
Acta Horticulturae Sinica ›› 2021, Vol. 48 ›› Issue (9): 1680-1694.doi: 10.16420/j.issn.0513-353x.2021- 0219
• Research Papers • Previous Articles Next Articles
XU Hongxia1, ZHOU Huifen2, LI Xiaoying1, JIANG Luhua3, CHEN Junwei1,*( )
)
Received:2021-05-24
Revised:2021-08-25
Online:2021-09-25
Published:2021-09-30
Contact:
CHEN Junwei
E-mail:chenjunwei@zaas.ac.cn
CLC Number:
XU Hongxia, ZHOU Huifen, LI Xiaoying, JIANG Luhua, CHEN Junwei. Comparative Transcriptome Analysis of Different Developmental Stages of Flowers and Fruits in Loquat Under Low Temperature Stress[J]. Acta Horticulturae Sinica, 2021, 48(9): 1680-1694.
Add to citation manager EndNote|Ris|BibTeX
URL: https://www.ahs.ac.cn/EN/10.16420/j.issn.0513-353x.2021- 0219
| 基因名称 Gene name | 基因 ID Gene ID | 引物序列(5′-3′) Primer sequence | |
|---|---|---|---|
| Ejactin | c158550.graph_c1 | F:GGATTTGCTGGTGATGATGC; | R:CCGTGCTCAATGGGATACTT |
| ERF37 | c106174.graph_c0 | F:GCTGGGTGAGGAGATAACGG | R:AAGATGCGAAGACGGTGGTG |
| ERF73 | c123742.graph_c0 | F:GCTTTCCTGAGGCGGACGAT | R:AATTGAGCTTGGAGGCGATT |
| ERF35 | c125424.graph_c0 | F:GGGGAGAGCATAGCGATACA | R:GCTTACGTCATTTTCTGGGC |
| ERF105 | c126492.graph_c0 | F:TCGCTCGTACTTGTCACACC | R:GCTGACCATCAAAGGCAACT |
| RAV2 | c126935.graph_c1 | F:GAGGATGATCGGCTGGGAGT | R:AGCTGCTGCAAGGCACTACA |
| ERF60 | c129543.graph_c0 | F:TGGCCAATAAGAATCACCAA | R:CCAAAAAGGATCTGACGAGG |
| ERF33 | c46214.graph_c0 | F:GCCTGTTCTTTGATTTTGGT | R:TCATGAGCTGATCTTCCCTC |
| WRKY33 | c142932.graph_c0 | F:CCACTGAAAACCAATCGTAC | R:CTTAGACTTCTTATCGCAAC |
| NAC72 | c51471.graph_c0 | F:ATGTTGGGGTTGGATAAGAA | R:CGGAATAGAAGAAGCAGAAGAG |
| NAC74 | c136256.graph_c0 | F:AGTCCATAGTATGCGCCACC | R:TGACTGCGGTTTTCTTTTGA |
| NAC29 | c130635.graph_c0 | F:GAAACGGGAACATTGGTAGT | R:AATGAATGAGATGCTGGTGT |
| NAC91 | c136368.graph_c0 | F:ATCATCGCTAAATTCATTCC | R:ACTTTACCACCTCTACCAAC |
Table 1 Differentially expressed genes and their primer sequences in qRT-PCR
| 基因名称 Gene name | 基因 ID Gene ID | 引物序列(5′-3′) Primer sequence | |
|---|---|---|---|
| Ejactin | c158550.graph_c1 | F:GGATTTGCTGGTGATGATGC; | R:CCGTGCTCAATGGGATACTT |
| ERF37 | c106174.graph_c0 | F:GCTGGGTGAGGAGATAACGG | R:AAGATGCGAAGACGGTGGTG |
| ERF73 | c123742.graph_c0 | F:GCTTTCCTGAGGCGGACGAT | R:AATTGAGCTTGGAGGCGATT |
| ERF35 | c125424.graph_c0 | F:GGGGAGAGCATAGCGATACA | R:GCTTACGTCATTTTCTGGGC |
| ERF105 | c126492.graph_c0 | F:TCGCTCGTACTTGTCACACC | R:GCTGACCATCAAAGGCAACT |
| RAV2 | c126935.graph_c1 | F:GAGGATGATCGGCTGGGAGT | R:AGCTGCTGCAAGGCACTACA |
| ERF60 | c129543.graph_c0 | F:TGGCCAATAAGAATCACCAA | R:CCAAAAAGGATCTGACGAGG |
| ERF33 | c46214.graph_c0 | F:GCCTGTTCTTTGATTTTGGT | R:TCATGAGCTGATCTTCCCTC |
| WRKY33 | c142932.graph_c0 | F:CCACTGAAAACCAATCGTAC | R:CTTAGACTTCTTATCGCAAC |
| NAC72 | c51471.graph_c0 | F:ATGTTGGGGTTGGATAAGAA | R:CGGAATAGAAGAAGCAGAAGAG |
| NAC74 | c136256.graph_c0 | F:AGTCCATAGTATGCGCCACC | R:TGACTGCGGTTTTCTTTTGA |
| NAC29 | c130635.graph_c0 | F:GAAACGGGAACATTGGTAGT | R:AATGAATGAGATGCTGGTGT |
| NAC91 | c136368.graph_c0 | F:ATCATCGCTAAATTCATTCC | R:ACTTTACCACCTCTACCAAC |
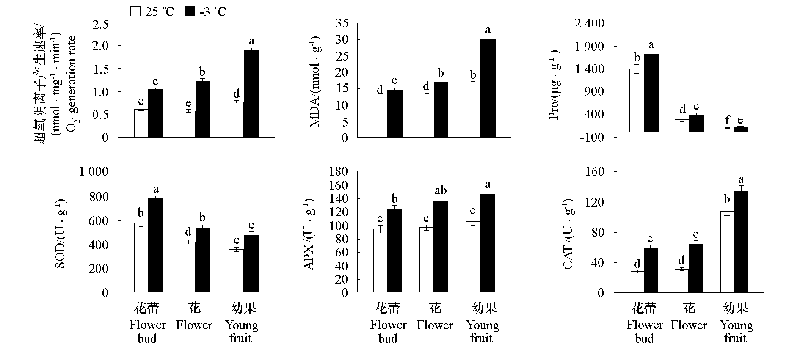
Fig. 1 Changes in superoxide anion generation rate,MDA content,proline content,SOD activity,APX activity,and CAT activity in response to low temperature stress in different developmental stages of flowers and young fruits * Different letters are significantly different at P ≤ 0.05.
| 数据库 Database | 注释数量 Number of annotated unigene | 比例/% Percentage of annotated unigene |
|---|---|---|
| Nr | 38 093 | 97.80 |
| Swiss-Prot | 22 119 | 56.79 |
| KEGG | 14 813 | 38.03 |
| eggNOG | 35 024 | 89.92 |
| KOG | 20 989 | 53.89 |
| GO | 25 436 | 65.30 |
| Pfam | 27 156 | 69.72 |
Table 2 The statistical result of unigene functional annotation
| 数据库 Database | 注释数量 Number of annotated unigene | 比例/% Percentage of annotated unigene |
|---|---|---|
| Nr | 38 093 | 97.80 |
| Swiss-Prot | 22 119 | 56.79 |
| KEGG | 14 813 | 38.03 |
| eggNOG | 35 024 | 89.92 |
| KOG | 20 989 | 53.89 |
| GO | 25 436 | 65.30 |
| Pfam | 27 156 | 69.72 |
| GO分类 GO classification | GO序列号 GO accession | 注释功能(词条) Term | 差异基因数 DEGs number | -3 ℃ vs 25 ℃ | ||
|---|---|---|---|---|---|---|
| 花蕾 Flower bud | 花 Flower | 幼果 Young fruit | ||||
| 生物学过程 Biological process | GO:0008152 | 代谢过程Metabolic process | 2 193 | 298 | 781 | 1 239 |
| GO:0009987 | 细胞过程Cellular process | 2 127 | 248 | 661 | 1 075 | |
| GO:0044699 | 单一生物过程Single-organism process | 1 863 | 230 | 585 | 1 033 | |
| GO:0051179 | 定位Localization | 790 | 108 | 276 | 546 | |
| GO:0065007 | 生物调控Biological regulation | 616 | 142 | 270 | 427 | |
| GO:0071840 | 细胞组分或生物合成 Cellular component organization or biogenesis | 465 | 40 | 132 | 293 | |
| GO:0050896 | 应激反应Response to stimulus | 457 | 94 | 193 | 279 | |
| GO:0023052 | 信号Signaling | 122 | 24 | 47 | 77 | |
| GO:0051704 | 多生物过程Multi-organism process | 114 | 11 | 39 | 73 | |
| GO:0032502 | 发育过程Developmental process | 94 | 25 | 35 | 46 | |
| GO:0040007 | 生长Growth | 75 | 2 | 16 | 41 | |
| GO:0022414 | 生殖过程Reproductive process | 59 | 15 | 25 | 44 | |
| GO:0000003 | 再生Reproduction | 55 | 0 | 26 | 46 | |
| GO:0032501 | 多细胞生物过程Multicellular organismal process | 48 | 17 | 22 | 17 | |
| GO:0022610 | 生物粘附Biological adhesion | 17 | 0 | 3 | 6 | |
| GO:0002376 | 免疫系统过程Immune system process | 7 | 0 | 2 | 0 | |
| GO:0048511 | 节律过程Rhythmic process | 3 | 0 | 1 | 1 | |
| 细胞组分 Cellular component | GO:0005623 | 细胞Cell | 1 808 | 283 | 718 | 1 229 |
| GO:0044464 | 细胞部分Cell part | 1 796 | 280 | 710 | 1 222 | |
| GO:0043226 | 细胞器Organelle | 1 260 | 201 | 485 | 858 | |
| GO:0016020 | 膜Membrane | 1 642 | 294 | 623 | 1 024 | |
| GO:0044425 | 膜部分Membrane part | 1 293 | 234 | 495 | 805 | |
| GO:0044422 | 细胞器部分Organelle part | 621 | 67 | 218 | 418 | |
| GO:0032991 | 大分子复合物Macromolecular complex | 552 | 42 | 208 | 361 | |
| GO:0031974 | 膜封闭的管腔Membrane-enclosed lumen | 160 | 11 | 40 | 112 | |
| GO:0005576 | 细胞外区域Extracellular region | 87 | 18 | 45 | 61 | |
| GO:0030054 | 细胞连接Cell junction | 15 | 4 | 6 | 5 | |
| GO:0055044 | 共质体Symplast | 14 | 4 | 5 | 5 | |
| GO:0019012 | 病毒Virion | 4 | 0 | 1 | 4 | |
| GO:0044423 | 病毒部分Virion part | 4 | 0 | 1 | 4 | |
| GO:0009295 | 内核Nucleoid | 11 | 0 | 5 | 6 | |
| GO:0044421 | 细胞外区域Extracellular region part | 28 | 3 | 21 | 26 | |
| 分子功能 | GO:0003824 | 催化活性Catalytic activity | 1 949 | 308 | 725 | 1 222 |
| Molecular | GO:0005488 | 联结Binding | 1 670 | 321 | 688 | 1 024 |
| function | GO:0005215 | 转运活动Transporter activity | 402 | 53 | 144 | 277 |
| GO:0005198 | 结构分子活动Structural molecule activity | 178 | 9 | 102 | 85 | |
| GO:0001071 | 核酸联结转录因子活性 Nucleic acid binding transcription factor activity | 109 | 40 | 64 | 68 | |
| GO:0016209 | 抗氧化活性Antioxidant activity | 40 | 10 | 21 | 21 | |
| GO:0009055 | 电子载体活动Electron carrier activity | 35 | 5 | 13 | 20 | |
| GO:0060089 | 分子传感器活动Molecular transducer activity | 36 | 5 | 7 | 8 | |
| GO:0004871 | 信号传导活动Signal transducer activity | 36 | 10 | 18 | 23 | |
| GO:0000988 | 转录因子活性,蛋白质结合 Transcription factor activity,protein binding | 16 | 4 | 3 | 12 | |
| GO:0045735 | 营养库活性Nutrient reservoir activity | 5 | 1 | 3 | 2 | |
| GO:0031386 | 蛋白质标签Protein tag | 5 | 0 | 0 | 2 | |
| GO:0016530 | 金属伴侣活性Metallochaperone activity | 1 | 1 | 1 | 1 | |
| GO:0045182 | 翻译调控活性Translation regulator activity | 2 | 0 | 0 | 2 | |
Table 3 The number of DEGs in GO classification in low-temperature treatment and control comparison groups of three developmental stages of flowers and young fruits in loquat
| GO分类 GO classification | GO序列号 GO accession | 注释功能(词条) Term | 差异基因数 DEGs number | -3 ℃ vs 25 ℃ | ||
|---|---|---|---|---|---|---|
| 花蕾 Flower bud | 花 Flower | 幼果 Young fruit | ||||
| 生物学过程 Biological process | GO:0008152 | 代谢过程Metabolic process | 2 193 | 298 | 781 | 1 239 |
| GO:0009987 | 细胞过程Cellular process | 2 127 | 248 | 661 | 1 075 | |
| GO:0044699 | 单一生物过程Single-organism process | 1 863 | 230 | 585 | 1 033 | |
| GO:0051179 | 定位Localization | 790 | 108 | 276 | 546 | |
| GO:0065007 | 生物调控Biological regulation | 616 | 142 | 270 | 427 | |
| GO:0071840 | 细胞组分或生物合成 Cellular component organization or biogenesis | 465 | 40 | 132 | 293 | |
| GO:0050896 | 应激反应Response to stimulus | 457 | 94 | 193 | 279 | |
| GO:0023052 | 信号Signaling | 122 | 24 | 47 | 77 | |
| GO:0051704 | 多生物过程Multi-organism process | 114 | 11 | 39 | 73 | |
| GO:0032502 | 发育过程Developmental process | 94 | 25 | 35 | 46 | |
| GO:0040007 | 生长Growth | 75 | 2 | 16 | 41 | |
| GO:0022414 | 生殖过程Reproductive process | 59 | 15 | 25 | 44 | |
| GO:0000003 | 再生Reproduction | 55 | 0 | 26 | 46 | |
| GO:0032501 | 多细胞生物过程Multicellular organismal process | 48 | 17 | 22 | 17 | |
| GO:0022610 | 生物粘附Biological adhesion | 17 | 0 | 3 | 6 | |
| GO:0002376 | 免疫系统过程Immune system process | 7 | 0 | 2 | 0 | |
| GO:0048511 | 节律过程Rhythmic process | 3 | 0 | 1 | 1 | |
| 细胞组分 Cellular component | GO:0005623 | 细胞Cell | 1 808 | 283 | 718 | 1 229 |
| GO:0044464 | 细胞部分Cell part | 1 796 | 280 | 710 | 1 222 | |
| GO:0043226 | 细胞器Organelle | 1 260 | 201 | 485 | 858 | |
| GO:0016020 | 膜Membrane | 1 642 | 294 | 623 | 1 024 | |
| GO:0044425 | 膜部分Membrane part | 1 293 | 234 | 495 | 805 | |
| GO:0044422 | 细胞器部分Organelle part | 621 | 67 | 218 | 418 | |
| GO:0032991 | 大分子复合物Macromolecular complex | 552 | 42 | 208 | 361 | |
| GO:0031974 | 膜封闭的管腔Membrane-enclosed lumen | 160 | 11 | 40 | 112 | |
| GO:0005576 | 细胞外区域Extracellular region | 87 | 18 | 45 | 61 | |
| GO:0030054 | 细胞连接Cell junction | 15 | 4 | 6 | 5 | |
| GO:0055044 | 共质体Symplast | 14 | 4 | 5 | 5 | |
| GO:0019012 | 病毒Virion | 4 | 0 | 1 | 4 | |
| GO:0044423 | 病毒部分Virion part | 4 | 0 | 1 | 4 | |
| GO:0009295 | 内核Nucleoid | 11 | 0 | 5 | 6 | |
| GO:0044421 | 细胞外区域Extracellular region part | 28 | 3 | 21 | 26 | |
| 分子功能 | GO:0003824 | 催化活性Catalytic activity | 1 949 | 308 | 725 | 1 222 |
| Molecular | GO:0005488 | 联结Binding | 1 670 | 321 | 688 | 1 024 |
| function | GO:0005215 | 转运活动Transporter activity | 402 | 53 | 144 | 277 |
| GO:0005198 | 结构分子活动Structural molecule activity | 178 | 9 | 102 | 85 | |
| GO:0001071 | 核酸联结转录因子活性 Nucleic acid binding transcription factor activity | 109 | 40 | 64 | 68 | |
| GO:0016209 | 抗氧化活性Antioxidant activity | 40 | 10 | 21 | 21 | |
| GO:0009055 | 电子载体活动Electron carrier activity | 35 | 5 | 13 | 20 | |
| GO:0060089 | 分子传感器活动Molecular transducer activity | 36 | 5 | 7 | 8 | |
| GO:0004871 | 信号传导活动Signal transducer activity | 36 | 10 | 18 | 23 | |
| GO:0000988 | 转录因子活性,蛋白质结合 Transcription factor activity,protein binding | 16 | 4 | 3 | 12 | |
| GO:0045735 | 营养库活性Nutrient reservoir activity | 5 | 1 | 3 | 2 | |
| GO:0031386 | 蛋白质标签Protein tag | 5 | 0 | 0 | 2 | |
| GO:0016530 | 金属伴侣活性Metallochaperone activity | 1 | 1 | 1 | 1 | |
| GO:0045182 | 翻译调控活性Translation regulator activity | 2 | 0 | 0 | 2 | |
| COG 分类 COG classification | 分类名称 Classification name | -3 ℃ vs 25 ℃ | ||
|---|---|---|---|---|
| 花蕾 Flower bud | 花 Flower | 幼果 Young fruit | ||
| A | RNA 加工和修饰RNA processing and modification | 0 | 0 | 1 |
| B | 染色质结构与动态Chromatin structure and dynamics | 0 | 0 | 1 |
| C | 能量产生与转换Energy production and conversion | 41 | 81 | 132 |
| D | 细胞周期控制,细胞分裂,染色体分割 Cell cycle control,cell division,chromosome partitioning | 7 | 7 | 13 |
| E | 氨基酸转运与代谢Amino acid transport and metabolism | 27 | 72 | 184 |
| F | 核苷酸转运与代谢Nucleotide transport and metabolism | 7 | 19 | 56 |
| G | 碳水化合物转运与代谢Carbohydrate transport and metabolism | 120 | 150 | 274 |
| H | 辅酶转运与代谢Coenzyme transport and metabolism | 26 | 36 | 73 |
| I | 脂质转运与代谢Lipid transport and metabolism | 66 | 82 | 156 |
| J | 翻译,核糖体结构与生物合成Translation,ribosomal structure and biogenesis | 33 | 89 | 156 |
| K | 转录Transcription | 26 | 25 | 33 |
| L | 复制,重组与修复Replication,recombination and repair | 15 | 16 | 35 |
| M | 细胞壁/膜/包膜生物合成Cell wall/membrane/envelope biogenesis | 36 | 43 | 73 |
| N | 细胞运动Cell motility | 2 | 1 | 4 |
| O | 翻译后修饰,蛋白质折叠,分子伴侣 Posttranslational modification,protein turnover,chaperones | 59 | 71 | 141 |
| P | 无机离子转运与代谢Inorganic ion transport and metabolism | 46 | 80 | 122 |
| Q | 次生代谢物生物合成,转运与代谢 Secondary metabolites biosynthesis,transport and catabolism | 42 | 69 | 123 |
| R | 一般功能预测General function prediction only | 85 | 126 | 261 |
| S | 功能未知Function unknown | 29 | 40 | 48 |
| T | 信号转导机制Signal transduction mechanisms | 67 | 70 | 101 |
| U | 细胞内运输,分泌和囊泡运输 Intracellular trafficking,secretion,and vesicular transport | 0 | 1 | 8 |
| V | 防御机制Defense mechanisms | 10 | 22 | 47 |
| W | 细胞外结构Extracellular structures | 0 | 0 | 1 |
| X | 移动元件:原噬菌体,转座子Mobilome:prophages,transposons | 7 | 9 | 7 |
| Y | 细胞核结构Nuclear structure | 0 | 0 | 0 |
| Z | 细胞骨架Cytoskeleton | 2 | 3 | 2 |
Table 4 The number of DEGs in COG classification in low-temperature treatment and control comparison groups of three developmental stages of flowers and young fruits in loquat
| COG 分类 COG classification | 分类名称 Classification name | -3 ℃ vs 25 ℃ | ||
|---|---|---|---|---|
| 花蕾 Flower bud | 花 Flower | 幼果 Young fruit | ||
| A | RNA 加工和修饰RNA processing and modification | 0 | 0 | 1 |
| B | 染色质结构与动态Chromatin structure and dynamics | 0 | 0 | 1 |
| C | 能量产生与转换Energy production and conversion | 41 | 81 | 132 |
| D | 细胞周期控制,细胞分裂,染色体分割 Cell cycle control,cell division,chromosome partitioning | 7 | 7 | 13 |
| E | 氨基酸转运与代谢Amino acid transport and metabolism | 27 | 72 | 184 |
| F | 核苷酸转运与代谢Nucleotide transport and metabolism | 7 | 19 | 56 |
| G | 碳水化合物转运与代谢Carbohydrate transport and metabolism | 120 | 150 | 274 |
| H | 辅酶转运与代谢Coenzyme transport and metabolism | 26 | 36 | 73 |
| I | 脂质转运与代谢Lipid transport and metabolism | 66 | 82 | 156 |
| J | 翻译,核糖体结构与生物合成Translation,ribosomal structure and biogenesis | 33 | 89 | 156 |
| K | 转录Transcription | 26 | 25 | 33 |
| L | 复制,重组与修复Replication,recombination and repair | 15 | 16 | 35 |
| M | 细胞壁/膜/包膜生物合成Cell wall/membrane/envelope biogenesis | 36 | 43 | 73 |
| N | 细胞运动Cell motility | 2 | 1 | 4 |
| O | 翻译后修饰,蛋白质折叠,分子伴侣 Posttranslational modification,protein turnover,chaperones | 59 | 71 | 141 |
| P | 无机离子转运与代谢Inorganic ion transport and metabolism | 46 | 80 | 122 |
| Q | 次生代谢物生物合成,转运与代谢 Secondary metabolites biosynthesis,transport and catabolism | 42 | 69 | 123 |
| R | 一般功能预测General function prediction only | 85 | 126 | 261 |
| S | 功能未知Function unknown | 29 | 40 | 48 |
| T | 信号转导机制Signal transduction mechanisms | 67 | 70 | 101 |
| U | 细胞内运输,分泌和囊泡运输 Intracellular trafficking,secretion,and vesicular transport | 0 | 1 | 8 |
| V | 防御机制Defense mechanisms | 10 | 22 | 47 |
| W | 细胞外结构Extracellular structures | 0 | 0 | 1 |
| X | 移动元件:原噬菌体,转座子Mobilome:prophages,transposons | 7 | 9 | 7 |
| Y | 细胞核结构Nuclear structure | 0 | 0 | 0 |
| Z | 细胞骨架Cytoskeleton | 2 | 3 | 2 |

Fig. 4 Pathway assignment based on Kyoto Encyclopedia of Genes and Genomes(KEGG)in low-temperature treatment and control comparison group of flower bud in loquat

Fig. 3 Pathway assignment based on Kyoto Encyclopedia of Genes and Genomes(KEGG)in low-temperature treatment and control comparison group of flower in loquat

Fig. 4 Pathway assignment based on Kyoto Encyclopedia of Genes and Genomes(KEGG)in low-temperature treatment and control comparison group of young fruit in loquat
| Unigene 编号 Unigene ID | 转录组log2值 RNA-seq log2 FC | qRT-PCR相对表达量 qRT-PCR relative expression | 表达 Express | Nr注释 Nr_ annotation | 来源 Source | ||||
|---|---|---|---|---|---|---|---|---|---|
| 花蕾 Flower bud | 花 Flower | 幼果 Young fruit | 花蕾 Flower bud | 花 Flower | 幼果 Young fruit | ||||
| c106174.graph_c0 | 1.88 | 1.90 | 1.56 | 2.86 ± 0.32 | 3.45 ± 0.46 | 2.26 ± 0.34 | 上调Up | ERF37 | 枇杷Eriobotrya japonica |
| c123742.graph_c0 | 3.34 | 2.37 | 1.39 | 1.59 ± 0.23 | 4.78 ± 0.35 | 2.56 ± 0.21 | 上调Up | ERF73 | 苹果Malus × domestica |
| c125424.graph_c0 | 3.29 | 3.67 | 3.91 | 6.57 ± 0.58 | 4.23 ± 0.52 | 3.12 ± 0.29 | 上调Up | ERF35 | 枇杷Eriobotrya japonica |
| c126935.graph_c1 | 2.85 | 2.16 | 1.42 | 1.23 ± 0.15 | 1.85 ± 0.20 | 1.36 ± 0.16 | 上调Up | ERF105 | 苹果Malus × domestica |
| c129543.graph_c0 | 2.75 | 2.63 | 1.57 | 1.28 ± 0.12 | 2.35 ± 0.24 | 3.42 ± 0.21 | 上调Up | RAV2 | 枇杷Eriobotrya japonica |
| c46214.graph_c0 | 1.55 | 3.52 | 4.65 | 0.79 ± 0.12 | 5.48 ± 0.35 | 2.59 ± 0.38 | 上调Up | ERF60 | 苹果Malus × domestica |
| c126492.graph_c0 | -1.90 | -2.04 | -2.54 | -1.02 ± 0.11 | -3.05± 0.36 | -2.13± 0.22 | 下调Down | ERF33 | 苹果Malus × domestica |
| c142932.graph_c0 | 2.13 | 2.87 | 1.42 | 1.36 ± 0.21 | 1.25 ± 0.15 | 1.49 ± 0.23 | 上调Up | WRKY33 | 苹果Malus × domestica |
| c51471.graph_c0 | 4.92 | 4.59 | 4.00 | 6.89 ± 0.71 | 7.68 ± 0.68 | 6.21 ± 0.52 | 上调Up | NAC 72 | 苹果Malus × domestica |
| c130635.graph_c | 1.92 | 2.95 | 1.78 | 2.06 ± 0.16 | 3.46 ± 0.42 | 2.76 ± 0.31 | 上调Up | NAC74 | 梨Pyrus bretschneideri |
| c136256.graph_c0 | 3.65 | 2.69 | 2.57 | 4.69 ± 0.53 | 3.76 ± 0.47 | 2.63 ± 0.18 | 上调Up | NAC29 | 苹果Malus × domestica |
| c136368.graph_c0 | 1.22 | 1.01 | 1.00 | 0.69 ± 0.10 | 1.38 ± 0.20 | 1.54 ± 0.23 | 上调Up | NAC91 | 苹果Malus × domestica |
Table 6 Relative expression of partial low temperature responding transcription factors in loquat
| Unigene 编号 Unigene ID | 转录组log2值 RNA-seq log2 FC | qRT-PCR相对表达量 qRT-PCR relative expression | 表达 Express | Nr注释 Nr_ annotation | 来源 Source | ||||
|---|---|---|---|---|---|---|---|---|---|
| 花蕾 Flower bud | 花 Flower | 幼果 Young fruit | 花蕾 Flower bud | 花 Flower | 幼果 Young fruit | ||||
| c106174.graph_c0 | 1.88 | 1.90 | 1.56 | 2.86 ± 0.32 | 3.45 ± 0.46 | 2.26 ± 0.34 | 上调Up | ERF37 | 枇杷Eriobotrya japonica |
| c123742.graph_c0 | 3.34 | 2.37 | 1.39 | 1.59 ± 0.23 | 4.78 ± 0.35 | 2.56 ± 0.21 | 上调Up | ERF73 | 苹果Malus × domestica |
| c125424.graph_c0 | 3.29 | 3.67 | 3.91 | 6.57 ± 0.58 | 4.23 ± 0.52 | 3.12 ± 0.29 | 上调Up | ERF35 | 枇杷Eriobotrya japonica |
| c126935.graph_c1 | 2.85 | 2.16 | 1.42 | 1.23 ± 0.15 | 1.85 ± 0.20 | 1.36 ± 0.16 | 上调Up | ERF105 | 苹果Malus × domestica |
| c129543.graph_c0 | 2.75 | 2.63 | 1.57 | 1.28 ± 0.12 | 2.35 ± 0.24 | 3.42 ± 0.21 | 上调Up | RAV2 | 枇杷Eriobotrya japonica |
| c46214.graph_c0 | 1.55 | 3.52 | 4.65 | 0.79 ± 0.12 | 5.48 ± 0.35 | 2.59 ± 0.38 | 上调Up | ERF60 | 苹果Malus × domestica |
| c126492.graph_c0 | -1.90 | -2.04 | -2.54 | -1.02 ± 0.11 | -3.05± 0.36 | -2.13± 0.22 | 下调Down | ERF33 | 苹果Malus × domestica |
| c142932.graph_c0 | 2.13 | 2.87 | 1.42 | 1.36 ± 0.21 | 1.25 ± 0.15 | 1.49 ± 0.23 | 上调Up | WRKY33 | 苹果Malus × domestica |
| c51471.graph_c0 | 4.92 | 4.59 | 4.00 | 6.89 ± 0.71 | 7.68 ± 0.68 | 6.21 ± 0.52 | 上调Up | NAC 72 | 苹果Malus × domestica |
| c130635.graph_c | 1.92 | 2.95 | 1.78 | 2.06 ± 0.16 | 3.46 ± 0.42 | 2.76 ± 0.31 | 上调Up | NAC74 | 梨Pyrus bretschneideri |
| c136256.graph_c0 | 3.65 | 2.69 | 2.57 | 4.69 ± 0.53 | 3.76 ± 0.47 | 2.63 ± 0.18 | 上调Up | NAC29 | 苹果Malus × domestica |
| c136368.graph_c0 | 1.22 | 1.01 | 1.00 | 0.69 ± 0.10 | 1.38 ± 0.20 | 1.54 ± 0.23 | 上调Up | NAC91 | 苹果Malus × domestica |
| [1] |
An D, Yang J, Zhang P. 2012. Transcriptome profiling of low temperature treated cassava apical shoots showed dynamic responses of tropical plant to cold stress. BMC Genomics, 13:64.
doi: 10.1186/1471-2164-13-64 URL |
| [2] |
Anders S, Huber W. 2010. Differential expression analysis for sequence count data. Genome Biology, 11:R106.
doi: 10.1186/gb-2010-11-10-r106 URL |
| [3] |
Bates L S, Waldren R P, Teare I D. 1973. Rapid determination of free proline for water-stress studies. Plant Soil, 39:205-207.
doi: 10.1007/BF00018060 URL |
| [4] | Beauchamp C O, Fridovich I. 1981. Superoxide dismutase:improved assays and an assay applicable to acrylamide gels. Anal Biochem, 44:867-880. |
| [5] | Berg J M, Tymoczko J L, Stryer L. 2010. Glycolysis and gluconeogenesis. Biochemistry,7th edn. New York:WH Freeman. |
| [6] |
Bocian A, Zwierzykowski Z, Rapacz M, Koczyk G, Ciesiołka D, Kosmala A. 2015. Metabolite profiling during cold acclimation of Lolium perenne genotypes distinct in the level of frost tolerance. J Appl Genet, 56:439-449.
doi: 10.1007/s13353-015-0293-6 URL |
| [7] |
Bradford M M. 1976. A rapid and sensitive method for the quantitation of microgram quantities of protein utilizing the principle of protein-dye binding. Anal Biochem, 72:248-254.
pmid: 942051 |
| [8] |
Chen J H, Tian Q A, Pang T, Jiang L B, Wu R L, Xia X L, Yin W L. 2014. Deep-sequencing transcriptome analysis of low temperature perception in a desert tree,Populus euphratica. BMC Genomics, 15(1):326.
doi: 10.1186/1471-2164-15-326 URL |
| [9] | Chen Jun-wei, Sun Jun, Li Xiao-ying, Xu Hong-xia, Zhou Xiao-yin, Jiang Lu-hua. 2017. Study on later flowering cultivation techniques to avoid freezing in white loquat. Journal of Zhejiang Agricultural Sciences, 58(3):417-419,425. (in Chinese) |
| 陈俊伟, 孙钧, 李晓颖, 徐红霞, 周晓音, 姜路花. 2017. 白肉枇杷晚花避冻栽培技术探讨. 浙江农业科学, 58(3):417-419,425. | |
| [10] |
Chen X, Wang Y, Lv B, Li J, Luo L, Lu S, Zhang X, Ma H, Ming F. 2014. The NAC family transcription factor OsNAP confers abiotic stress response through the ABA pathway. Plant Cell Physiol, 55:604-619.
doi: 10.1093/pcp/pct204 URL pmid: 24399239 |
| [11] |
Cheng H Y, Song S Q. 2008. Possible involvement of reactive oxygen species scavenging enzymes in desiccation sensitivity of Antiaris toxicaria seeds and axes. J Integr Plant Biol, 50:1549-1556.
doi: 10.1111/jipb.2008.50.issue-12 URL |
| [12] | Cong Qing, Ni Xiao-xiang, Cheng Long-jun. 2020. Ectopic express of EgrNAC1 enhances cold tolerance and sensitivity to drought and salt in Arabidopsis thaliana. Journal of Nuclear Agriculture Sciences, 34(7):567-575. (in Chinese) |
| 从青, 倪晓祥, 程龙军. 2020. 异源表达EgrNAC1提高拟南芥抗寒性和对干旱、高盐的敏感性. 核农学报, 34(7):567-575. | |
| [13] | Degenkolbe T, Giavalisco P, Zuther E, Seiwert B, Hincha D K, Willmitzer L. 2012. Differential remodeling of the lipidome during cold acclimation in natural accessions of Arabidopsis thaliana. Plant J, 32:972-982. |
| [14] |
Dhindsa R S, Plumb-Dhindsa P, Thorpe T A. 1981. Leaf senescence:correlated with increased levels of membrane-permeability and lipid peroxidation,and decreased levels of superoxide dismutase and catalase. J Exp Bot, 32:93-101.
doi: 10.1093/jxb/32.1.93 URL |
| [15] | Du Wen-li, Chen Zhong-shan, Xu Rui-xiang, Xu Tong-wei, Gao Shan, Wen Qing-fang. 2021. Physiological response and differentially expressed genes analysis of transcriptome in Momordica charantia L. leaf under cold stress. Journal of Nuclear Agriculture Sciences, 35(2):338-348. (in Chinese) |
| 杜文丽, 陈中钐, 许端祥, 徐同伟, 高山, 温庆放. 2021. 低温胁迫下苦瓜叶片转录组差异基因分析及生理响应特征. 核农学报, 35(2):338-348. | |
| [16] |
Elstner E F, Heupel A. 1976. Inhibition of nitrite formation from hydroxylammonium chloride:a simple assay for superoxide dismutase. Anal Biochem, 70:616-620.
pmid: 817618 |
| [17] | Fujikawa S, Kasuga J, Takata N, Arakawa K. 2009. Factors related to change of deep supercooling capability in xylem parenchyma cells of tress// Gusta L,Wisnieswski M,Tanino K K. Plant cold hardiness from the laboratory to the field. Wallingford: CABI Press. |
| [18] |
Hamilton C A, Good A G, Taylor G J. 2001. Induction of vacuolar ATPase and mitochondrial ATP synthase by aluminum in an aluminum-resistant cultivar of wheat. Plant Physiol, 125:2068-2077.
pmid: 11299386 |
| [19] |
Kawamura Y, Uemura M. 2003. Mass spectrometric approach for identifying putative plasma membrane proteins of Arabidopsis leaves associated with cold acclimation. Plant J, 36:141-154.
pmid: 14535880 |
| [20] |
Kim C Y, Vo K, Nguyen C D, Jeong D H, Lee S K, Kumar M, Kim S R, Park S H, Kim J J, Jeon J S. 2016. Functional analysis of a cold-responsive rice WRKY gene,OsWRKY71. Plant Biotechnol Rep, 10(1):13-23.
doi: 10.1007/s11816-015-0383-2 URL |
| [21] |
Klotke J, Kopka J, Gatzke N, Heyer A. 2004. Impact of soluble sugar concentrations on the acquisition of freezing tolerance in accessions of Arabidopsis thaliana with contrasting cold adaptation-evidence for a role of raffinose in cold acclimation. Plant Cell Environ, 27:1395-1404
doi: 10.1111/pce.2004.27.issue-11 URL |
| [22] | Klowden M J. 2007. Physiological systems in insects. Burlington: Academic Press. |
| [23] |
Kovi M R, Ergon A, Rognli O A. 2016. Freezing tolerance revisited effects of variable temperatures on gene regulation in temperate grasses and legumes. Curr Opin Plant Biol, 33:140-146.
doi: 10.1016/j.pbi.2016.07.006 URL |
| [24] |
Lei X, Xiao Y, Xia W, Mason A S, Yang Y, Ma Z, Peng M. 2014. RNA-seq analysis of oil palm under cold stress reveals a different C-repeat binding factor(CBF)mediated gene expression pattern in Elaeis guineensis compared to other species. PLoS ONE, 9:e114482.
doi: 10.1371/journal.pone.0114482 URL |
| [25] | Liu Jiao-jiao, Wang Xue-min, Ma Lin, Cui Miao-miao, Cao Xiao-yu, Zhao Wei. 2020. Isolation,identification,and response to abiotic stress of MsWRKY42 gene from Medicago sativa L. Scientia Agricultura Sinica, 53(17):3455-3466. (in Chinese) |
| 刘佼佼, 王学敏, 马琳, 崔苗苗, 曹晓宇, 赵威. 2020. 紫花苜蓿MsWRKY42的分离、鉴定及其对非生物胁迫的响应. 中国农业科学, 53(17):3455-3466. | |
| [26] |
Liu J Y, Men J L, Chang M C, Feng C P, Yuan L G. 2017. iTRAQ based quantitative proteome revealed metabolic changes of Flammulina velutipes mycelia in response to cold stress. J Proteom, 156:75-84.
doi: 10.1016/j.jprot.2017.01.009 URL |
| [27] |
Liu Q, Kasuga M, Sakuma Y, Abe H, Miura S, Yamaguchi-Shinozaki K, Shinozaki K. 1998. Two transcription factors,DREB1 and DREB2,with an EREBP/AP2 DNA binding domain separate two cellular signal transduction pathways in drought- and low-temperature-responsive gene expression,respectively,in Arabidopsis. Plant Cell, 10:1391-1406.
pmid: 9707537 |
| [28] |
Lv J H, Liu Z B, Liu Y H, Ou L J, Deng M H, Wang J, Song J S, Ma Y Q, Chen W C, Zhang Z Q, Dai X Z, Zou X X. 2020. Comparative transcriptome analysis between cytoplasmic male sterile line and its maintainer during the floral bud development of pepper. Horticultural Plant Journal, 6(2):89-98.
doi: 10.1016/j.hpj.2020.01.004 URL |
| [29] |
Mittler R. 2002. Oxidative stress,antioxidants and stress tolerance. Trends Plant Sci, 7:405-410.
pmid: 12234732 |
| [30] |
Moellering E R, Muthan B, Benning C. 2010. Freezing tolerance in plants requires lipid remodeling at the outer chloroplast membrane. Science, 330:226-228.
doi: 10.1126/science.1191803 pmid: 20798281 |
| [31] | Nakano Y, Asada K. 1981. Hydrogen peroxide is scavenged by ascorbate specific peroxidase in spinach chloroplasts. Plant Cell Physiol, 22:867-880. |
| [32] | Nakashima K, Takasaki H, Mizoi J, Shinozaki K, Yamaguchi- Shinozaki. 2012. NAC transcription factors in plant abiotic stress responses. BBA Gene Regul Mech, 1819:97-103. |
| [33] |
Nakashima K, Tran L S, Van Nguyen D, Fujita M, Maruyama K, Todaka D, Ito Y, Hayashi N, Shinozaki K, Yamaguchi-Shinozaki K. 2007. Functional analysis of a NAC-type transcription factor OsNAC6 involved in abiotic and biotic stress-responsive gene expression in rice. Plant J, 51:617-630.
pmid: 17587305 |
| [34] |
Qu Y T, Mei D, Zhang Z Q, Dong J L, Tao W. 2016. Overexpression of the Medicago falcata NAC transcription factor MfNAC3 enhances cold tolerance in Medicago truncatula. Environ Exp Bot, 129:67-76.
doi: 10.1016/j.envexpbot.2015.12.012 URL |
| [35] |
Takasaki H, Maruyama K, Kidokoro S, Ito Y, Fujita Y, Shinozaki K, Yamaguchi-Shinozaki K, Nakashima K. 2010. The abiotic stress-responsive NAC-type transcription factor OsNAC5 regulates stress-inducible genes and stress tolerance in rice. Mol Genet Genom, 284:173-183.
doi: 10.1007/s00438-010-0557-0 URL |
| [36] |
Trapnell C, Williams B A, Pertea G, Mortazavi A, Kwan G, Baren M J, Salzberg S L, Wold B J, Pachter L. 2010. Transcript assembly and quantification by RNA-Seq reveals unannotated transcripts and isoform switching during cell differentiation. Nat Biotech, 28(5):511-515.
doi: 10.1038/nbt.1621 URL |
| [37] |
Wang Z Q, Xu X Y, Gong Q Q, Xie C, Fan W, Yang J L, Lin Q S, Zheng S J. 2014. Root proteome of rice studied by iTRAQ provides integrated insight into aluminum stress tolerance mechanisms in plants. J Proteom, 98:189-205.
doi: 10.1016/j.jprot.2013.12.023 URL |
| [38] |
Xu H X, Li X Y, Chen J W. 2017. Comparative transcriptome profiling of freezing stress responses in loquat(Eriobotrya japonica) fruitlets. J Plant Res, 130:893-907.
doi: 10.1007/s10265-017-0942-4 URL |
| [39] | Zhang F, Wang Y Q, Zhu K, Zhang Z P, Zhu Z X, Lu F, Zou J Q. 2019. Comparative transcriptome analysis of different salt tolerance sorghum (Sorghum bicolor L. Moench)under salt stress. Scientia Agricultura Sinica, 52(22):4002-4015. (in Chinese) |
| 张飞, 王艳秋, 朱凯, 张志鹏, 朱振兴, 卢峰, 邹剑秋. 2019. 不同耐盐性高粱在盐逆境下的比较转录组分析. 中国农业科学, 52(22):4002-4015. | |
| [40] |
Zhang Y, Dong W, Zhao X H, Song A, Guo K W, Liu Z J, Zhang L S. 2019. Transcriptomic analysis of differentially expressed genes and alternative splicing events associated with crassulacean acid metabolism in orchids. Horticultural Plant Journal, 5(6):268-280.
doi: 10.1016/j.hpj.2019.12.001 |
| [41] |
Zheng X, Chen B, Lu G, Han B. 2009. Overexpression of a NAC transcription factor enhances rice drought and salt tolerance. Biochem Biophys Res Commun, 379:985-999.
doi: 10.1016/j.bbrc.2008.12.163 URL |
| [42] |
Zou C, Jiang W, Yu D. 2010. Male gametophyte-specific WRKY34 transcription factor mediates cold sensitivity of mature pollen in Arabidopsis. J Exp Bot, 61(14):3901-3914.
doi: 10.1093/jxb/erq204 URL |
| [43] | Zou C, Yu J. 2010. Analysis of the cold-responsive transcriptome in the mature pollen of Arabidopsis. Plant Biol, 53:400-416. |
| [1] | YE Zimao, SHEN Wanxia, LIU Mengyu, WANG Tong, ZHANG Xiaonan, YU Xin, LIU Xiaofeng, and ZHAO Xiaochun, . Effect of R2R3-MYB Transcription Factor CitMYB21 on Flavonoids Biosynthesis in Citrus [J]. Acta Horticulturae Sinica, 2023, 50(2): 250-264. |
| [2] | SONG Yanhong, CHEN Yaduo, ZHANG Xiaoyu, SONG Pan, LIU Lifeng, LI Gang, ZHAO Xia, and ZHOU Houcheng, . The Transcription Factor FvbHLH130 Activates Flowering in Fragaria vesca [J]. Acta Horticulturae Sinica, 2023, 50(2): 295-306. |
| [3] | REN Hailong, XU Donglin, ZHANG Jing, ZOU Jiwen, LI Guangguang, ZHOU Xianyu, XIAO Wanyu, and SUN Yijia. Establishment of SNP Fingerprinting and Identification of Chinese Flowering Cabbage Varieties Based on KASP Genotyping [J]. Acta Horticulturae Sinica, 2023, 50(2): 307-318. |
| [4] | HAN Rui, ZHONG Xionghui, CHEN Denghui, CUI Jian, YUE Xiangqing, XIE Jianming, and KANG Jungen, . Cloning and Functional Analysis of BobHLH34 Gene in Cabbage that Interacts with XopR from Xanthomonas [J]. Acta Horticulturae Sinica, 2023, 50(2): 319-330. |
| [5] | LI Ruiya, SONG Chengwei, NIU Tongfei, WEI Zhenzhen, GUO Lili, and HOU Xiaogai. The Emitted Pattern Analysis of Flower Volatiles and Cloning of PsGDS Gene in Tree Peony Cultivar‘High Noon’ [J]. Acta Horticulturae Sinica, 2023, 50(2): 331-344. |
| [6] | ZHANG Ailing, TU Hongyan, XIAO Wang, ZHONG Xiaoqing, LU Qiuchan, CHENG Liping, LIN Xiaoping, and MAI Yuling. Morphology and Microstructure Characteristics of Diploid and Tetraploid Hedychium coronarium Cut Flowers [J]. Acta Horticulturae Sinica, 2023, 50(2): 345-358. |
| [7] | JIANG Yu, TU Xunliang, and HE Junrong. Analysis of Differential Expression Genes in Leaves of Leaf Color Mutant of Chinese Orchid [J]. Acta Horticulturae Sinica, 2023, 50(2): 371-381. |
| [8] | TIAN Mingkang, XU Zhixiang, LIU Xiuqun, SUI Shunzhao, LI Mingyang, and LI Zhineng, . Identification of the AP2 Subfamily Transcription Factors in Chimonanthus praecox and the Functional Study of CpAP2-L11 [J]. Acta Horticulturae Sinica, 2023, 50(2): 382-396. |
| [9] | WANG Mengmeng, SUN Deling, CHEN Rui, YANG Yingxia, ZHANG Guan, LÜ Mingjie, WANG Qian, XIE Tianyu, NIU Guobao, SHAN Xiaozheng, TAN Jin, and YAO Xingwei, . Construction and Evaluation of Cauliflower Core Collection [J]. Acta Horticulturae Sinica, 2023, 50(2): 421-431. |
| [10] | YANG Zhi, ZHANG Chuanjiang, YANG Xinfang, DONG Mengyi, WANG Zhenlei, YAN Fenfen, WU Cuiyun, WANG Jiurui, LIU Mengjun, LIN Minjuan. Analysis of Fruit Genetic Tendency and Mixed Inheritance in Hybrid Progeny of Jujube and Wild Jujube [J]. Acta Horticulturae Sinica, 2023, 50(1): 36-52. |
| [11] | YUAN Xin, XU Yunhe, ZHANG Yupei, SHAN Nan, CHEN Chuying, WAN Chunpeng, KAI Wenbin, ZHAI Xiawan, CHEN Jinyin, GAN Zengyu. Studies on AcAREB1 Regulating the Expression of AcGH3.1 During Postharvest Ripening of Kiwifruit [J]. Acta Horticulturae Sinica, 2023, 50(1): 53-64. |
| [12] | LIN Haijiao, LIANG Yuchen, LI Ling, MA Jun, ZHANG Lu, LAN Zhenying, YUAN Zening. Exploration and Regulation Network Analysis of CBF Pathway Related Cold Tolerance Genes in Lavandula angustifolia [J]. Acta Horticulturae Sinica, 2023, 50(1): 131-144. |
| [13] | ZHAO Xueyan, WANG Qi, WANG Li, WANG Fangyuan, WANG Qing, LI Yan. Comparative Transcriptome Analysis of Differential Expression in Different Tissues of Corydalis yanhusuo [J]. Acta Horticulturae Sinica, 2023, 50(1): 177-187. |
| [14] | SHAO Fengqing, LUO Xiurong, WANG Qi, ZHANG Xianzhi, WANG Wencai. Advances in Research of DNA Methylation Regulation During Fruit Ripening [J]. Acta Horticulturae Sinica, 2023, 50(1): 197-208. |
| [15] | WANG Yingying, LIU Lichang, LIU Zhiwu, YANG Xingwang, LIU Wanchun, and WANG Xiaodi, . A New Little Nectarine Cultivar‘Zhongnong Zhenzhu’ [J]. Acta Horticulturae Sinica, 2022, 49(S2): 25-26. |
| Viewed | ||||||
|
Full text |
|
|||||
|
Abstract |
|
|||||
Copyright © 2012 Acta Horticulturae Sinica 京ICP备10030308号-2 国际联网备案号 11010802023439
Tel: 010-82109523 E-Mail: yuanyixuebao@126.com
Support by: Beijing Magtech Co.Ltd