富集率 Rich ratio | Q值 Q value |
|---|---|---|---|---|
| OE 0 d vs. OE 7 d(2 003) | 苯丙烷生物合成 Phenylpropanoid biosynthesis | 89 | 0.13 | 5.35E-10 |
| 二萜生物合成 Diterpenoid biosynthesis | 21 | 0.23 | 2.83E-06 | |
| 植物—病原菌互作 Plant-pathogen interaction | 94 | 0.10 | 2.83E-06 | |
| 植物MAPK信号途径 MAPK signaling pathway - plant | 96 | 0.10 | 7.54E-06 | |
| RNA聚合酶 RNA polymerase | 55 | 0.12 | 1.14E-05 | |
| 黄酮生物合成 Flavonoid biosynthesis | 22 | 0.15 | 9.43E-04 | |
| ABC转运蛋白 ABC transporters | 26 | 0.13 | 2.85E-03 | |
| 芪类、二苯基庚烷和姜酚生物合成 Stilbenoid,diarylheptanoid and gingerol biosynthesis | 13 | 0.17 | 5.75E-03 | |
| 角质、软木脂和蜡质生物合成 Cutin,suberine and wax biosynthesis | 14 | 0.16 | 6.14E-03 | |
| 甘露糖类O-聚糖生物合成 Mannose type O-glycan biosynthesis | 8 | 0.22 | 1.13E-02 | |
| WT 0 d vs. WT 7 d(1 145) | 光合作用天线蛋白 Photosynthesis-antenna proteins | 10 | 0.29 | 2.41E-05 |
| 植物激素信号传导 Plant hormone signal transduction | 65 | 0.05 | 1.08E-02 | |
| 淀粉和蔗糖代谢 Starch and sucrose metabolism | 34 | 0.06 | 2.49E-02 | |
| 倍半萜和三萜生物合成 Sesquiterpenoid and triterpenoid biosynthesis | 11 | 0.10 | 3.60E-02 | |
| 甘油酯代谢 Glycerolipid metabolism | 17 | 0.07 | 4.77E-02 | |
| OE 0 d vs. OE7 d & WT 0 d vs. WT 7 d(1 897) | 植物昼夜节律 Circadian rhythm-plant | 67 | 0.16 | 2.43E-13 |
| 泛醌和其他萜—醌生物合成 Ubiquinone and other terpenoid-quinone biosynthesis | 14 | 0.16 | 7.13E-03 | |
| 类胡萝卜素生物合成 Carotenoid biosynthesis | 16 | 0.15 | 7.13E-03 | |
| 花生四烯酸代谢 Arachidonic acid metabolism | 13 | 0.17 | 8.61E-03 | |
| 类固醇生物合成 Steroid biosynthesis | 11 | 0.15 | 3.71E-02 | |
| 淀粉和蔗糖代谢 Starch and sucrose metabolism | 48 | 0.09 | 3.71E-02 | |
| OE 7 d vs. OE 14 d(154) | 戊糖和葡萄糖醛酸酯相互转化 Pentose and glucuronate interconversions | 6 | 0.02 | 9.66E-02 |
| 苯丙烷生物合成 Phenylpropanoid biosynthesis | 9 | 0.01 | 9.66E-02 | |
| 硒化合物代谢 Selenocompound metabolism | 3 | 0.04 | 1.01E-01 | |
| 黄酮和黄酮醇生物合成 Flavone and flavonol biosynthesis | 2 | 0.03 | 2.78E-01 | |
| OE 7 d vs. OE 14 d & WT 7 d vs. WT 14 d(264) | 黄酮生物合成 Flavonoid biosynthesis | 11 | 0.07 | 1.86E-05 |
| 内质网蛋白质加工 Protein processing in endoplasmic reticulum | 15 | 0.02 | 4.52E-02 | |
| 植物—病原菌互作 Plant-pathogen interaction | 19 | 0.02 | 4.52E-02 | |
| WT 7 d vs. WT 14 d(3 407) | 植物—病原菌互作 Plant-pathogen interaction | 169 | 0.19 | 1.45E-11 |
| 植物MAPK信号途径 MAPK signaling pathway-plant | 166 | 0.17 | 5.42E-09 | |
| 苯丙氨酸代谢 Phenylalanine metabolism | 37 | 0.27 | 1.47E-06 | |
| 黄酮生物合成 Flavonoid biosynthesis | 34 | 0.23 | 2.25E-04 | |
| 苯丙烷生物合成 Phenylpropanoid biosynthesis | 110 | 0.16 | 3.48E-04 | |
| 光合作用 Photosynthesis | 32 | 0.23 | 4.57E-04 | |
| 光合作用天线蛋白 Photosynthesis-antenna proteins | 12 | 0.34 | 2.50E-03 | |
| 异喹啉生物碱生物合成 Isoquinoline alkaloid biosynthesis | 14 | 0.29 | 5.82E-03 | |
| 谷胱甘肽代谢 Glutathione metabolism | 30 | 0.20 | 6.99E-03 | |
| 芪类、二苯基庚烷和姜酚生物合成 Stilbenoid,diarylheptanoid and gingerol biosynthesis | 18 | 0.24 | 8.25E-03 | |
| ABC转运蛋白 ABC transporters | 37 | 0.18 | 9.11E-03 | |
| 光合生物碳固定 Carbon fixation in photosynthetic organisms | 33 | 0.18 | 1.43E-02 | |
| 淀粉和蔗糖代谢 Starch and sucrose metabolism | 80 | 0.14 | 2.84E-02 | |
| 黄酮和黄酮醇生物合成 Flavone and flavonol biosynthesis | 14 | 0.23 | 4.18E-02 | |
| 精氨酸生物合成 Arginine biosynthesis | 18 | 0.20 | 4.37E-02 | |
| 酪氨酸代谢 Tyrosine metabolism | 16 | 0.21 | 4.92E-02 |
Table 5 KEGG pathways enriched different express genes of wild(WT)and transgenic(OE)bananas before and after inoculation(0,7 and 14 d)based venn analysis
| 比较组 Pair comparison | KEGG通路 KEGG pathway | 基因数量 Gene number | 富集率 Rich ratio | Q值 Q value |
|---|---|---|---|---|
| OE 0 d vs. OE 7 d(2 003) | 苯丙烷生物合成 Phenylpropanoid biosynthesis | 89 | 0.13 | 5.35E-10 |
| 二萜生物合成 Diterpenoid biosynthesis | 21 | 0.23 | 2.83E-06 | |
| 植物—病原菌互作 Plant-pathogen interaction | 94 | 0.10 | 2.83E-06 | |
| 植物MAPK信号途径 MAPK signaling pathway - plant | 96 | 0.10 | 7.54E-06 | |
| RNA聚合酶 RNA polymerase | 55 | 0.12 | 1.14E-05 | |
| 黄酮生物合成 Flavonoid biosynthesis | 22 | 0.15 | 9.43E-04 | |
| ABC转运蛋白 ABC transporters | 26 | 0.13 | 2.85E-03 | |
| 芪类、二苯基庚烷和姜酚生物合成 Stilbenoid,diarylheptanoid and gingerol biosynthesis | 13 | 0.17 | 5.75E-03 | |
| 角质、软木脂和蜡质生物合成 Cutin,suberine and wax biosynthesis | 14 | 0.16 | 6.14E-03 | |
| 甘露糖类O-聚糖生物合成 Mannose type O-glycan biosynthesis | 8 | 0.22 | 1.13E-02 | |
| WT 0 d vs. WT 7 d(1 145) | 光合作用天线蛋白 Photosynthesis-antenna proteins | 10 | 0.29 | 2.41E-05 |
| 植物激素信号传导 Plant hormone signal transduction | 65 | 0.05 | 1.08E-02 | |
| 淀粉和蔗糖代谢 Starch and sucrose metabolism | 34 | 0.06 | 2.49E-02 | |
| 倍半萜和三萜生物合成 Sesquiterpenoid and triterpenoid biosynthesis | 11 | 0.10 | 3.60E-02 | |
| 甘油酯代谢 Glycerolipid metabolism | 17 | 0.07 | 4.77E-02 | |
| OE 0 d vs. OE7 d & WT 0 d vs. WT 7 d(1 897) | 植物昼夜节律 Circadian rhythm-plant | 67 | 0.16 | 2.43E-13 |
| 泛醌和其他萜—醌生物合成 Ubiquinone and other terpenoid-quinone biosynthesis | 14 | 0.16 | 7.13E-03 | |
| 类胡萝卜素生物合成 Carotenoid biosynthesis | 16 | 0.15 | 7.13E-03 | |
| 花生四烯酸代谢 Arachidonic acid metabolism | 13 | 0.17 | 8.61E-03 | |
| 类固醇生物合成 Steroid biosynthesis | 11 | 0.15 | 3.71E-02 | |
| 淀粉和蔗糖代谢 Starch and sucrose metabolism | 48 | 0.09 | 3.71E-02 | |
| OE 7 d vs. OE 14 d(154) | 戊糖和葡萄糖醛酸酯相互转化 Pentose and glucuronate interconversions | 6 | 0.02 | 9.66E-02 |
| 苯丙烷生物合成 Phenylpropanoid biosynthesis | 9 | 0.01 | 9.66E-02 | |
| 硒化合物代谢 Selenocompound metabolism | 3 | 0.04 | 1.01E-01 | |
| 黄酮和黄酮醇生物合成 Flavone and flavonol biosynthesis | 2 | 0.03 | 2.78E-01 | |
| OE 7 d vs. OE 14 d & WT 7 d vs. WT 14 d(264) | 黄酮生物合成 Flavonoid biosynthesis | 11 | 0.07 | 1.86E-05 |
| 内质网蛋白质加工 Protein processing in endoplasmic reticulum | 15 | 0.02 | 4.52E-02 | |
| 植物—病原菌互作 Plant-pathogen interaction | 19 | 0.02 | 4.52E-02 | |
| WT 7 d vs. WT 14 d(3 407) | 植物—病原菌互作 Plant-pathogen interaction | 169 | 0.19 | 1.45E-11 |
| 植物MAPK信号途径 MAPK signaling pathway-plant | 166 | 0.17 | 5.42E-09 | |
| 苯丙氨酸代谢 Phenylalanine metabolism | 37 | 0.27 | 1.47E-06 | |
| 黄酮生物合成 Flavonoid biosynthesis | 34 | 0.23 | 2.25E-04 | |
| 苯丙烷生物合成 Phenylpropanoid biosynthesis | 110 | 0.16 | 3.48E-04 | |
| 光合作用 Photosynthesis | 32 | 0.23 | 4.57E-04 | |
| 光合作用天线蛋白 Photosynthesis-antenna proteins | 12 | 0.34 | 2.50E-03 | |
| 异喹啉生物碱生物合成 Isoquinoline alkaloid biosynthesis | 14 | 0.29 | 5.82E-03 | |
| 谷胱甘肽代谢 Glutathione metabolism | 30 | 0.20 | 6.99E-03 | |
| 芪类、二苯基庚烷和姜酚生物合成 Stilbenoid,diarylheptanoid and gingerol biosynthesis | 18 | 0.24 | 8.25E-03 | |
| ABC转运蛋白 ABC transporters | 37 | 0.18 | 9.11E-03 | |
| 光合生物碳固定 Carbon fixation in photosynthetic organisms | 33 | 0.18 | 1.43E-02 | |
| 淀粉和蔗糖代谢 Starch and sucrose metabolism | 80 | 0.14 | 2.84E-02 | |
| 黄酮和黄酮醇生物合成 Flavone and flavonol biosynthesis | 14 | 0.23 | 4.18E-02 | |
| 精氨酸生物合成 Arginine biosynthesis | 18 | 0.20 | 4.37E-02 | |
| 酪氨酸代谢 Tyrosine metabolism | 16 | 0.21 | 4.92E-02 |
| 基因 Gene ID | 基因表达量(FPKM) | 基因功能注释 Gene function annotation | |||||
|---|---|---|---|---|---|---|---|
| 野生型 Wild type | 转基因香蕉 DX11 Transgenic bananas | ||||||
| 0 d | 7 d | 14 d | 0 d | 7 d | 14 d | ||
| Ma11_g19450 | 8.72 | 7.66 | 125.14 | 645.73 | 85.73 | 76.26 | 泛素羧基末端水解酶36/42 Ubiquitin carboxyl- terminal hydrolase 36/42 |
| Ma10_g12250 | 0.48 | 0.76 | 0.29 | 491.73 | 326.85 | 317.63 | 转录因子ICE1 Transcription factor ICE1 |
| Ma06_g16670 | 52.83 | 15.99 | 30.77 | 490.96 | 79.46 | 12.19 | 甘露糖特异类凝集素Mannose-specific lectin-like |
| Ma04_g33410 | 2.01 | 0.98 | 3.80 | 220.36 | 19.60 | 0.60 | 泛素羧基末端水解酶36/42 Ubiquitin carboxyl- terminal hydrolase 36/42 |
| Ma11_g22720 | 11.51 | 35.87 | 52.02 | 179.83 | 117.17 | 27.65 | 类金属硫蛋白2型Metallothionein-like protein type 2 |
| Ma06_g03820 | 1.65 | 7.76 | 82.79 | 170.87 | 45.08 | 16.56 | 类EXORDIUM 蛋白2 Protein EXORDIUM-like 2 |
| Ma10_g15940 | 48.26 | 55.42 | 226.86 | 145.97 | 240.12 | 88.15 | 过氧化物酶 Peroxidase |
| Ma04_g40060 | 17.93 | 38.03 | 16.79 | 128.60 | 196.36 | 51.97 | 果胶酯酶 Pectinesterase |
| Ma02_g16400 | 9.11 | 11.60 | 36.76 | 73.16 | 60.69 | 34.71 | 非特异性脂转运蛋白2 Non-specific lipid-transfer protein 2-like |
| Ma00_g02560 | 4.43 | 0.72 | 2.53 | 53.73 | 3.23 | 0.62 | UDP-葡萄糖醛酸酯4-异构酶 UDP-glucuronate 4-epimerase |
| Ma04_g31200 | 0.05 | 0.10 | 15.01 | 52.53 | 2.20 | 0.05 | 果胶酯酶Pectinesterase |
| Ma03_g33000 | 8.73 | 12.50 | 108.45 | 39.39 | 43.65 | 36.67 | DNA指导的RNA聚合酶Ⅱ亚基RPB1 DNA- directed RNA polymerase II subunit RPB1 |
| Ma10 g01930 | 5.74 | 4.03 | 9.69 | 38.51 | 14.91 | 4.65 | 1-脱氧-D-木酮糖-5-磷酸合成酶 1-Deoxy-D-xylulose- 5-phosphate synthase |
| Ma09_g10470 | 3.11 | 1.51 | 16.58 | 37.15 | 23.73 | 27.74 | 类GOS9蛋白Protein GOS9-like |
| Ma08_g31490 | 2.09 | 0.71 | 1.19 | 36.48 | 4.28 | 1.23 | 富半胱氨酸和甘氨酸蛋白 Cysteine and glycine-rich protein |
| Ma09_g25000 | 9.62 | 9.09 | 9.12 | 35.19 | 44.91 | 19.02 | 糖转运蛋白C Sugar carrier protein C |
| Ma08_g05750 | 5.34 | 18.13 | 205.01 | 33.95 | 91.41 | 47.89 | 赤霉素受体GID1 Gibberellin receptor GID1 |
| Ma08_g09150 | 1.01 | 0.91 | 8.82 | 32.42 | 11.90 | 6.70 | 多酚氧化酶polyphenol oxidase |
| Ma02 g13020 | 7.09 | 4.53 | 617.71 | 30.60 | 30.74 | 49.95 | 磷脂:二酰甘油酰基转移酶 Phospholipid:diacylglycerol acyltransferase |
| Ma03_g28020 | 8.34 | 2.91 | 159.63 | 25.75 | 10.80 | 11.54 | 几丁质酶1亚型X1 Chitinase 1 isoform X1 |
| Ma11_g25080 | 5.53 | 22.31 | 77.52 | 19.21 | 114.19 | 48.29 | 糖转运家族32 Solute carrier family 32 |
| Ma08_g12790 | 0.09 | 1.53 | 63.49 | 19.07 | 97.90 | 34.95 | DNA连接酶1 DNA ligase 1 |
| Ma09_g20710 | 1.58 | 2.09 | 194.79 | 11.40 | 20.77 | 12.12 | 几丁质酶 chitinase |
| Ma09 g11910 | 3.01 | 2.83 | 30.79 | 12.41 | 9.83 | 5.99 | 白介素-1受体相关激酶4 Interleukin-1 receptor- associated kinase 4 |
| Ma04_g09070 | 0.00 | 0.04 | 85.83 | 13.85 | 5.26 | 0.12 | 糖转运家族50 Solute carrier family 50 |
| Ma06_g31980 | 1.13 | 0.99 | 124.83 | 3.44 | 19.23 | 6.20 | 几丁质酶Chitinase |
| Ma11_g21980 | 0.08 | 0.16 | 35.66 | 3.66 | 15.75 | 0.08 | 扩张蛋白Expansin |
| Ma00_g04060 | 0.12 | 0.12 | 0.11 | 3.61 | 2.23 | 0.57 | 丝裂原激活蛋白激酶激酶4/5 Mitogen-activated protein kinase kinase 4/5 |
| Ma01_g04570 | 0.00 | 0 | 0.05 | 3.49 | 1.95 | 0.16 | 丝裂原激活蛋白激酶激酶4/5 Mitogen-activated protein kinase kinase 4/5 |
| Ma06_g18840 | 21.60 | 109.27 | 228.42 | 0.08 | 0.28 | 0.20 | 12-氧-植物二烯酸氧化还原酶 12-Oxophytodienoic acid reductase |
Table 6 Thirty common different express genes of wild and transgenic bananas before inoculation(0 d)and after inoculation(7 d)
| 基因 Gene ID | 基因表达量(FPKM) | 基因功能注释 Gene function annotation | |||||
|---|---|---|---|---|---|---|---|
| 野生型 Wild type | 转基因香蕉 DX11 Transgenic bananas | ||||||
| 0 d | 7 d | 14 d | 0 d | 7 d | 14 d | ||
| Ma11_g19450 | 8.72 | 7.66 | 125.14 | 645.73 | 85.73 | 76.26 | 泛素羧基末端水解酶36/42 Ubiquitin carboxyl- terminal hydrolase 36/42 |
| Ma10_g12250 | 0.48 | 0.76 | 0.29 | 491.73 | 326.85 | 317.63 | 转录因子ICE1 Transcription factor ICE1 |
| Ma06_g16670 | 52.83 | 15.99 | 30.77 | 490.96 | 79.46 | 12.19 | 甘露糖特异类凝集素Mannose-specific lectin-like |
| Ma04_g33410 | 2.01 | 0.98 | 3.80 | 220.36 | 19.60 | 0.60 | 泛素羧基末端水解酶36/42 Ubiquitin carboxyl- terminal hydrolase 36/42 |
| Ma11_g22720 | 11.51 | 35.87 | 52.02 | 179.83 | 117.17 | 27.65 | 类金属硫蛋白2型Metallothionein-like protein type 2 |
| Ma06_g03820 | 1.65 | 7.76 | 82.79 | 170.87 | 45.08 | 16.56 | 类EXORDIUM 蛋白2 Protein EXORDIUM-like 2 |
| Ma10_g15940 | 48.26 | 55.42 | 226.86 | 145.97 | 240.12 | 88.15 | 过氧化物酶 Peroxidase |
| Ma04_g40060 | 17.93 | 38.03 | 16.79 | 128.60 | 196.36 | 51.97 | 果胶酯酶 Pectinesterase |
| Ma02_g16400 | 9.11 | 11.60 | 36.76 | 73.16 | 60.69 | 34.71 | 非特异性脂转运蛋白2 Non-specific lipid-transfer protein 2-like |
| Ma00_g02560 | 4.43 | 0.72 | 2.53 | 53.73 | 3.23 | 0.62 | UDP-葡萄糖醛酸酯4-异构酶 UDP-glucuronate 4-epimerase |
| Ma04_g31200 | 0.05 | 0.10 | 15.01 | 52.53 | 2.20 | 0.05 | 果胶酯酶Pectinesterase |
| Ma03_g33000 | 8.73 | 12.50 | 108.45 | 39.39 | 43.65 | 36.67 | DNA指导的RNA聚合酶Ⅱ亚基RPB1 DNA- directed RNA polymerase II subunit RPB1 |
| Ma10 g01930 | 5.74 | 4.03 | 9.69 | 38.51 | 14.91 | 4.65 | 1-脱氧-D-木酮糖-5-磷酸合成酶 1-Deoxy-D-xylulose- 5-phosphate synthase |
| Ma09_g10470 | 3.11 | 1.51 | 16.58 | 37.15 | 23.73 | 27.74 | 类GOS9蛋白Protein GOS9-like |
| Ma08_g31490 | 2.09 | 0.71 | 1.19 | 36.48 | 4.28 | 1.23 | 富半胱氨酸和甘氨酸蛋白 Cysteine and glycine-rich protein |
| Ma09_g25000 | 9.62 | 9.09 | 9.12 | 35.19 | 44.91 | 19.02 | 糖转运蛋白C Sugar carrier protein C |
| Ma08_g05750 | 5.34 | 18.13 | 205.01 | 33.95 | 91.41 | 47.89 | 赤霉素受体GID1 Gibberellin receptor GID1 |
| Ma08_g09150 | 1.01 | 0.91 | 8.82 | 32.42 | 11.90 | 6.70 | 多酚氧化酶polyphenol oxidase |
| Ma02 g13020 | 7.09 | 4.53 | 617.71 | 30.60 | 30.74 | 49.95 | 磷脂:二酰甘油酰基转移酶 Phospholipid:diacylglycerol acyltransferase |
| Ma03_g28020 | 8.34 | 2.91 | 159.63 | 25.75 | 10.80 | 11.54 | 几丁质酶1亚型X1 Chitinase 1 isoform X1 |
| Ma11_g25080 | 5.53 | 22.31 | 77.52 | 19.21 | 114.19 | 48.29 | 糖转运家族32 Solute carrier family 32 |
| Ma08_g12790 | 0.09 | 1.53 | 63.49 | 19.07 | 97.90 | 34.95 | DNA连接酶1 DNA ligase 1 |
| Ma09_g20710 | 1.58 | 2.09 | 194.79 | 11.40 | 20.77 | 12.12 | 几丁质酶 chitinase |
| Ma09 g11910 | 3.01 | 2.83 | 30.79 | 12.41 | 9.83 | 5.99 | 白介素-1受体相关激酶4 Interleukin-1 receptor- associated kinase 4 |
| Ma04_g09070 | 0.00 | 0.04 | 85.83 | 13.85 | 5.26 | 0.12 | 糖转运家族50 Solute carrier family 50 |
| Ma06_g31980 | 1.13 | 0.99 | 124.83 | 3.44 | 19.23 | 6.20 | 几丁质酶Chitinase |
| Ma11_g21980 | 0.08 | 0.16 | 35.66 | 3.66 | 15.75 | 0.08 | 扩张蛋白Expansin |
| Ma00_g04060 | 0.12 | 0.12 | 0.11 | 3.61 | 2.23 | 0.57 | 丝裂原激活蛋白激酶激酶4/5 Mitogen-activated protein kinase kinase 4/5 |
| Ma01_g04570 | 0.00 | 0 | 0.05 | 3.49 | 1.95 | 0.16 | 丝裂原激活蛋白激酶激酶4/5 Mitogen-activated protein kinase kinase 4/5 |
| Ma06_g18840 | 21.60 | 109.27 | 228.42 | 0.08 | 0.28 | 0.20 | 12-氧-植物二烯酸氧化还原酶 12-Oxophytodienoic acid reductase |
| [1] |
doi: 10.1093/pcp/pcn100 URL |
| [2] |
doi: 10.1371/journal.pone.0073945 URL |
| [3] |
doi: 10.1186/1741-7007-11-121 pmid: 24350981 |
| [4] |
doi: 10.1093/jee/tox374 pmid: 29420735 |
| [5] |
doi: 10.1038/s41467-017-01670-6 pmid: 29133817 |
| [6] |
|
|
邓秀新, 束怀瑞, 郝玉金, 徐强, 韩明玉, 张绍铃, 段常青, 姜全, 易干军, 陈厚彬. 2018. 果树学科百年发展回顾. 农学学报, 8 (1):24-34.
|
|
| [7] |
doi: S1674-2052(20)30034-4 pmid: 32068158 |
| [8] |
doi: 10.1111/pbi.v18.1 URL |
| [9] |
doi: 10.1016/j.plaphy.2013.09.014 URL |
| [10] |
doi: 10.1186/s12870-021-02868-z |
| [11] |
doi: 10.1093/mp/ssu098 URL |
| [12] |
doi: 10.1111/jipb.v60.9 URL |
| [13] |
doi: 10.1038/s41598-019-53229-8 |
| [14] |
doi: 10.3389/fpls.2018.00282 URL |
| [15] |
doi: 10.1105/tpc.18.00825 URL |
| [16] |
doi: 10.1093/jxb/erv138 URL |
| [17] |
doi: 10.1016/j.hpj.2022.02.004 URL |
| [18] |
doi: 10.3390/ijms22063002 URL |
| [19] |
doi: 10.1016/j.scienta.2021.110590 URL |
| [20] |
|
|
李华平, 李云锋, 聂燕芳. 2019. 香蕉枯萎病的发生及防控研究现状. 华南农业大学学报, 40 (5):128-136.
|
|
| [21] |
doi: 10.1016/j.xplc.2021.100231 URL |
| [22] |
doi: 10.3389/fpls.2019.01069 URL |
| [23] |
doi: 10.1016/j.plaphy.2017.02.002 URL |
| [24] |
doi: 10.1002/cbf.v38.5 URL |
| [25] |
|
| [26] |
|
| [27] |
doi: 10.1046/j.1364-3703.2003.00180.x URL |
| [28] |
doi: 10.1016/j.cropro.2015.01.007 URL |
| [29] |
doi: 10.1105/tpc.12.7.1041 pmid: 10899973 |
| [30] |
doi: 10.1111/tpj.2015.81.issue-2 URL |
| [31] |
|
| [32] |
|
|
邵秀红, 窦同心, 林雪茜, 盛鸥, 邓贵明, 毕方铖, 胡春华, 李春雨, 易干军, 杨乔松. 2018. 香蕉对枯萎病的抗性机制研究现状与展望. 园艺学报, 45 (9):1661-1674.
doi: 10.16420/j.issn.0513-353x.2018-0579 URL |
|
| [33] |
doi: 10.1016/j.hpj.2018.08.001 URL |
| [34] |
doi: 10.1111/jipb.v62.3 URL |
| [35] |
doi: 10.1105/tpc.19.00487 pmid: 31690653 |
| [36] |
doi: 10.1371/journal.pone.0102303 URL |
| [37] |
|
|
许奕, 徐碧玉, 宋顺, 刘菊华, 张建斌, 贾彩红, 金志强. 2013. 香蕉茉莉酸合成关键酶基因MaOPR的克隆和表达分析. 园艺学报, 40 (2):237-246.
|
|
| [38] |
doi: 10.1111/tpj.2007.51.issue-3 URL |
| [39] |
|
|
张欣, 漆艳香, 曾凡云, 王艳玮, 谢培兰, 谢艺贤, 彭军. 2023. 香蕉枯萎病菌Dicer-like基因的功能分析. 园艺学报, 50 (2):279-294.
doi: 10.16420/j.issn.0513-353x.2021-1081 |
|
| [40] |
doi: 10.1016/j.devcel.2017.11.016 URL |
| [41] |
doi: 10.1104/pp.16.00533 URL |
| [42] |
|
| [43] |
doi: 10.1111/plb.13095 pmid: 32009285 |
| [1] | ZHANG Xin, QI Yanxiang, ZENG Fanyun, WANG Yanwei, XIE Peilan, XIE Yixian, PENG Jun. Functional Analysis of Dicer-like Genes in Fusarium oxysporum f. sp. cubense Race 4 [J]. Acta Horticulturae Sinica, 2023, 50(2): 279-294. |
| [2] | LIANG Jiali, WU Qisong, CHEN Guangquan, ZHANG Rong, XU Chunxiang, FENG Shujie. Identification of the Neopestalotiopsis musae Pathogen of Banana Leaf Spot Disease [J]. Acta Horticulturae Sinica, 2023, 50(2): 410-420. |
| [3] | RAO Xueqin, LI Huaping. Research Progress on Endogenous Banana Streak Virus [J]. Acta Horticulturae Sinica, 2023, 50(10): 2288-2296. |
| [4] | YANG Xingyu, XU Linbing, WU Yuanli, QIU Diyang, FAN Linlin, HUANG Bingzhi. Genome(ABBB)Identification of a Hybrid Banana Cultivar‘Fenza 1’ [J]. Acta Horticulturae Sinica, 2022, 49(9): 1991-1997. |
| [5] | MENG Zhen, ZHANG Weiping, WANG Ying, LI Long, JI Xiaoxue, DONG Bei, QIAO Kang. Establishment and Application of Real-Time PCR for Quantitative Detection of Fusarium oxysporum f. sp. lycopersici [J]. Acta Horticulturae Sinica, 2022, 49(11): 2479-2488. |
| [6] | HUANG Weijian, LI Meng. Status and Prospects Whole Genome Sequencing in Fruit Trees [J]. Acta Horticulturae Sinica, 2021, 48(4): 733-748. |
| [7] | ZHUANG Yueying, ZHOU Lijun, CHENG Bixuan, YU Chao, LUO Le, PAN Huitang, ZHANG Qixiang. Study on the Fragrance Metabolic Genes of Rosa odorata Based on Transcriptome Sequencing [J]. Acta Horticulturae Sinica, 2021, 48(11): 2262-2274. |
| [8] | YANG Xingyu, XIAO Weiqiang, XU Linbing, LI Huaping, HUANG Bingzhi, YE Chunhai, CHEN Biao, CHEN Gu, LÜ Qingfang, LIANG Jiaxian, WU Yuanli, and HU Lingyu. A New Banana Cultivar‘Nantianhong’ [J]. Acta Horticulturae Sinica, 2020, 47(S2): 2961-2962. |
| [9] | WU Yuanli1,HUANG Bingzhi1,*,ZHANG Zhisheng2,and Yang Xingyu1. Modification of in vitro Bioassay for Screening Musa Species Against Fusarium oxysporum f. sp. cubense [J]. ACTA HORTICULTURAE SINICA, 2020, 47(8): 1577-1584. |
| [10] | LIU Yanying,NI Shanshan,XIANG Leilei,CHEN Yukun,and LAI Zhongxiong*. Genome-wide Identification of the Laccase Gene Family and Its Expression Analysis Under Low Temperature Stress in Musa accuminata [J]. ACTA HORTICULTURAE SINICA, 2020, 47(5): 837-852. |
| [11] | ZHAI Zixiang, LI Deming, DENG Tao, DENG Dahao, YANG Laying, ZHOU You, GUO Lijia, and HUANG Junsheng, . Inhibition of Fusarium oxysporum by Different Phenolic Acids Secreted from Roots of Different Resistant Banana Varieties [J]. Acta Horticulturae Sinica, 2020, 47(11): 2207-2214. |
| [12] | LIU Fan, TIAN Na, SUN Xueli, LIU Jiapeng, WU Junwei, HUANG Yuji, and CHENG Chunzhen. Genome-wide Identification and Expression Analysis of Banana GLP Gene Family [J]. Acta Horticulturae Sinica, 2020, 47(10): 1930-1946. |
| [13] | KOU Dandan,ZHANG Ye,WANG Pengfei,Li Dongdong,ZHANG Xueying*,and CHEN Haijiang*. Differences in Sucrose and Malic Acid Accumulation and The Related Gene Expression in‘Kurakato Wase’Peach and Its Early-ripening Mutant [J]. ACTA HORTICULTURAE SINICA, 2019, 46(12): 2286-2298. |
| [14] | SUN Xueli,LIU Fan,TIAN Na,XIANG Leilei,HAO Xiangyang,WANG Yun,PENG Liyun,WANG Tianchi,CHENG Chunzhen*,and LAI Zhongxiong*. Genome-wide Identification and Expression Analysis of Aux/IAA Gene Family in Banana [J]. ACTA HORTICULTURAE SINICA, 2019, 46(10): 1919-1935. |
| [15] | SHAO Xiuhong1,2,DOU Tongxin2,LIN Xuexi1,2,SHENG Ou2,DENG Guiming2,BI Fangcheng2,HU Chunhua2,LI Chunyu2,YI Ganjun2,and YANG Qiaosong2,*. Research Status and Prospects on Resistance Mechanisms to Fusarium Wilt of Banana [J]. ACTA HORTICULTURAE SINICA, 2018, 45(9): 1661-1674. |
| Viewed | ||||||
|
Full text |
|
|||||
|
Abstract |
|
|||||
Copyright © 2012 Acta Horticulturae Sinica 京ICP备10030308号-2 国际联网备案号 11010802023439
Tel: 010-82109523 E-Mail: yuanyixuebao@126.com
Support by: Beijing Magtech Co.Ltd