
园艺学报 ›› 2023, Vol. 50 ›› Issue (10): 2242-2256.doi: 10.16420/j.issn.0513-353x.2022-0682
史敬芳1,3, 胡春华1, 李昊宸1, 杨乔松1,4, 盛鸥1, 毕方铖1,2, 董涛1, 李春雨1, 邓贵明1, 高慧君1, 何维弟1, 刘思文1, 易干军1,*( ), 窦同心1,*(
), 窦同心1,*( )
)
收稿日期:2023-03-21
修回日期:2023-08-13
出版日期:2023-10-25
发布日期:2023-10-31
通讯作者:
*(E-mail:doutongxin@gdaas.cn,yiganjun@vip.163.com)
基金资助:
SHI Jingfang1,3, HU Chunhua1, LI Haochen1, YANG Qiaosong1,4, SHENG Ou1, BI Fangcheng1,2, DONG Tao1, LI Chunyu1, DENG Guiming1, GAO Huijun1, HE Weidi1, LIU Siwen1, YI Ganjun1,*( ), DOU Tongxin1,*(
), DOU Tongxin1,*( )
)
Received:2023-03-21
Revised:2023-08-13
Published:2023-10-25
Online:2023-10-31
Contact:
*(E-mail:doutongxin@gdaas.cn,yiganjun@vip.163.com)
摘要:
前期研究发现在香蕉中超表达大蕉(Musa spp.‘Lingchuan Dajiao’)MpICE1能显著提高香蕉抗枯萎病能力,为进一步揭示其分子机理,通过转录组对接种Foc TR4前0 d和接种后7、14 d的野生型和超表达MpICE1香蕉(Musa spp.‘Brazil’)的根部中差异表达基因进行分析。结果表明,在接种前超表达MpICE1香蕉可诱导一系列基因的高表达,且这些高表达的基因富集在苯丙烷合成、黄酮合成、ABC转运蛋白、MAPK信号传导和戊糖、葡萄糖醛酸转换等途径中,与细胞壁结构和功能相关。MpICE1的超表达可能会增强香蕉的基础免疫反应,从而对Foc TR4的抗性增强。另外,接种Foc TR4后,超表达MpICE1香蕉接种前0 d高表达的大部分基因表达量开始下降,到7 d时达到相对稳定的状态。对差异表达基因进一步分析发现,编码12-氧-植物二烯酸还原酶的基因(Ma06_g18840)可能是MpICE1的靶基因,且其表达量受MpICE1的抑制,与氧化脂质(JA和OPDA)代谢相关,可能通过JA或OPDA信号途径调节根部细胞壁结构。泛素碳端水解酶(Ubiquitin carboxyl-terminal hydrolase 36/42)也受MpICE1的影响,但具体的作用机制需进一步研究。
史敬芳, 胡春华, 李昊宸, 杨乔松, 盛鸥, 毕方铖, 董涛, 李春雨, 邓贵明, 高慧君, 何维弟, 刘思文, 易干军, 窦同心. 转录组分析超表达MpICE1香蕉抗枯萎病机理[J]. 园艺学报, 2023, 50(10): 2242-2256.
SHI Jingfang, HU Chunhua, LI Haochen, YANG Qiaosong, SHENG Ou, BI Fangcheng, DONG Tao, LI Chunyu, DENG Guiming, GAO Huijun, HE Weidi, LIU Siwen, YI Ganjun, DOU Tongxin. Molecular Mechanism of MpICE1 Overexpressing Banana Resistant to Fusarium oxysporum Based on Transcriptome Analysis[J]. Acta Horticulturae Sinica, 2023, 50(10): 2242-2256.
| 基因 Gene | 引物(5′-3′) Primer |
|---|---|
| Ma06_g18840 | F:TATGGCAATGTCCCCCAACC;R:GACCAGTGTCTGAGACGCAA |
| Ma06_g16670 | F:ATCAACGTCGAGATCGGCAA;R:GGCTGAGGCTGATGGGTA |
| Ma11_g19450 | F:ATGCTCTCGGGGATCAATGG;R:TCATGACACTGTATTATCG |
| Ma04_g40060 | F:ATGGCGTCCAAGACCACAAG;R:GCTCTTTTCCTCTTGTTCAAC |
| Ma10_g15940 | F:GTCCTCACCAGCGCCTGCGG;R:CCCCGAGCCCTGCCCCCTCCG |
| Ma10_g12250 | F:TGGGTTTGCCATGGATGTTT;R:AGAACACCAGGGCCTTCCTT |
| Ma08_g05750 | F:AGCGGAGAATGTCATAGGTG;R:TGGATGCTCTGTCCATCTTG |
| Ma09_g10470 | F:ACAGTCGTCAGGAGTCGTGCTT;R:CGCTATCGATTCATGACACTGTA |
表1 qRT-PCR验证基因及引物信息
Table 1 Validation gene and primer information for qRT-PCR
| 基因 Gene | 引物(5′-3′) Primer |
|---|---|
| Ma06_g18840 | F:TATGGCAATGTCCCCCAACC;R:GACCAGTGTCTGAGACGCAA |
| Ma06_g16670 | F:ATCAACGTCGAGATCGGCAA;R:GGCTGAGGCTGATGGGTA |
| Ma11_g19450 | F:ATGCTCTCGGGGATCAATGG;R:TCATGACACTGTATTATCG |
| Ma04_g40060 | F:ATGGCGTCCAAGACCACAAG;R:GCTCTTTTCCTCTTGTTCAAC |
| Ma10_g15940 | F:GTCCTCACCAGCGCCTGCGG;R:CCCCGAGCCCTGCCCCCTCCG |
| Ma10_g12250 | F:TGGGTTTGCCATGGATGTTT;R:AGAACACCAGGGCCTTCCTT |
| Ma08_g05750 | F:AGCGGAGAATGTCATAGGTG;R:TGGATGCTCTGTCCATCTTG |
| Ma09_g10470 | F:ACAGTCGTCAGGAGTCGTGCTT;R:CGCTATCGATTCATGACACTGTA |
| 样本 Sample | 原始序列/Mb Raw reads | 过滤后序列/Mb Clean reads | Q20/% | Q30/% | 序列比值 Clean reads ratio/% | 总基因比对率 Total mapping/% |
|---|---|---|---|---|---|---|
| WT0d A | 43.82 | 42.72 | 97.26 | 93.03 | 97.49 | 87.00 |
| WT0d B | 43.82 | 42.84 | 97.22 | 92.84 | 97.76 | 86.83 |
| WT0d C | 42.61 | 41.87 | 96.76 | 91.99 | 98.26 | 86.20 |
| WT7d A | 43.82 | 43.29 | 97.12 | 92.54 | 98.80 | 88.15 |
| WT7d B | 43.82 | 43.38 | 97.03 | 92.41 | 98.99 | 87.82 |
| WT7d C | 43.82 | 42.56 | 97.15 | 92.71 | 97.12 | 87.81 |
| WT14d A | 43.82 | 42.61 | 97.19 | 92.84 | 97.23 | 87.19 |
| WT14d B | 43.82 | 43.05 | 97.22 | 92.91 | 98.25 | 87.56 |
| WT14d C | 43.82 | 42.87 | 97.38 | 93.25 | 97.84 | 87.58 |
| OE0d A | 43.82 | 42.64 | 97.17 | 92.88 | 97.32 | 87.11 |
| OE0d B | 43.82 | 43.22 | 97.12 | 92.67 | 98.62 | 87.54 |
| OE0d C | 43.82 | 43.26 | 96.99 | 92.31 | 98.72 | 86.53 |
| OE7d A | 43.82 | 43.20 | 97.19 | 92.76 | 98.59 | 87.44 |
| OE7d B | 43.82 | 42.95 | 97.31 | 93.15 | 98.02 | 87.17 |
| OE7d C | 43.82 | 42.83 | 97.02 | 92.45 | 97.74 | 87.59 |
| OE14d A | 43.82 | 42.94 | 96.97 | 92.27 | 97.99 | 87.17 |
| OE14d B | 43.82 | 42.94 | 97.00 | 92.41 | 97.98 | 87.41 |
| OE14d C | 43.82 | 43.09 | 96.99 | 92.32 | 98.33 | 86.77 |
表2 香蕉野生型(WT)和超表达MpICE1(OE)根接种枯萎病菌(0、7、14 d)的RNA-seq数据统计
Table 2 Summary of RNA-seq data from roots of wild(WT)and ICE1 overexpress(OE)banana before and after inoculation(0,7 and 14 d)
| 样本 Sample | 原始序列/Mb Raw reads | 过滤后序列/Mb Clean reads | Q20/% | Q30/% | 序列比值 Clean reads ratio/% | 总基因比对率 Total mapping/% |
|---|---|---|---|---|---|---|
| WT0d A | 43.82 | 42.72 | 97.26 | 93.03 | 97.49 | 87.00 |
| WT0d B | 43.82 | 42.84 | 97.22 | 92.84 | 97.76 | 86.83 |
| WT0d C | 42.61 | 41.87 | 96.76 | 91.99 | 98.26 | 86.20 |
| WT7d A | 43.82 | 43.29 | 97.12 | 92.54 | 98.80 | 88.15 |
| WT7d B | 43.82 | 43.38 | 97.03 | 92.41 | 98.99 | 87.82 |
| WT7d C | 43.82 | 42.56 | 97.15 | 92.71 | 97.12 | 87.81 |
| WT14d A | 43.82 | 42.61 | 97.19 | 92.84 | 97.23 | 87.19 |
| WT14d B | 43.82 | 43.05 | 97.22 | 92.91 | 98.25 | 87.56 |
| WT14d C | 43.82 | 42.87 | 97.38 | 93.25 | 97.84 | 87.58 |
| OE0d A | 43.82 | 42.64 | 97.17 | 92.88 | 97.32 | 87.11 |
| OE0d B | 43.82 | 43.22 | 97.12 | 92.67 | 98.62 | 87.54 |
| OE0d C | 43.82 | 43.26 | 96.99 | 92.31 | 98.72 | 86.53 |
| OE7d A | 43.82 | 43.20 | 97.19 | 92.76 | 98.59 | 87.44 |
| OE7d B | 43.82 | 42.95 | 97.31 | 93.15 | 98.02 | 87.17 |
| OE7d C | 43.82 | 42.83 | 97.02 | 92.45 | 97.74 | 87.59 |
| OE14d A | 43.82 | 42.94 | 96.97 | 92.27 | 97.99 | 87.17 |
| OE14d B | 43.82 | 42.94 | 97.00 | 92.41 | 97.98 | 87.41 |
| OE14d C | 43.82 | 43.09 | 96.99 | 92.32 | 98.33 | 86.77 |
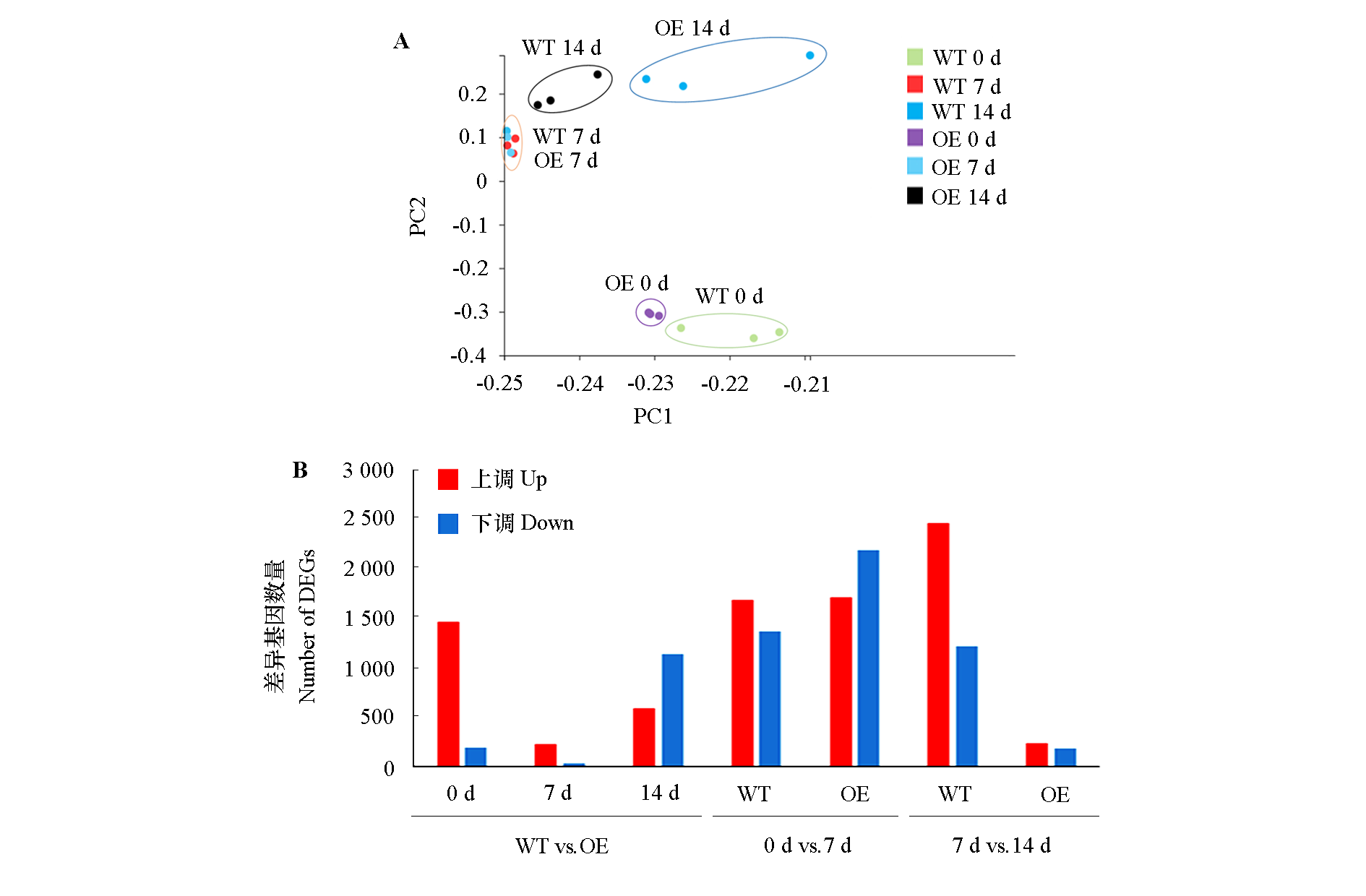
图1 野生型(WT)和超表达MpICE1(OE)香蕉根接种枯萎病菌(0、7、14 d)的PCA得分图(A)和差异表达基因数统计(B)
Fig. 1 PCA results(A)and statistics on number of different express genes(B)of WT and overexpress MpICE1(OE)banana root before and after inoculation(0,7 and 14 d)
| 比较组 Pair comparison | GO功能 GO term | 基因数量 Gene number | 富集率 Rich ratio | Q值 Q value |
|---|---|---|---|---|
| WT 0 d vs. OE 0 d | 细胞外区域Extracellular region | 56 | 0.19 | 6.84E-19 |
| 细胞壁组织或生物发生Cell wall organization or biogenesis | 39 | 0.16 | 6.10E-10 | |
| 细胞壁组织Cell wall organization | 34 | 0.16 | 2.23E-08 | |
| 表面封装结构组织External encapsulating structure organization | 34 | 0.15 | 3.20E-08 | |
| 细胞壁Cell wall | 27 | 0.18 | 3.66E-08 | |
| 表面封装结构External encapsulating structure | 27 | 0.18 | 3.66E-08 | |
| 糖基水解酶活性Hydrolase activity,acting on glycosyl bonds | 41 | 0.15 | 3.98E-08 | |
| O-糖基水解酶活性 Hydrolase activity,hydrolyzing O-glycosyl compounds | 39 | 0.15 | 4.60E-08 | |
| 细胞碳水化合物代谢过程Cellular carbohydrate metabolic process | 28 | 0.17 | 1.20E-07 | |
| 碳水化合物代谢过程Carbohydrate metabolic process | 59 | 0.10 | 1.44E-07 | |
| WT 7 d vs. OE 7 d | 细胞外区域Extracellular region | 18 | 0.06 | 1.68E-09 |
| 植物细胞壁组织Plant-type cell wall organization | 6 | 0.13 | 1.47E-04 | |
| 氧化还原酶活性Oxidoreductase activity | 25 | 0.02 | 2.39E-04 | |
| 植物细胞壁组织或生物发生 Plant-type cell wall organization or biogenesis | 6 | 0.10 | 2.56E-04 | |
| 细胞外周Cell periphery | 13 | 0.03 | 9.29E-03 | |
| 细胞壁Cell wall | 7 | 0.05 | 9.73E-03 | |
| 表面封装结构External encapsulating structure | 7 | 0.05 | 9.73E-03 | |
| 细胞壁组织或生物发生Cell wall organization or biogenesis | 8 | 0.03 | 9.73E-03 | |
| 质外体Apoplast | 5 | 0.06 | 2.32E-02 | |
| 细胞壁组织Cell wall organization | 7 | 0.03 | 2.32E-02 | |
| WT 14 d vs. OE 14 d | 逆转录转座子核衣壳Retrotransposon nucleocapsid | 20 | 0.40 | 7.45E-11 |
| DNA整合DNA integration | 23 | 0.29 | 5.50E-10 | |
| RNA指导的DNA聚合酶活性RNA-directed DNA polymerase activity | 10 | 0.43 | 2.49E-05 | |
| 碳水化合物代谢过程Carbohydrate metabolic process | 57 | 0.10 | 3.29E-05 | |
| 氧化还原酶活性Oxidoreductase activity | 93 | 0.08 | 2.83E-04 | |
| 细胞外区域Extracellular region | 34 | 0.11 | 5.48E-04 | |
| DNA聚合酶活性DNA polymerase activity | 10 | 0.30 | 5.48E-04 | |
| 甲基丙二酸半醛脱氢酶活性 Methylmalonate-semialdehyde dehydrogenase(acylating)activity | 5 | 0.63 | 2.20E-03 | |
| 质膜组成成分Integral component of membrane | 243 | 0.06 | 2.20E-03 | |
| 质膜固有成分Intrinsic component of membrane | 244 | 0.06 | 2.20E-03 | |
| 催化活性Catalytic activity | 353 | 0.06 | 4.09E-03 |
表3 野生型(WT)和超表达MpICE1(OE)香蕉根接种枯萎病菌(0、7、14 d)差异表达基因GO富集
Table 3 GO enrichment of different express genes in wild(WT)and transgenic(OE)bananas before and after inoculation(0,7,14 d)
| 比较组 Pair comparison | GO功能 GO term | 基因数量 Gene number | 富集率 Rich ratio | Q值 Q value |
|---|---|---|---|---|
| WT 0 d vs. OE 0 d | 细胞外区域Extracellular region | 56 | 0.19 | 6.84E-19 |
| 细胞壁组织或生物发生Cell wall organization or biogenesis | 39 | 0.16 | 6.10E-10 | |
| 细胞壁组织Cell wall organization | 34 | 0.16 | 2.23E-08 | |
| 表面封装结构组织External encapsulating structure organization | 34 | 0.15 | 3.20E-08 | |
| 细胞壁Cell wall | 27 | 0.18 | 3.66E-08 | |
| 表面封装结构External encapsulating structure | 27 | 0.18 | 3.66E-08 | |
| 糖基水解酶活性Hydrolase activity,acting on glycosyl bonds | 41 | 0.15 | 3.98E-08 | |
| O-糖基水解酶活性 Hydrolase activity,hydrolyzing O-glycosyl compounds | 39 | 0.15 | 4.60E-08 | |
| 细胞碳水化合物代谢过程Cellular carbohydrate metabolic process | 28 | 0.17 | 1.20E-07 | |
| 碳水化合物代谢过程Carbohydrate metabolic process | 59 | 0.10 | 1.44E-07 | |
| WT 7 d vs. OE 7 d | 细胞外区域Extracellular region | 18 | 0.06 | 1.68E-09 |
| 植物细胞壁组织Plant-type cell wall organization | 6 | 0.13 | 1.47E-04 | |
| 氧化还原酶活性Oxidoreductase activity | 25 | 0.02 | 2.39E-04 | |
| 植物细胞壁组织或生物发生 Plant-type cell wall organization or biogenesis | 6 | 0.10 | 2.56E-04 | |
| 细胞外周Cell periphery | 13 | 0.03 | 9.29E-03 | |
| 细胞壁Cell wall | 7 | 0.05 | 9.73E-03 | |
| 表面封装结构External encapsulating structure | 7 | 0.05 | 9.73E-03 | |
| 细胞壁组织或生物发生Cell wall organization or biogenesis | 8 | 0.03 | 9.73E-03 | |
| 质外体Apoplast | 5 | 0.06 | 2.32E-02 | |
| 细胞壁组织Cell wall organization | 7 | 0.03 | 2.32E-02 | |
| WT 14 d vs. OE 14 d | 逆转录转座子核衣壳Retrotransposon nucleocapsid | 20 | 0.40 | 7.45E-11 |
| DNA整合DNA integration | 23 | 0.29 | 5.50E-10 | |
| RNA指导的DNA聚合酶活性RNA-directed DNA polymerase activity | 10 | 0.43 | 2.49E-05 | |
| 碳水化合物代谢过程Carbohydrate metabolic process | 57 | 0.10 | 3.29E-05 | |
| 氧化还原酶活性Oxidoreductase activity | 93 | 0.08 | 2.83E-04 | |
| 细胞外区域Extracellular region | 34 | 0.11 | 5.48E-04 | |
| DNA聚合酶活性DNA polymerase activity | 10 | 0.30 | 5.48E-04 | |
| 甲基丙二酸半醛脱氢酶活性 Methylmalonate-semialdehyde dehydrogenase(acylating)activity | 5 | 0.63 | 2.20E-03 | |
| 质膜组成成分Integral component of membrane | 243 | 0.06 | 2.20E-03 | |
| 质膜固有成分Intrinsic component of membrane | 244 | 0.06 | 2.20E-03 | |
| 催化活性Catalytic activity | 353 | 0.06 | 4.09E-03 |
| 比较组 Pair comparison | KEGG 通路 KEGG pathway | 基因数量 Gene number | 富集率 Rich ratio | Q值 Q value |
|---|---|---|---|---|
| WT 0 d vs. OE 0 d | 苯丙烷合成途径Phenylpropanoid biosynthesis | 80 | 0.11 | 1.43E-09 |
| 黄酮生物合成Flavonoid biosynthesis | 23 | 0.16 | 7.14E-05 | |
| 氰基氨基酸代谢Cyanoamino acid metabolism | 21 | 0.15 | 4.09E-04 | |
| ABC转运蛋白ABC transporters | 26 | 0.13 | 4.45E-04 | |
| 油菜素内酯生物合成Brassinosteroid biosynthesis | 9 | 0.27 | 6.06E-04 | |
| 单萜生物合成Monoterpenoid biosynthesis | 7 | 0.32 | 1.36E-03 | |
| 植物MAPK信号途径MAPK signaling pathway-plant | 76 | 0.08 | 1.85E-03 | |
| 戊糖和葡萄糖醛酸酯互换 Pentose and glucuronate interconversions | 34 | 0.10 | 2.16E-03 | |
| 角质、软木脂和蜡质生物合成 Cutin,suberine and wax biosynthesis | 13 | 0.15 | 4.69E-03 | |
| RNA聚合酶RNA polymerase | 40 | 0.09 | 8.29E-03 | |
| 其他聚糖降解Other glycan degradation | 17 | 0.11 | 2.11E-02 | |
| 异喹啉生物碱生物合成Isoquinoline alkaloid biosynthesis | 8 | 0.16 | 2.77E-02 | |
| WT 7 d vs. OE 7 d | 黄酮生物合成Flavonoid biosynthesis | 12 | 0.08 | 1.23E-06 |
| 类胡萝卜素生物合成Carotenoid biosynthesis | 6 | 0.06 | 6.80E-03 | |
| 苯丙烷合成途径Phenylpropanoid biosynthesis | 17 | 0.02 | 6.80E-03 | |
| 植物—病原菌互作Plant-pathogen interaction | 20 | 0.02 | 6.80E-03 | |
| ABC转运蛋白ABC transporters | 8 | 0.04 | 9.06E-03 | |
| 植物MAPK信号途径MAPK signaling pathway-plant | 20 | 0.02 | 9.06E-03 | |
| 二萜生物合成Diterpenoid biosynthesis | 5 | 0.05 | 1.45E-02 | |
| WT 14 d vs. OE 14 d | 黄酮生物合成Flavonoid biosynthesis | 23 | 0.16 | 2.03E-04 |
| 淀粉和蔗糖代谢Starch and sucrose metabolism | 56 | 0.10 | 2.09E-04 | |
| 植物MAPK信号途径MAPK signaling pathway-plant | 81 | 0.08 | 1.52E-03 | |
| ABC转运蛋白ABC transporters | 25 | 0.12 | 2.80E-03 | |
| 苯丙烷合成途径Phenylpropanoid biosynthesis | 61 | 0.09 | 3.20E-03 | |
| 植物—病原菌互作Plant-pathogen interaction | 75 | 0.08 | 3.20E-03 | |
| Alpha-亚油酸代谢alpha-Linolenic acid metabolism | 15 | 0.14 | 1.33E-02 | |
| 二萜生物合成Diterpenoid biosynthesis | 13 | 0.14 | 1.85E-02 | |
| 芪类、二苯基庚烷和姜酚生物合成 Stilbenoid,diarylheptanoid and gingerol biosynthesis | 11 | 0.15 | 2.58E-02 | |
| RNA聚合酶RNA polymerase | 40 | 0.09 | 2.58E-02 |
表4 香蕉野生型(WT)和超表达MpICE1(OE)根接种枯萎病菌(0、7、14 d)差异基因KEGG代谢通路富集
Table 4 KEGG enrichment of different express genes in wild(WT)and transgenic(OE)bananas before and after inoculation(0,7 and 14 d)
| 比较组 Pair comparison | KEGG 通路 KEGG pathway | 基因数量 Gene number | 富集率 Rich ratio | Q值 Q value |
|---|---|---|---|---|
| WT 0 d vs. OE 0 d | 苯丙烷合成途径Phenylpropanoid biosynthesis | 80 | 0.11 | 1.43E-09 |
| 黄酮生物合成Flavonoid biosynthesis | 23 | 0.16 | 7.14E-05 | |
| 氰基氨基酸代谢Cyanoamino acid metabolism | 21 | 0.15 | 4.09E-04 | |
| ABC转运蛋白ABC transporters | 26 | 0.13 | 4.45E-04 | |
| 油菜素内酯生物合成Brassinosteroid biosynthesis | 9 | 0.27 | 6.06E-04 | |
| 单萜生物合成Monoterpenoid biosynthesis | 7 | 0.32 | 1.36E-03 | |
| 植物MAPK信号途径MAPK signaling pathway-plant | 76 | 0.08 | 1.85E-03 | |
| 戊糖和葡萄糖醛酸酯互换 Pentose and glucuronate interconversions | 34 | 0.10 | 2.16E-03 | |
| 角质、软木脂和蜡质生物合成 Cutin,suberine and wax biosynthesis | 13 | 0.15 | 4.69E-03 | |
| RNA聚合酶RNA polymerase | 40 | 0.09 | 8.29E-03 | |
| 其他聚糖降解Other glycan degradation | 17 | 0.11 | 2.11E-02 | |
| 异喹啉生物碱生物合成Isoquinoline alkaloid biosynthesis | 8 | 0.16 | 2.77E-02 | |
| WT 7 d vs. OE 7 d | 黄酮生物合成Flavonoid biosynthesis | 12 | 0.08 | 1.23E-06 |
| 类胡萝卜素生物合成Carotenoid biosynthesis | 6 | 0.06 | 6.80E-03 | |
| 苯丙烷合成途径Phenylpropanoid biosynthesis | 17 | 0.02 | 6.80E-03 | |
| 植物—病原菌互作Plant-pathogen interaction | 20 | 0.02 | 6.80E-03 | |
| ABC转运蛋白ABC transporters | 8 | 0.04 | 9.06E-03 | |
| 植物MAPK信号途径MAPK signaling pathway-plant | 20 | 0.02 | 9.06E-03 | |
| 二萜生物合成Diterpenoid biosynthesis | 5 | 0.05 | 1.45E-02 | |
| WT 14 d vs. OE 14 d | 黄酮生物合成Flavonoid biosynthesis | 23 | 0.16 | 2.03E-04 |
| 淀粉和蔗糖代谢Starch and sucrose metabolism | 56 | 0.10 | 2.09E-04 | |
| 植物MAPK信号途径MAPK signaling pathway-plant | 81 | 0.08 | 1.52E-03 | |
| ABC转运蛋白ABC transporters | 25 | 0.12 | 2.80E-03 | |
| 苯丙烷合成途径Phenylpropanoid biosynthesis | 61 | 0.09 | 3.20E-03 | |
| 植物—病原菌互作Plant-pathogen interaction | 75 | 0.08 | 3.20E-03 | |
| Alpha-亚油酸代谢alpha-Linolenic acid metabolism | 15 | 0.14 | 1.33E-02 | |
| 二萜生物合成Diterpenoid biosynthesis | 13 | 0.14 | 1.85E-02 | |
| 芪类、二苯基庚烷和姜酚生物合成 Stilbenoid,diarylheptanoid and gingerol biosynthesis | 11 | 0.15 | 2.58E-02 | |
| RNA聚合酶RNA polymerase | 40 | 0.09 | 2.58E-02 |
| 比较组 Pair comparison | KEGG通路 KEGG pathway | 基因数量 Gene number | 富集率 Rich ratio | Q值 Q value |
|---|---|---|---|---|
| OE 0 d vs. OE 7 d(2 003) | 苯丙烷生物合成 Phenylpropanoid biosynthesis | 89 | 0.13 | 5.35E-10 |
| 二萜生物合成 Diterpenoid biosynthesis | 21 | 0.23 | 2.83E-06 | |
| 植物—病原菌互作 Plant-pathogen interaction | 94 | 0.10 | 2.83E-06 | |
| 植物MAPK信号途径 MAPK signaling pathway - plant | 96 | 0.10 | 7.54E-06 | |
| RNA聚合酶 RNA polymerase | 55 | 0.12 | 1.14E-05 | |
| 黄酮生物合成 Flavonoid biosynthesis | 22 | 0.15 | 9.43E-04 | |
| ABC转运蛋白 ABC transporters | 26 | 0.13 | 2.85E-03 | |
| 芪类、二苯基庚烷和姜酚生物合成 Stilbenoid,diarylheptanoid and gingerol biosynthesis | 13 | 0.17 | 5.75E-03 | |
| 角质、软木脂和蜡质生物合成 Cutin,suberine and wax biosynthesis | 14 | 0.16 | 6.14E-03 | |
| 甘露糖类O-聚糖生物合成 Mannose type O-glycan biosynthesis | 8 | 0.22 | 1.13E-02 | |
| WT 0 d vs. WT 7 d(1 145) | 光合作用天线蛋白 Photosynthesis-antenna proteins | 10 | 0.29 | 2.41E-05 |
| 植物激素信号传导 Plant hormone signal transduction | 65 | 0.05 | 1.08E-02 | |
| 淀粉和蔗糖代谢 Starch and sucrose metabolism | 34 | 0.06 | 2.49E-02 | |
| 倍半萜和三萜生物合成 Sesquiterpenoid and triterpenoid biosynthesis | 11 | 0.10 | 3.60E-02 | |
| 甘油酯代谢 Glycerolipid metabolism | 17 | 0.07 | 4.77E-02 | |
| OE 0 d vs. OE7 d & WT 0 d vs. WT 7 d(1 897) | 植物昼夜节律 Circadian rhythm-plant | 67 | 0.16 | 2.43E-13 |
| 泛醌和其他萜—醌生物合成 Ubiquinone and other terpenoid-quinone biosynthesis | 14 | 0.16 | 7.13E-03 | |
| 类胡萝卜素生物合成 Carotenoid biosynthesis | 16 | 0.15 | 7.13E-03 | |
| 花生四烯酸代谢 Arachidonic acid metabolism | 13 | 0.17 | 8.61E-03 | |
| 类固醇生物合成 Steroid biosynthesis | 11 | 0.15 | 3.71E-02 | |
| 淀粉和蔗糖代谢 Starch and sucrose metabolism | 48 | 0.09 | 3.71E-02 | |
| OE 7 d vs. OE 14 d(154) | 戊糖和葡萄糖醛酸酯相互转化 Pentose and glucuronate interconversions | 6 | 0.02 | 9.66E-02 |
| 苯丙烷生物合成 Phenylpropanoid biosynthesis | 9 | 0.01 | 9.66E-02 | |
| 硒化合物代谢 Selenocompound metabolism | 3 | 0.04 | 1.01E-01 | |
| 黄酮和黄酮醇生物合成 Flavone and flavonol biosynthesis | 2 | 0.03 | 2.78E-01 | |
| OE 7 d vs. OE 14 d & WT 7 d vs. WT 14 d(264) | 黄酮生物合成 Flavonoid biosynthesis | 11 | 0.07 | 1.86E-05 |
| 内质网蛋白质加工 Protein processing in endoplasmic reticulum | 15 | 0.02 | 4.52E-02 | |
| 植物—病原菌互作 Plant-pathogen interaction | 19 | 0.02 | 4.52E-02 | |
| WT 7 d vs. WT 14 d(3 407) | 植物—病原菌互作 Plant-pathogen interaction | 169 | 0.19 | 1.45E-11 |
| 植物MAPK信号途径 MAPK signaling pathway-plant | 166 | 0.17 | 5.42E-09 | |
| 苯丙氨酸代谢 Phenylalanine metabolism | 37 | 0.27 | 1.47E-06 | |
| 黄酮生物合成 Flavonoid biosynthesis | 34 | 0.23 | 2.25E-04 | |
| 苯丙烷生物合成 Phenylpropanoid biosynthesis | 110 | 0.16 | 3.48E-04 | |
| 光合作用 Photosynthesis | 32 | 0.23 | 4.57E-04 | |
| 光合作用天线蛋白 Photosynthesis-antenna proteins | 12 | 0.34 | 2.50E-03 | |
| 异喹啉生物碱生物合成 Isoquinoline alkaloid biosynthesis | 14 | 0.29 | 5.82E-03 | |
| 谷胱甘肽代谢 Glutathione metabolism | 30 | 0.20 | 6.99E-03 | |
| 芪类、二苯基庚烷和姜酚生物合成 Stilbenoid,diarylheptanoid and gingerol biosynthesis | 18 | 0.24 | 8.25E-03 | |
| ABC转运蛋白 ABC transporters | 37 | 0.18 | 9.11E-03 | |
| 光合生物碳固定 Carbon fixation in photosynthetic organisms | 33 | 0.18 | 1.43E-02 | |
| 淀粉和蔗糖代谢 Starch and sucrose metabolism | 80 | 0.14 | 2.84E-02 | |
| 黄酮和黄酮醇生物合成 Flavone and flavonol biosynthesis | 14 | 0.23 | 4.18E-02 | |
| 精氨酸生物合成 Arginine biosynthesis | 18 | 0.20 | 4.37E-02 | |
| 酪氨酸代谢 Tyrosine metabolism | 16 | 0.21 | 4.92E-02 |
表5 香蕉野生型(WT)和超表达MpICE1(OE)根接种枯萎病菌(0、7、14 d)基于venn分析的差异表达基因KEGG富集
Table 5 KEGG pathways enriched different express genes of wild(WT)and transgenic(OE)bananas before and after inoculation(0,7 and 14 d)based venn analysis
| 比较组 Pair comparison | KEGG通路 KEGG pathway | 基因数量 Gene number | 富集率 Rich ratio | Q值 Q value |
|---|---|---|---|---|
| OE 0 d vs. OE 7 d(2 003) | 苯丙烷生物合成 Phenylpropanoid biosynthesis | 89 | 0.13 | 5.35E-10 |
| 二萜生物合成 Diterpenoid biosynthesis | 21 | 0.23 | 2.83E-06 | |
| 植物—病原菌互作 Plant-pathogen interaction | 94 | 0.10 | 2.83E-06 | |
| 植物MAPK信号途径 MAPK signaling pathway - plant | 96 | 0.10 | 7.54E-06 | |
| RNA聚合酶 RNA polymerase | 55 | 0.12 | 1.14E-05 | |
| 黄酮生物合成 Flavonoid biosynthesis | 22 | 0.15 | 9.43E-04 | |
| ABC转运蛋白 ABC transporters | 26 | 0.13 | 2.85E-03 | |
| 芪类、二苯基庚烷和姜酚生物合成 Stilbenoid,diarylheptanoid and gingerol biosynthesis | 13 | 0.17 | 5.75E-03 | |
| 角质、软木脂和蜡质生物合成 Cutin,suberine and wax biosynthesis | 14 | 0.16 | 6.14E-03 | |
| 甘露糖类O-聚糖生物合成 Mannose type O-glycan biosynthesis | 8 | 0.22 | 1.13E-02 | |
| WT 0 d vs. WT 7 d(1 145) | 光合作用天线蛋白 Photosynthesis-antenna proteins | 10 | 0.29 | 2.41E-05 |
| 植物激素信号传导 Plant hormone signal transduction | 65 | 0.05 | 1.08E-02 | |
| 淀粉和蔗糖代谢 Starch and sucrose metabolism | 34 | 0.06 | 2.49E-02 | |
| 倍半萜和三萜生物合成 Sesquiterpenoid and triterpenoid biosynthesis | 11 | 0.10 | 3.60E-02 | |
| 甘油酯代谢 Glycerolipid metabolism | 17 | 0.07 | 4.77E-02 | |
| OE 0 d vs. OE7 d & WT 0 d vs. WT 7 d(1 897) | 植物昼夜节律 Circadian rhythm-plant | 67 | 0.16 | 2.43E-13 |
| 泛醌和其他萜—醌生物合成 Ubiquinone and other terpenoid-quinone biosynthesis | 14 | 0.16 | 7.13E-03 | |
| 类胡萝卜素生物合成 Carotenoid biosynthesis | 16 | 0.15 | 7.13E-03 | |
| 花生四烯酸代谢 Arachidonic acid metabolism | 13 | 0.17 | 8.61E-03 | |
| 类固醇生物合成 Steroid biosynthesis | 11 | 0.15 | 3.71E-02 | |
| 淀粉和蔗糖代谢 Starch and sucrose metabolism | 48 | 0.09 | 3.71E-02 | |
| OE 7 d vs. OE 14 d(154) | 戊糖和葡萄糖醛酸酯相互转化 Pentose and glucuronate interconversions | 6 | 0.02 | 9.66E-02 |
| 苯丙烷生物合成 Phenylpropanoid biosynthesis | 9 | 0.01 | 9.66E-02 | |
| 硒化合物代谢 Selenocompound metabolism | 3 | 0.04 | 1.01E-01 | |
| 黄酮和黄酮醇生物合成 Flavone and flavonol biosynthesis | 2 | 0.03 | 2.78E-01 | |
| OE 7 d vs. OE 14 d & WT 7 d vs. WT 14 d(264) | 黄酮生物合成 Flavonoid biosynthesis | 11 | 0.07 | 1.86E-05 |
| 内质网蛋白质加工 Protein processing in endoplasmic reticulum | 15 | 0.02 | 4.52E-02 | |
| 植物—病原菌互作 Plant-pathogen interaction | 19 | 0.02 | 4.52E-02 | |
| WT 7 d vs. WT 14 d(3 407) | 植物—病原菌互作 Plant-pathogen interaction | 169 | 0.19 | 1.45E-11 |
| 植物MAPK信号途径 MAPK signaling pathway-plant | 166 | 0.17 | 5.42E-09 | |
| 苯丙氨酸代谢 Phenylalanine metabolism | 37 | 0.27 | 1.47E-06 | |
| 黄酮生物合成 Flavonoid biosynthesis | 34 | 0.23 | 2.25E-04 | |
| 苯丙烷生物合成 Phenylpropanoid biosynthesis | 110 | 0.16 | 3.48E-04 | |
| 光合作用 Photosynthesis | 32 | 0.23 | 4.57E-04 | |
| 光合作用天线蛋白 Photosynthesis-antenna proteins | 12 | 0.34 | 2.50E-03 | |
| 异喹啉生物碱生物合成 Isoquinoline alkaloid biosynthesis | 14 | 0.29 | 5.82E-03 | |
| 谷胱甘肽代谢 Glutathione metabolism | 30 | 0.20 | 6.99E-03 | |
| 芪类、二苯基庚烷和姜酚生物合成 Stilbenoid,diarylheptanoid and gingerol biosynthesis | 18 | 0.24 | 8.25E-03 | |
| ABC转运蛋白 ABC transporters | 37 | 0.18 | 9.11E-03 | |
| 光合生物碳固定 Carbon fixation in photosynthetic organisms | 33 | 0.18 | 1.43E-02 | |
| 淀粉和蔗糖代谢 Starch and sucrose metabolism | 80 | 0.14 | 2.84E-02 | |
| 黄酮和黄酮醇生物合成 Flavone and flavonol biosynthesis | 14 | 0.23 | 4.18E-02 | |
| 精氨酸生物合成 Arginine biosynthesis | 18 | 0.20 | 4.37E-02 | |
| 酪氨酸代谢 Tyrosine metabolism | 16 | 0.21 | 4.92E-02 |
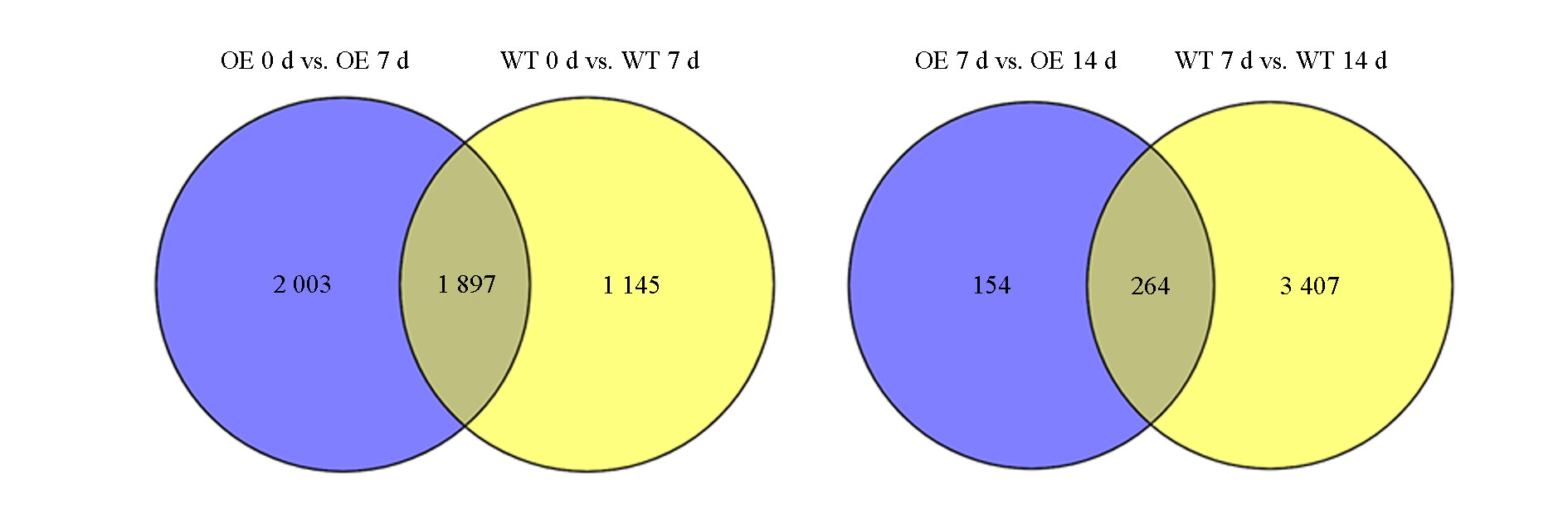
图2 香蕉野生型(WT)和超表达MpICE1(OE)根接种枯萎病菌(0、7、14 d)差异表达基因venn图
Fig. 2 Venn analysis ofdifferent express genes of wild(WT)and transgenic(OE)bananas before and after inoculation(0,7 and 14 d)

图3 野生型(WT)和超表达MpICE1(OE)香蕉根接种接种枯萎病菌前0 d、接种后7 d差异表达基因venn图
Fig. 3 Venn analysis of wild(WT)and transgenic(OE)bananas before(0 d)and after(14 d)inoculation
| 基因 Gene ID | 基因表达量(FPKM) | 基因功能注释 Gene function annotation | |||||
|---|---|---|---|---|---|---|---|
| 野生型 Wild type | 转基因香蕉 DX11 Transgenic bananas | ||||||
| 0 d | 7 d | 14 d | 0 d | 7 d | 14 d | ||
| Ma11_g19450 | 8.72 | 7.66 | 125.14 | 645.73 | 85.73 | 76.26 | 泛素羧基末端水解酶36/42 Ubiquitin carboxyl- terminal hydrolase 36/42 |
| Ma10_g12250 | 0.48 | 0.76 | 0.29 | 491.73 | 326.85 | 317.63 | 转录因子ICE1 Transcription factor ICE1 |
| Ma06_g16670 | 52.83 | 15.99 | 30.77 | 490.96 | 79.46 | 12.19 | 甘露糖特异类凝集素Mannose-specific lectin-like |
| Ma04_g33410 | 2.01 | 0.98 | 3.80 | 220.36 | 19.60 | 0.60 | 泛素羧基末端水解酶36/42 Ubiquitin carboxyl- terminal hydrolase 36/42 |
| Ma11_g22720 | 11.51 | 35.87 | 52.02 | 179.83 | 117.17 | 27.65 | 类金属硫蛋白2型Metallothionein-like protein type 2 |
| Ma06_g03820 | 1.65 | 7.76 | 82.79 | 170.87 | 45.08 | 16.56 | 类EXORDIUM 蛋白2 Protein EXORDIUM-like 2 |
| Ma10_g15940 | 48.26 | 55.42 | 226.86 | 145.97 | 240.12 | 88.15 | 过氧化物酶 Peroxidase |
| Ma04_g40060 | 17.93 | 38.03 | 16.79 | 128.60 | 196.36 | 51.97 | 果胶酯酶 Pectinesterase |
| Ma02_g16400 | 9.11 | 11.60 | 36.76 | 73.16 | 60.69 | 34.71 | 非特异性脂转运蛋白2 Non-specific lipid-transfer protein 2-like |
| Ma00_g02560 | 4.43 | 0.72 | 2.53 | 53.73 | 3.23 | 0.62 | UDP-葡萄糖醛酸酯4-异构酶 UDP-glucuronate 4-epimerase |
| Ma04_g31200 | 0.05 | 0.10 | 15.01 | 52.53 | 2.20 | 0.05 | 果胶酯酶Pectinesterase |
| Ma03_g33000 | 8.73 | 12.50 | 108.45 | 39.39 | 43.65 | 36.67 | DNA指导的RNA聚合酶Ⅱ亚基RPB1 DNA- directed RNA polymerase II subunit RPB1 |
| Ma10 g01930 | 5.74 | 4.03 | 9.69 | 38.51 | 14.91 | 4.65 | 1-脱氧-D-木酮糖-5-磷酸合成酶 1-Deoxy-D-xylulose- 5-phosphate synthase |
| Ma09_g10470 | 3.11 | 1.51 | 16.58 | 37.15 | 23.73 | 27.74 | 类GOS9蛋白Protein GOS9-like |
| Ma08_g31490 | 2.09 | 0.71 | 1.19 | 36.48 | 4.28 | 1.23 | 富半胱氨酸和甘氨酸蛋白 Cysteine and glycine-rich protein |
| Ma09_g25000 | 9.62 | 9.09 | 9.12 | 35.19 | 44.91 | 19.02 | 糖转运蛋白C Sugar carrier protein C |
| Ma08_g05750 | 5.34 | 18.13 | 205.01 | 33.95 | 91.41 | 47.89 | 赤霉素受体GID1 Gibberellin receptor GID1 |
| Ma08_g09150 | 1.01 | 0.91 | 8.82 | 32.42 | 11.90 | 6.70 | 多酚氧化酶polyphenol oxidase |
| Ma02 g13020 | 7.09 | 4.53 | 617.71 | 30.60 | 30.74 | 49.95 | 磷脂:二酰甘油酰基转移酶 Phospholipid:diacylglycerol acyltransferase |
| Ma03_g28020 | 8.34 | 2.91 | 159.63 | 25.75 | 10.80 | 11.54 | 几丁质酶1亚型X1 Chitinase 1 isoform X1 |
| Ma11_g25080 | 5.53 | 22.31 | 77.52 | 19.21 | 114.19 | 48.29 | 糖转运家族32 Solute carrier family 32 |
| Ma08_g12790 | 0.09 | 1.53 | 63.49 | 19.07 | 97.90 | 34.95 | DNA连接酶1 DNA ligase 1 |
| Ma09_g20710 | 1.58 | 2.09 | 194.79 | 11.40 | 20.77 | 12.12 | 几丁质酶 chitinase |
| Ma09 g11910 | 3.01 | 2.83 | 30.79 | 12.41 | 9.83 | 5.99 | 白介素-1受体相关激酶4 Interleukin-1 receptor- associated kinase 4 |
| Ma04_g09070 | 0.00 | 0.04 | 85.83 | 13.85 | 5.26 | 0.12 | 糖转运家族50 Solute carrier family 50 |
| Ma06_g31980 | 1.13 | 0.99 | 124.83 | 3.44 | 19.23 | 6.20 | 几丁质酶Chitinase |
| Ma11_g21980 | 0.08 | 0.16 | 35.66 | 3.66 | 15.75 | 0.08 | 扩张蛋白Expansin |
| Ma00_g04060 | 0.12 | 0.12 | 0.11 | 3.61 | 2.23 | 0.57 | 丝裂原激活蛋白激酶激酶4/5 Mitogen-activated protein kinase kinase 4/5 |
| Ma01_g04570 | 0.00 | 0 | 0.05 | 3.49 | 1.95 | 0.16 | 丝裂原激活蛋白激酶激酶4/5 Mitogen-activated protein kinase kinase 4/5 |
| Ma06_g18840 | 21.60 | 109.27 | 228.42 | 0.08 | 0.28 | 0.20 | 12-氧-植物二烯酸氧化还原酶 12-Oxophytodienoic acid reductase |
表6 香蕉野生型(WT)和超表达MpICE1(OE)根接种枯萎病菌前0d、接种后7 d共有的30个差异表达基因
Table 6 Thirty common different express genes of wild and transgenic bananas before inoculation(0 d)and after inoculation(7 d)
| 基因 Gene ID | 基因表达量(FPKM) | 基因功能注释 Gene function annotation | |||||
|---|---|---|---|---|---|---|---|
| 野生型 Wild type | 转基因香蕉 DX11 Transgenic bananas | ||||||
| 0 d | 7 d | 14 d | 0 d | 7 d | 14 d | ||
| Ma11_g19450 | 8.72 | 7.66 | 125.14 | 645.73 | 85.73 | 76.26 | 泛素羧基末端水解酶36/42 Ubiquitin carboxyl- terminal hydrolase 36/42 |
| Ma10_g12250 | 0.48 | 0.76 | 0.29 | 491.73 | 326.85 | 317.63 | 转录因子ICE1 Transcription factor ICE1 |
| Ma06_g16670 | 52.83 | 15.99 | 30.77 | 490.96 | 79.46 | 12.19 | 甘露糖特异类凝集素Mannose-specific lectin-like |
| Ma04_g33410 | 2.01 | 0.98 | 3.80 | 220.36 | 19.60 | 0.60 | 泛素羧基末端水解酶36/42 Ubiquitin carboxyl- terminal hydrolase 36/42 |
| Ma11_g22720 | 11.51 | 35.87 | 52.02 | 179.83 | 117.17 | 27.65 | 类金属硫蛋白2型Metallothionein-like protein type 2 |
| Ma06_g03820 | 1.65 | 7.76 | 82.79 | 170.87 | 45.08 | 16.56 | 类EXORDIUM 蛋白2 Protein EXORDIUM-like 2 |
| Ma10_g15940 | 48.26 | 55.42 | 226.86 | 145.97 | 240.12 | 88.15 | 过氧化物酶 Peroxidase |
| Ma04_g40060 | 17.93 | 38.03 | 16.79 | 128.60 | 196.36 | 51.97 | 果胶酯酶 Pectinesterase |
| Ma02_g16400 | 9.11 | 11.60 | 36.76 | 73.16 | 60.69 | 34.71 | 非特异性脂转运蛋白2 Non-specific lipid-transfer protein 2-like |
| Ma00_g02560 | 4.43 | 0.72 | 2.53 | 53.73 | 3.23 | 0.62 | UDP-葡萄糖醛酸酯4-异构酶 UDP-glucuronate 4-epimerase |
| Ma04_g31200 | 0.05 | 0.10 | 15.01 | 52.53 | 2.20 | 0.05 | 果胶酯酶Pectinesterase |
| Ma03_g33000 | 8.73 | 12.50 | 108.45 | 39.39 | 43.65 | 36.67 | DNA指导的RNA聚合酶Ⅱ亚基RPB1 DNA- directed RNA polymerase II subunit RPB1 |
| Ma10 g01930 | 5.74 | 4.03 | 9.69 | 38.51 | 14.91 | 4.65 | 1-脱氧-D-木酮糖-5-磷酸合成酶 1-Deoxy-D-xylulose- 5-phosphate synthase |
| Ma09_g10470 | 3.11 | 1.51 | 16.58 | 37.15 | 23.73 | 27.74 | 类GOS9蛋白Protein GOS9-like |
| Ma08_g31490 | 2.09 | 0.71 | 1.19 | 36.48 | 4.28 | 1.23 | 富半胱氨酸和甘氨酸蛋白 Cysteine and glycine-rich protein |
| Ma09_g25000 | 9.62 | 9.09 | 9.12 | 35.19 | 44.91 | 19.02 | 糖转运蛋白C Sugar carrier protein C |
| Ma08_g05750 | 5.34 | 18.13 | 205.01 | 33.95 | 91.41 | 47.89 | 赤霉素受体GID1 Gibberellin receptor GID1 |
| Ma08_g09150 | 1.01 | 0.91 | 8.82 | 32.42 | 11.90 | 6.70 | 多酚氧化酶polyphenol oxidase |
| Ma02 g13020 | 7.09 | 4.53 | 617.71 | 30.60 | 30.74 | 49.95 | 磷脂:二酰甘油酰基转移酶 Phospholipid:diacylglycerol acyltransferase |
| Ma03_g28020 | 8.34 | 2.91 | 159.63 | 25.75 | 10.80 | 11.54 | 几丁质酶1亚型X1 Chitinase 1 isoform X1 |
| Ma11_g25080 | 5.53 | 22.31 | 77.52 | 19.21 | 114.19 | 48.29 | 糖转运家族32 Solute carrier family 32 |
| Ma08_g12790 | 0.09 | 1.53 | 63.49 | 19.07 | 97.90 | 34.95 | DNA连接酶1 DNA ligase 1 |
| Ma09_g20710 | 1.58 | 2.09 | 194.79 | 11.40 | 20.77 | 12.12 | 几丁质酶 chitinase |
| Ma09 g11910 | 3.01 | 2.83 | 30.79 | 12.41 | 9.83 | 5.99 | 白介素-1受体相关激酶4 Interleukin-1 receptor- associated kinase 4 |
| Ma04_g09070 | 0.00 | 0.04 | 85.83 | 13.85 | 5.26 | 0.12 | 糖转运家族50 Solute carrier family 50 |
| Ma06_g31980 | 1.13 | 0.99 | 124.83 | 3.44 | 19.23 | 6.20 | 几丁质酶Chitinase |
| Ma11_g21980 | 0.08 | 0.16 | 35.66 | 3.66 | 15.75 | 0.08 | 扩张蛋白Expansin |
| Ma00_g04060 | 0.12 | 0.12 | 0.11 | 3.61 | 2.23 | 0.57 | 丝裂原激活蛋白激酶激酶4/5 Mitogen-activated protein kinase kinase 4/5 |
| Ma01_g04570 | 0.00 | 0 | 0.05 | 3.49 | 1.95 | 0.16 | 丝裂原激活蛋白激酶激酶4/5 Mitogen-activated protein kinase kinase 4/5 |
| Ma06_g18840 | 21.60 | 109.27 | 228.42 | 0.08 | 0.28 | 0.20 | 12-氧-植物二烯酸氧化还原酶 12-Oxophytodienoic acid reductase |

图4 野生型(WT)和超表达MpICE1(OE)香蕉根接种枯萎病菌(0、7、14 d)差异表达基因qRT-PCR验证
Fig. 4 Different express genes of wild(WT)and transgenic(OE)bananas before and after inoculation(0,7 and 14 d)were identified by qRT-PCR
| [1] |
doi: 10.1093/pcp/pcn100 URL |
| [2] |
doi: 10.1371/journal.pone.0073945 URL |
| [3] |
doi: 10.1186/1741-7007-11-121 pmid: 24350981 |
| [4] |
doi: 10.1093/jee/tox374 pmid: 29420735 |
| [5] |
doi: 10.1038/s41467-017-01670-6 pmid: 29133817 |
| [6] |
|
|
邓秀新, 束怀瑞, 郝玉金, 徐强, 韩明玉, 张绍铃, 段常青, 姜全, 易干军, 陈厚彬. 2018. 果树学科百年发展回顾. 农学学报, 8 (1):24-34.
|
|
| [7] |
doi: S1674-2052(20)30034-4 pmid: 32068158 |
| [8] |
doi: 10.1111/pbi.v18.1 URL |
| [9] |
doi: 10.1016/j.plaphy.2013.09.014 URL |
| [10] |
doi: 10.1186/s12870-021-02868-z |
| [11] |
doi: 10.1093/mp/ssu098 URL |
| [12] |
doi: 10.1111/jipb.v60.9 URL |
| [13] |
doi: 10.1038/s41598-019-53229-8 |
| [14] |
doi: 10.3389/fpls.2018.00282 URL |
| [15] |
doi: 10.1105/tpc.18.00825 URL |
| [16] |
doi: 10.1093/jxb/erv138 URL |
| [17] |
doi: 10.1016/j.hpj.2022.02.004 URL |
| [18] |
doi: 10.3390/ijms22063002 URL |
| [19] |
doi: 10.1016/j.scienta.2021.110590 URL |
| [20] |
|
|
李华平, 李云锋, 聂燕芳. 2019. 香蕉枯萎病的发生及防控研究现状. 华南农业大学学报, 40 (5):128-136.
|
|
| [21] |
doi: 10.1016/j.xplc.2021.100231 URL |
| [22] |
doi: 10.3389/fpls.2019.01069 URL |
| [23] |
doi: 10.1016/j.plaphy.2017.02.002 URL |
| [24] |
doi: 10.1002/cbf.v38.5 URL |
| [25] |
|
| [26] |
|
| [27] |
doi: 10.1046/j.1364-3703.2003.00180.x URL |
| [28] |
doi: 10.1016/j.cropro.2015.01.007 URL |
| [29] |
doi: 10.1105/tpc.12.7.1041 pmid: 10899973 |
| [30] |
doi: 10.1111/tpj.2015.81.issue-2 URL |
| [31] |
|
| [32] |
|
|
邵秀红, 窦同心, 林雪茜, 盛鸥, 邓贵明, 毕方铖, 胡春华, 李春雨, 易干军, 杨乔松. 2018. 香蕉对枯萎病的抗性机制研究现状与展望. 园艺学报, 45 (9):1661-1674.
doi: 10.16420/j.issn.0513-353x.2018-0579 URL |
|
| [33] |
doi: 10.1016/j.hpj.2018.08.001 URL |
| [34] |
doi: 10.1111/jipb.v62.3 URL |
| [35] |
doi: 10.1105/tpc.19.00487 pmid: 31690653 |
| [36] |
doi: 10.1371/journal.pone.0102303 URL |
| [37] |
|
|
许奕, 徐碧玉, 宋顺, 刘菊华, 张建斌, 贾彩红, 金志强. 2013. 香蕉茉莉酸合成关键酶基因MaOPR的克隆和表达分析. 园艺学报, 40 (2):237-246.
|
|
| [38] |
doi: 10.1111/tpj.2007.51.issue-3 URL |
| [39] |
|
|
张欣, 漆艳香, 曾凡云, 王艳玮, 谢培兰, 谢艺贤, 彭军. 2023. 香蕉枯萎病菌Dicer-like基因的功能分析. 园艺学报, 50 (2):279-294.
doi: 10.16420/j.issn.0513-353x.2021-1081 |
|
| [40] |
doi: 10.1016/j.devcel.2017.11.016 URL |
| [41] |
doi: 10.1104/pp.16.00533 URL |
| [42] |
|
| [43] |
doi: 10.1111/plb.13095 pmid: 32009285 |
| [1] | 卢艳清, 林燕金, 卢新坤. 果皮细胞壁物质代谢及果皮对高温和水分亏缺逆境的响应与‘度尾文旦柚’裂果相关[J]. 园艺学报, 2023, 50(8): 1747-1768. |
| [2] | 阮若昕, 骆慧枫, 张琛, 黄康康, 郗笃隽, 裴嘉博, 邢梦云, 刘辉. 杭州地区不同需冷量甜樱桃品种休眠阶段花芽转录组分析[J]. 园艺学报, 2023, 50(6): 1187-1202. |
| [3] | 徐悦, 李子雄, 陈婕, 孙亮. 2,4-D调控番茄果形的转录基础[J]. 园艺学报, 2023, 50(4): 802-814. |
| [4] | 王泽涵, 于文涛, 王鹏杰, 刘财国, 樊晓静, 谷梦雅, 蔡春平, 王攀, 叶乃兴. 茶树秃房与茸房种质花器官差异表达基因的WGCNA分析[J]. 园艺学报, 2023, 50(3): 620-634. |
| [5] | 张欣, 漆艳香, 曾凡云, 王艳玮, 谢培兰, 谢艺贤, 彭军. 香蕉枯萎病菌Dicer-like基因的功能分析[J]. 园艺学报, 2023, 50(2): 279-294. |
| [6] | 蒋彧, 涂勋良, 何俊蓉. 国兰叶色突变体叶片差异表达基因分析[J]. 园艺学报, 2023, 50(2): 371-381. |
| [7] | 梁嘉莉, 吴启松, 陈广全, 张荣, 徐春香, 冯淑杰. 香蕉叶斑病病原菌芭蕉新拟盘多毛孢的鉴定[J]. 园艺学报, 2023, 50(2): 410-420. |
| [8] | 王喜庆, 贾云鹤, 闫闻, 付永凯, 尤海波, 李冬艳, 赵景超. 高抗枯萎病西瓜新品种‘龙盛佳力’[J]. 园艺学报, 2023, 50(2): 455-456. |
| [9] | 谢 倩, 张诗艳, 江 来, 丁明月, 刘玲玲, 吴如健, 陈清西, . 橄榄转录组SSR信息分析及分子标记开发与应用[J]. 园艺学报, 2023, 50(11): 2350-2364. |
| [10] | 饶雪琴, 李华平. 整合态香蕉线条病毒研究进展[J]. 园艺学报, 2023, 50(10): 2288-2296. |
| [11] | 蔺海娇, 梁雨晨, 李玲, 马军, 张璐, 兰振颖, 苑泽宁. 薰衣草CBF途径相关耐寒基因挖掘与调控网络分析[J]. 园艺学报, 2023, 50(1): 131-144. |
| [12] | 赵雪艳, 王琪, 王莉, 王方圆, 王庆, 李艳. 基于比较转录组的延胡索组织差异性表达分析[J]. 园艺学报, 2023, 50(1): 177-187. |
| [13] | 杨兴玉, 许林兵, 吴元立, 邱迪洋, 范琳琳, 黄秉智. 香蕉杂交种‘粉杂1号’ABBB基因组鉴定[J]. 园艺学报, 2022, 49(9): 1991-1997. |
| [14] | 周徐子鑫, 杨威, 毛美琴, 薛彦斌, 马均. 金边红苞凤梨叶色突变体色素鉴定及类胡萝卜素合成限速基因筛选[J]. 园艺学报, 2022, 49(5): 1081-1091. |
| [15] | 沈楠, 张荆城, 王成晨, 边银丙, 肖扬. 香菇子实体发育过程中的转录组研究[J]. 园艺学报, 2022, 49(4): 801-815. |
| 阅读次数 | ||||||
|
全文 |
|
|||||
|
摘要 |
|
|||||
版权所有 © 2012 《园艺学报》编辑部 京ICP备10030308号-2 国际联网备案号 11010802023439
编辑部地址: 北京市海淀区中关村南大街12号中国农业科学院蔬菜花卉研究所 邮编: 100081
电话: 010-82109523 E-Mail: yuanyixuebao@126.com
技术支持:北京玛格泰克科技发展有限公司