
园艺学报 ›› 2025, Vol. 52 ›› Issue (7): 1758-1768.doi: 10.16420/j.issn.0513-353x.2024-0690
宋倩娜1,2, 宋陆帅1,3, 段永红1, 白小东3,*( ), 冯瑞云1,2,*(
), 冯瑞云1,2,*( )
)
收稿日期:2024-10-14
修回日期:2025-04-16
出版日期:2025-07-23
发布日期:2025-07-23
通讯作者:
基金资助:
SONG Qianna1,2, SONG Lushuai1,3, DUAN Yonghong1, BAI Xiaodong3,*( ), and FENG Ruiyun1,2,*(
), and FENG Ruiyun1,2,*( )
)
Received:2024-10-14
Revised:2025-04-16
Published:2025-07-23
Online:2025-07-23
摘要:
以四倍体栽培马铃薯品种‘Desiree’‘青薯9号’和‘晋薯16号’为研究材料,StFBH3、StbHLH52和StPDS为靶基因,构建5个GFP荧光筛选CRISPR/Cas9基因编辑载体。通过发根农杆菌介导的遗传转化法侵染茎段诱导产生毛状根,评估转化频率及编辑效率。进一步探索毛状根再生体系,并对再生马铃薯完整植株进行突变鉴定。测序检测表明5个靶位点在3个品种中均有编辑活性,StPDS单靶点在Desiree和晋薯16号毛状根中的敲除效率分别为87.8%和80.0%,StFBH3单靶点在青薯9号毛状根中的敲除效率为72.7%。StbHLH52的双靶点在Desiree毛状根中的片段删除效率可达19.4%。基因编辑的毛状根经组织培养再生形成植株,Desiree、青薯9号和晋薯16号的再生效率分别为60%、75%和50%,再生植株的编辑效率为100%,且可获得StPDS基因敲除的白化植株。该技术体系的建立可快速验证基因编辑靶位点活性,高效获得基因编辑马铃薯植株,为马铃薯基因功能研究及精准遗传育种提供技术支撑。
宋倩娜, 宋陆帅, 段永红, 白小东, 冯瑞云. 马铃薯CRISPR/Cas9介导的毛状根高效基因编辑体系的建立[J]. 园艺学报, 2025, 52(7): 1758-1768.
SONG Qianna, SONG Lushuai, DUAN Yonghong, BAI Xiaodong, and FENG Ruiyun. Establishment a System to Obtain CRISPR/Cas9 Genome Editing Plants Based on Hairy Root Transformation in Potatoes[J]. Acta Horticulturae Sinica, 2025, 52(7): 1758-1768.
| 引物名称 Prime name | 引物序列(5′-3′) Sequences of primes |
|---|---|
| StFBH3-sgRNA-F1 | ATTGAGTTGAGAGAATAAACCAGC |
| StFBH3-sgRNA-R1 | AAACGCTGGTTTATTCTCTCAACT |
| StFBH3-sgRNA-F2 | ATTGTCCATAGCTGCTATATCAGC |
| StFBH3-sgRNA-R2 | AAACGCTGATATAGCAGCTATGGA |
| StPDS-sgRNA-F | ATTGAAGGAGCGGGTAAAGCTTCG |
| StPDS-sgRNA-R | AAACCGAAGCTTTACCCGCTCCTT |
| StbHLH52-DT1-BsF | ATATATGGTCTCGATTGAATTGTCAATAGTTTCAACGTT |
| StbHLH52-DT1-F0 | TGAATTGTCAATAGTTTCAACGTTTTAGAGCTAGAAATAGC |
| StbHLH52-DT2-R0 | AACGAGTACTCTGTTTATGAGACAATCTCTTAGTCGACTCTAC |
| StbHLH52-DT2-BsR | ATTATTGGTCTCGAAACGAGTACTCTGTTTATGAGACAA |
| StbHLH52-DT3-BsF | ATATATGGTCTCGATTGTCTCAAAAGCAGAGTACTCGTT |
| StbHLH52-DT3-F0 | TGTCTCAAAAGCAGAGTACTCGTTTTAGAGCTAGAAATAGC |
| StbHLH52-DT4-R0 | AACGCCACTCACAATTCGTCATCAATCTCTTAGTCGACTCTAC |
| StbHLH52-DT4-BsR | TTATTGGTCTCGAAACGCCACTCACAATTCGTCATCAA |
| StFBH3-F1 | CACAGTTGCCACCTCAATATCCAAGG |
| StFBH3-R1 | ATAGTCCCTCAGGCTCAGCTCA |
| StFBH3-F2 | CAGCAGCACTCACAGGCCAC |
| StFBH3-R2 | GCTCTTGTAGCTTTCTCATTCGTTCAC |
| StPDS-F | GCCTTGCTTTTCCTTTCTCATCTTG |
| StPDS-R | GGCAGGTAGACAATCTAACACCTC |
| StbHLH52-F | GGAACCAAATGTTGAAGAATTCAAC |
| StbHLH52-R | CTCCAGCATTATTGCTGCTAAAA |
| Cas9-F | CCCTCAAAGAAGTTCAAGGTCCT |
| Cas9-R | AGATCCCCCTCGATCAGGAAATG |
表1 本研究中的引物序列
Table 1 Prime sequences in this study
| 引物名称 Prime name | 引物序列(5′-3′) Sequences of primes |
|---|---|
| StFBH3-sgRNA-F1 | ATTGAGTTGAGAGAATAAACCAGC |
| StFBH3-sgRNA-R1 | AAACGCTGGTTTATTCTCTCAACT |
| StFBH3-sgRNA-F2 | ATTGTCCATAGCTGCTATATCAGC |
| StFBH3-sgRNA-R2 | AAACGCTGATATAGCAGCTATGGA |
| StPDS-sgRNA-F | ATTGAAGGAGCGGGTAAAGCTTCG |
| StPDS-sgRNA-R | AAACCGAAGCTTTACCCGCTCCTT |
| StbHLH52-DT1-BsF | ATATATGGTCTCGATTGAATTGTCAATAGTTTCAACGTT |
| StbHLH52-DT1-F0 | TGAATTGTCAATAGTTTCAACGTTTTAGAGCTAGAAATAGC |
| StbHLH52-DT2-R0 | AACGAGTACTCTGTTTATGAGACAATCTCTTAGTCGACTCTAC |
| StbHLH52-DT2-BsR | ATTATTGGTCTCGAAACGAGTACTCTGTTTATGAGACAA |
| StbHLH52-DT3-BsF | ATATATGGTCTCGATTGTCTCAAAAGCAGAGTACTCGTT |
| StbHLH52-DT3-F0 | TGTCTCAAAAGCAGAGTACTCGTTTTAGAGCTAGAAATAGC |
| StbHLH52-DT4-R0 | AACGCCACTCACAATTCGTCATCAATCTCTTAGTCGACTCTAC |
| StbHLH52-DT4-BsR | TTATTGGTCTCGAAACGCCACTCACAATTCGTCATCAA |
| StFBH3-F1 | CACAGTTGCCACCTCAATATCCAAGG |
| StFBH3-R1 | ATAGTCCCTCAGGCTCAGCTCA |
| StFBH3-F2 | CAGCAGCACTCACAGGCCAC |
| StFBH3-R2 | GCTCTTGTAGCTTTCTCATTCGTTCAC |
| StPDS-F | GCCTTGCTTTTCCTTTCTCATCTTG |
| StPDS-R | GGCAGGTAGACAATCTAACACCTC |
| StbHLH52-F | GGAACCAAATGTTGAAGAATTCAAC |
| StbHLH52-R | CTCCAGCATTATTGCTGCTAAAA |
| Cas9-F | CCCTCAAAGAAGTTCAAGGTCCT |
| Cas9-R | AGATCCCCCTCGATCAGGAAATG |
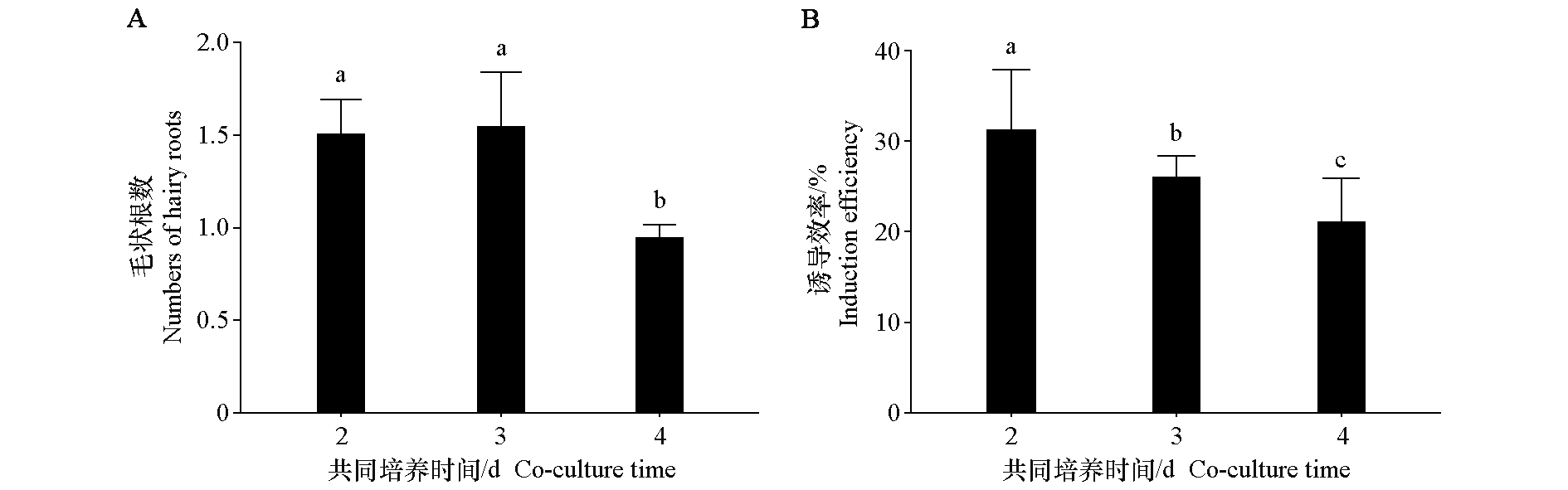
图1 不同共培养时间处理下马铃薯毛状根数(A)和荧光毛状根的诱导效率(B) 采用单因素方差分析,不同小写字母表示在0.05水平差异显著。下同
Fig. 1 Numbers of hairy roots(A)and induction efficiency of fluorescent hairy roots(B)under different co-culture time treatments in potato One-way ANOVA Duncan’s test is used for statistical analysis. Different lowercase letters indicate a signification difference at the 0.05 level. The same below
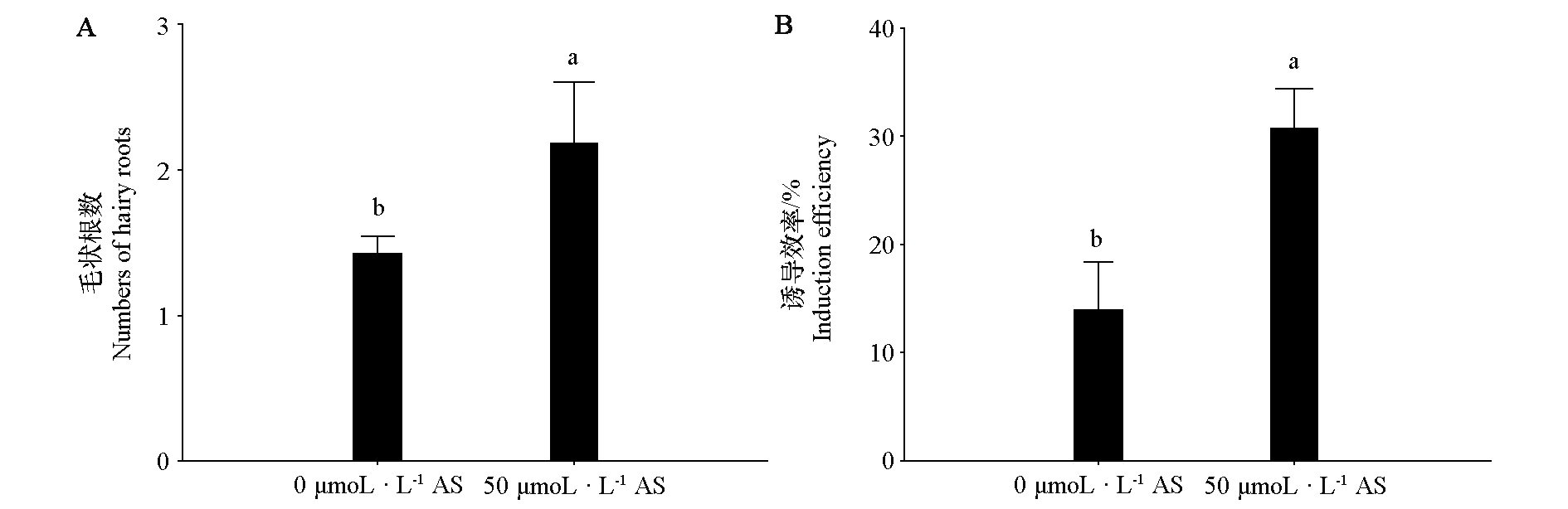
图2 两种AS浓度处理下马铃薯毛状根数(A)和荧光毛状根诱导效率(B)
Fig. 2 Numbers of hairy roots(A)and induction efficiency of fluorescent hairy roots(B)under two kinds of AS concentration treatments in potato
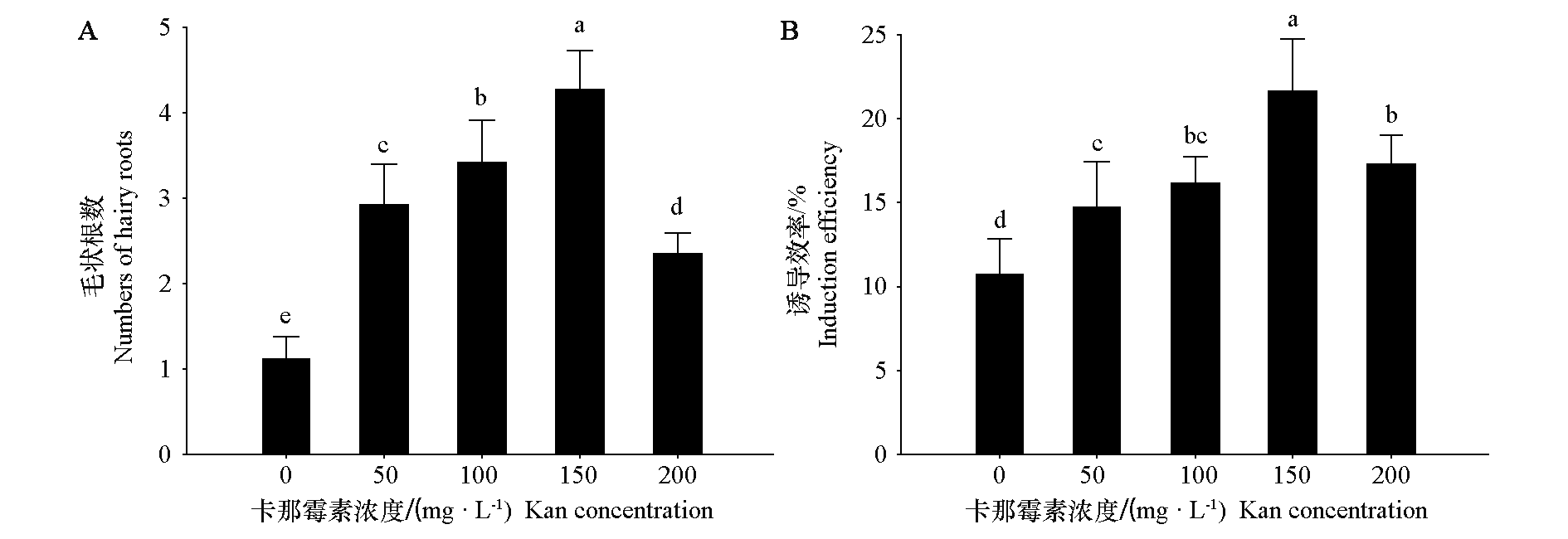
图3 不同卡那霉素浓度处理下的毛状根数量(A)和荧光毛状根诱导效率(B)
Fig. 3 Numbers of hairy roots(A)and induction efficiency of fluorescent hairy roots(B)under different Kan concentration treatments in potato

图4 StFBH3、StbHLH52和StPDS基因编辑载体转化发根农杆菌诱导马铃薯毛状根的鉴定 A:外植体的侵染;B:毛状根的诱导;C:毛状根的扩繁培养;D:荧光毛状根;E:荧光毛状根转化效率;F:CRISPR/Cas9载体示意图;G:StFBH3、StbHLH52和StPDS基因结构示意图和靶点序列;H:毛状根的PCR检测
Fig. 4 Identification of hairy roots induced by Agrobacterium rhizogenes strain with StFBH3,StbHLH52 and StPDS gene editing vectors A:The infected explants;B:Hairy root appear from explants;C:Hairy root further growth;D:Fluorescent hairy roots;E:The Percentage of fluorescent roots;F:Schematic illustration of the CRISPR/Cas9 construct;G:Schematic of the StFBH3,StbHLH52,StPDS gene and the selected target sequences;H:PCR screening of hairy roots
| 基因 Gene | 载体 Vectors | 材料 Materials | 总毛状根数 No. of total hairy roots | 荧光毛状根数 No. of fluorescent hairy roots | 突变毛状根数 No. of mutant hairy roots | 编辑效率/% Editing efficiency |
|---|---|---|---|---|---|---|
| StPDS | 青薯9号 Qingshu 9 | 62 | 36 | 30 | 83.3 | |
| StPDS-sgRNA | 晋薯16号 Jinshu 6 | 42 | 15 | 12 | 80.0 | |
| Desiree | 60 | 33 | 29 | 87.8 | ||
| StFBH3 | StFBH3-sgRNA1 | 青薯9号 Qingshu 9 | 68 | 17 | 12 | 70.6 |
| StFBH3-sgRNA2 | 30 | 11 | 8 | 72.7 | ||
| StbHLH52 | StbHLH52-sgRNA14 | Desiree | 173 | 42 | 7 | 16.7 |
| StbHLH52-sgRNA23 | 130 | 31 | 6 | 19.4 |
表2 荧光毛状根基因编辑事件的PCR鉴定
Table 2 Identification of gene editing events in fluorescent hairy roots based on PCR
| 基因 Gene | 载体 Vectors | 材料 Materials | 总毛状根数 No. of total hairy roots | 荧光毛状根数 No. of fluorescent hairy roots | 突变毛状根数 No. of mutant hairy roots | 编辑效率/% Editing efficiency |
|---|---|---|---|---|---|---|
| StPDS | 青薯9号 Qingshu 9 | 62 | 36 | 30 | 83.3 | |
| StPDS-sgRNA | 晋薯16号 Jinshu 6 | 42 | 15 | 12 | 80.0 | |
| Desiree | 60 | 33 | 29 | 87.8 | ||
| StFBH3 | StFBH3-sgRNA1 | 青薯9号 Qingshu 9 | 68 | 17 | 12 | 70.6 |
| StFBH3-sgRNA2 | 30 | 11 | 8 | 72.7 | ||
| StbHLH52 | StbHLH52-sgRNA14 | Desiree | 173 | 42 | 7 | 16.7 |
| StbHLH52-sgRNA23 | 130 | 31 | 6 | 19.4 |

图5 毛状根再生马铃薯植株系的突变检测 A:突变的毛状根;B:愈伤组织的诱导;C:苗的分化;D ~ E:再生苗的GFP表达;F:获取基因编辑马铃薯植株过程及每一步骤需要的时间;G:PCR/RE和一代测序分析突变类型;H:光漂白马铃薯植株表型
Fig. 5 Mutation detection of regeneration plants from mutated hairy root lines A:Mutated hairy roots;B:Callus induction;C:Seedings differentiation;D - E:GFP expression of regenerated plants;F:Schematic of the process for obtaining mutated potato plants;G:PCR/RE and sanger sequencing to analysis mutant types;H:Photobleaching phenotype of potato plants
| [1] |
|
| [2] |
|
| [3] |
|
| [4] |
doi: 10.1105/tpc.16.00922 pmid: 28522548 |
| [5] |
|
| [6] |
|
| [7] |
|
| [8] |
|
|
杜静雅, 陈凯园, 普金, 周会英, 祝光涛, 张春芝, 杜慧. 2023. 高效GFPuv荧光筛选基因编辑载体的改造及其在马铃薯遗传转化中的应用. 中国农业科学, 56 (11):2223-2236.
|
|
| [9] |
|
| [10] |
|
| [11] |
|
| [12] |
|
| [13] |
|
| [14] |
|
| [15] |
|
|
刘光宇, 徐晓静, 夏科科, 孙海汐, 陶月如, 崔震, 顾颖. 2022. 基于原生质体的谷子CRISPR/Cas9 基因编辑系统优化. 河南农业科学, 51 (1):34-42.
|
|
| [16] |
|
| [17] |
doi: 10.16420/j.issn.0513-353x.2021-0531 |
|
陆晨飞, 高月霞, 黄河, 戴思兰. 2022. 植物类胡萝卜素代谢及调控研究进展. 园艺学报, 49 (12):2559-2578.
doi: 10.16420/j.issn.0513-353x.2021-0531 |
|
| [18] |
doi: 10.1126/science.234.4778.856 pmid: 17758108 |
| [19] |
doi: 10.1186/s13007-020-00692-4 pmid: 33292393 |
| [20] |
doi: 10.1007/s11248-007-9083-1 pmid: 17436060 |
| [21] |
doi: 10.1104/pp.114.239392 |
| [22] |
doi: 10.13560/j.cnki.biotech.bull.1985.2024-0536 |
|
宋倩娜, 段永红, 冯瑞云. 2024. CRISPR/Cas9介导的高效四倍体马铃薯试管薯基因编辑体系的建立. 生物技术通报, 40 (9):33-41.
doi: 10.13560/j.cnki.biotech.bull.1985.2024-0536 |
|
| [23] |
pmid: 9526659 |
| [24] |
doi: 10.1016/j.jplph.2008.08.002 pmid: 18789557 |
| [28] |
|
| [29] |
doi: 10.1080/10543406.2015.1052475 pmid: 26010892 |
| [30] |
|
| [31] |
|
| [32] |
|
| [33] |
doi: 10.1186/s13059-019-1622-6 pmid: 30651124 |
| [25] |
doi: 10.1111/tpj.13217 pmid: 27227462 |
| [26] |
|
| [27] |
|
| [1] | 杨 利, 覃 亚, 边巴次仁, 珍 珍, 穷 吉, 白玛曲珍, 周 雅. 彩色马铃薯新品种‘喜格孜2号’[J]. 园艺学报, 2025, 52(S1): 145-146. |
| [2] | 刘丽锋, 宋艳红, 赵霞, 李刚, 周厚成. 利用CRISPR/Cas9编辑草莓FvPDS基因的研究[J]. 园艺学报, 2025, 52(7): 1687-1695. |
| [3] | 窦雪婷, 朱熙, 张宁, 司怀军. 马铃薯低温胁迫相关基因StHY5的功能分析[J]. 园艺学报, 2025, 52(7): 1745-1757. |
| [4] | 叶艳然, 刘建刚, 卞春松, 郭华春, 金黎平. 利用RGB影像量化分析不同氮素管理对马铃薯生长的影响[J]. 园艺学报, 2025, 52(7): 1870-1882. |
| [5] | 祝兴喆, 苏仙月, 苏旺, 张红林, 潘哲超, 刘涛, 徐笑宇. 两个马铃薯品种冬、春种植模式下叶片糖苷生物碱合成代谢差异分析[J]. 园艺学报, 2025, 52(7): 1883-1900. |
| [6] | 李姿燕, 陈炜曦, 李子涵, 黎茵, 梁峰铭, 曾祥利, 荐红举, 吕典秋. 基于RNA-Seq筛选调控马铃薯熟性的候选基因[J]. 园艺学报, 2025, 52(6): 1505-1518. |
| [7] | 王文龙, 刘照坤, 鲁文君, 王耀龙, 李晓锋, 朱红芳, 刘同坤, 李英, 侯喜林, 张昌伟. 利用基因编辑技术提高不结球白菜抗坏血酸的含量[J]. 园艺学报, 2025, 52(5): 1317-1325. |
| [8] | 马曦, 张金睿, 庄红梅, 陈巍, 张蜜, 袁晓琪, 易小芳, 王聪聪, 王海云, 王燕. 发根农杆菌介导的芜菁高效遗传转化体系建立[J]. 园艺学报, 2025, 52(5): 1389-1398. |
| [9] | 林茜, 邓振鹏, 阳新月, 周克友, 易小平, 王季春. 减施化肥配施有机肥对马铃薯产量及养分利用的影响[J]. 园艺学报, 2025, 52(4): 1007-1019. |
| [10] | 王中一, 刘熠, 胡博文, 朱凡, 刘峰, 杨莎, 熊程, 欧立军, 戴雄泽, 邹学校. 基于RUBY及CaREF1的辣椒高效遗传转化体系构建[J]. 园艺学报, 2025, 52(4): 1093-1104. |
| [11] | 周进华, 白磊, 张锐, 郭华春. 秸秆覆盖降温栽培对马铃薯生长的影响[J]. 园艺学报, 2025, 52(2): 453-466. |
| [12] | 李敏, 李思雨, 施紫涵, 陈爽, 徐炎, 刘国甜. 葡萄GRF/GIF家族基因对遗传转化再生效率的影响[J]. 园艺学报, 2025, 52(1): 51-65. |
| [13] | 黄勋, 刘霞, 邓琳梅, 王兴国, 徐亚锦, 杨艳丽. 马铃薯疮痂病生防菌1X1Y的鉴定及其生防促生特性研究[J]. 园艺学报, 2025, 52(1): 229-246. |
| [14] | 仲阳, 秦亚芝, 罗帅, 荆玉玲, 王万兴, 李广存, 胡新喜, 秦玉芝, 程旭, 熊兴耀. 泥炭和蘑菇渣改土对马铃薯根源微生物组及产量的影响[J]. 园艺学报, 2024, 51(9): 2143-2154. |
| [15] | 王晨雨, 刘孟军, 王立新, 刘志国. CRISPR/Cas9技术研究进展及其在园艺植物中的应用进展[J]. 园艺学报, 2024, 51(7): 1439-1454. |
| 阅读次数 | ||||||
|
全文 |
|
|||||
|
摘要 |
|
|||||
版权所有 © 2012 《园艺学报》编辑部 京ICP备10030308号-2 国际联网备案号 11010802023439
编辑部地址: 北京市海淀区中关村南大街12号中国农业科学院蔬菜花卉研究所 邮编: 100081
电话: 010-82109523 E-Mail: yuanyixuebao@126.com
技术支持:北京玛格泰克科技发展有限公司