
园艺学报 ›› 2025, Vol. 52 ›› Issue (12): 3288-3302.doi: 10.16420/j.issn.0513-353x.2024-0781
彭朝凤1,2, 齐帅征2, 耿喜宁2, 姚鹏强2, 谢丽华2, 张钰2,3, 陈明辉2, 施江1,*( ), 程世平2,*(
), 程世平2,*( )
)
收稿日期:2025-05-29
修回日期:2025-09-26
出版日期:2025-12-25
发布日期:2025-12-20
通讯作者:
基金资助:
PENG Chaofeng1,2, QI Shuaizheng2, GENG Xining2, YAO Pengqiang2, XIE Lihua2, ZHANG Yu2,3, CHEN Minghui2, SHI Jiang1,*( ), CHENG Shiping2,*(
), CHENG Shiping2,*( )
)
Received:2025-05-29
Revised:2025-09-26
Published:2025-12-25
Online:2025-12-20
摘要:
以不同倍性‘凤丹’牡丹(Paeonia ostii‘Fengdan’)盛花期和花衰败期叶片为试验材料,分析其叶片表型、解剖结构和转录组变化。结果表明,叶面积大小顺序为三倍体 > 四倍体 > 二倍体;叶片厚度与叶脉直径大小为四倍体 > 三倍体 > 二倍体。与二倍体叶片相比,盛开期三倍体叶片中SAUR23、SAUR32、ARR4、IPT3和IPT5等19个基因上调表达,四倍体叶片中SAUR78、CYCD3、ARP5、ZOG2、HEMD和HEME等18个基因上调表达;衰败期三倍体叶片中ARF10、IPT9和HEMB等10个基因上调表达,四倍体叶片中AHP4、CKX7、ZOG、UGT83A1、PAO和SGR等13个基因上调表达。对光合生物的固碳作用途径中的差异基因分析表明,GAPC、MDH在盛开期三倍体叶片中的表达量较高。加权基因共表达网络分析(weighted gene co-expression network analysis,WGCNA)表明,TCTP、BTB、BRH1和PsMYC2等18个核心基因在三倍体中的表达量较高,GLY、NmrA-like和WAT1等15个核心基因在四倍体中高表达, qRT-PCR结果证明转录组测序数据可靠。表明这些基因的差异表达影响了多倍体‘凤丹’牡丹叶片的生长发育。
彭朝凤, 齐帅征, 耿喜宁, 姚鹏强, 谢丽华, 张钰, 陈明辉, 施江, 程世平. 不同倍性‘凤丹’牡丹叶片转录组差异表达分析[J]. 园艺学报, 2025, 52(12): 3288-3302.
PENG Chaofeng, QI Shuaizheng, GENG Xining, YAO Pengqiang, XIE Lihua, ZHANG Yu, CHEN Minghui, SHI Jiang, CHENG Shiping. Differential Expressed Analysis by Transcriptome Sequencing in Leaves of Different Ploidy Paeonia ostii‘Fengdan’[J]. Acta Horticulturae Sinica, 2025, 52(12): 3288-3302.

图1 叶片取样位置示意图 a,b,c表示‘凤丹’牡丹复叶中小叶的取样位置
Fig. 1 Schematic diagram of leaf sampling location a,b,and c indicate the sampling positions of the leaflets in the compound leaf of‘Fengdan’
| 倍性 Ploidy | 叶长/cm Leaf length | 叶宽/cm Leaf width | 叶面积/cm2 Leaf area | 叶片厚度/μm Leaf thickness | 叶脉直径/μm Vein diameter |
|---|---|---|---|---|---|
| 二倍体 Diploid | 9.56 ± 0.50 a | 2.93 ± 0.25 b | 23.38 ± 2.08 b | 152.70 ± 15.34 c | 828.08 ± 46.42 b |
| 三倍体 Triploid | 9.03 ± 0.85 a | 5.80 ± 0.87 a | 34.86 ± 5.76 a | 206.76 ± 14.19 b | 840.70 ± 34.38 b |
| 四倍体 Tetraploid | 7.23 ± 0.25 b | 3.80 ± 0.50 b | 25.01 ± 3.01 b | 233.81 ± 18.95 a | 976.92 ± 56.21 a |
表 1 不同倍性‘凤丹’牡丹叶片生长差异
Table 1 Growth characteristics of different ploidy Paeonia ostii‘Fengdan’
| 倍性 Ploidy | 叶长/cm Leaf length | 叶宽/cm Leaf width | 叶面积/cm2 Leaf area | 叶片厚度/μm Leaf thickness | 叶脉直径/μm Vein diameter |
|---|---|---|---|---|---|
| 二倍体 Diploid | 9.56 ± 0.50 a | 2.93 ± 0.25 b | 23.38 ± 2.08 b | 152.70 ± 15.34 c | 828.08 ± 46.42 b |
| 三倍体 Triploid | 9.03 ± 0.85 a | 5.80 ± 0.87 a | 34.86 ± 5.76 a | 206.76 ± 14.19 b | 840.70 ± 34.38 b |
| 四倍体 Tetraploid | 7.23 ± 0.25 b | 3.80 ± 0.50 b | 25.01 ± 3.01 b | 233.81 ± 18.95 a | 976.92 ± 56.21 a |
| 样品 倍性 Sample Ploidy | 原始数据 Raw reads | 质控数据 Clean reads | 质控数据碱基数 Clean base(G) | Q20/% | Q30/% | GC含量/% GC Content | |
|---|---|---|---|---|---|---|---|
| 盛花期 Full flowering period | 二倍体 Diploid | 59 143 608 | 55 527 696 | 8.33 | 98.96 | 96.66 | 45.23 |
| 53 698 588 | 50 179 058 | 7.53 | 98.82 | 96.25 | 45.10 | ||
| 54 181 490 | 50 801 050 | 7.62 | 98.77 | 96.10 | 45.07 | ||
| 三倍体 Triploid | 43 391 280 | 41 104 402 | 6.17 | 98.85 | 96.31 | 45.15 | |
| 53 750 618 | 50 548 380 | 7.58 | 99.01 | 96.79 | 45.17 | ||
| 50 647 764 | 47 605 318 | 7.14 | 98.98 | 96.73 | 45.17 | ||
| 四倍体 Tetraploid | 57 476 282 | 54 146 594 | 8.12 | 98.96 | 96.66 | 45.05 | |
| 43 140 704 | 40 339 548 | 6.05 | 98.92 | 96.54 | 44.98 | ||
| 49 974 664 | 47 103 112 | 7.07 | 98.84 | 96.29 | 44.90 | ||
| 花衰 败期 Flower decay period | 二倍体 Diploid | 57 079 036 | 53 328 478 | 8.00 | 99.01 | 96.81 | 44.77 |
| 52 656 282 | 49 232 680 | 7.38 | 98.83 | 96.22 | 44.75 | ||
| 52 691 956 | 49 349 472 | 7.40 | 98.79 | 96.10 | 44.82 | ||
| 三倍体 Triploid | 58 114 202 | 54 795 360 | 8.22 | 98.91 | 96.47 | 44.95 | |
| 48 957 976 | 45 976 076 | 6.90 | 98.90 | 96.43 | 44.95 | ||
| 53 091 772 | 50 658 874 | 7.60 | 98.86 | 96.32 | 44.90 | ||
| 四倍体 Tetraploid | 57 574 644 | 55 121 600 | 8.27 | 98.76 | 96.04 | 44.88 | |
| 55 111 304 | 52 274 248 | 7.84 | 99.00 | 96.78 | 45.01 | ||
| 54 178 576 | 50 654 914 | 7.60 | 99.00 | 96.79 | 45.03 | ||
表2 转录组测序数据质量分析
Table 2 Quality analysis of transcriptome sequencing data
| 样品 倍性 Sample Ploidy | 原始数据 Raw reads | 质控数据 Clean reads | 质控数据碱基数 Clean base(G) | Q20/% | Q30/% | GC含量/% GC Content | |
|---|---|---|---|---|---|---|---|
| 盛花期 Full flowering period | 二倍体 Diploid | 59 143 608 | 55 527 696 | 8.33 | 98.96 | 96.66 | 45.23 |
| 53 698 588 | 50 179 058 | 7.53 | 98.82 | 96.25 | 45.10 | ||
| 54 181 490 | 50 801 050 | 7.62 | 98.77 | 96.10 | 45.07 | ||
| 三倍体 Triploid | 43 391 280 | 41 104 402 | 6.17 | 98.85 | 96.31 | 45.15 | |
| 53 750 618 | 50 548 380 | 7.58 | 99.01 | 96.79 | 45.17 | ||
| 50 647 764 | 47 605 318 | 7.14 | 98.98 | 96.73 | 45.17 | ||
| 四倍体 Tetraploid | 57 476 282 | 54 146 594 | 8.12 | 98.96 | 96.66 | 45.05 | |
| 43 140 704 | 40 339 548 | 6.05 | 98.92 | 96.54 | 44.98 | ||
| 49 974 664 | 47 103 112 | 7.07 | 98.84 | 96.29 | 44.90 | ||
| 花衰 败期 Flower decay period | 二倍体 Diploid | 57 079 036 | 53 328 478 | 8.00 | 99.01 | 96.81 | 44.77 |
| 52 656 282 | 49 232 680 | 7.38 | 98.83 | 96.22 | 44.75 | ||
| 52 691 956 | 49 349 472 | 7.40 | 98.79 | 96.10 | 44.82 | ||
| 三倍体 Triploid | 58 114 202 | 54 795 360 | 8.22 | 98.91 | 96.47 | 44.95 | |
| 48 957 976 | 45 976 076 | 6.90 | 98.90 | 96.43 | 44.95 | ||
| 53 091 772 | 50 658 874 | 7.60 | 98.86 | 96.32 | 44.90 | ||
| 四倍体 Tetraploid | 57 574 644 | 55 121 600 | 8.27 | 98.76 | 96.04 | 44.88 | |
| 55 111 304 | 52 274 248 | 7.84 | 99.00 | 96.78 | 45.01 | ||
| 54 178 576 | 50 654 914 | 7.60 | 99.00 | 96.79 | 45.03 | ||
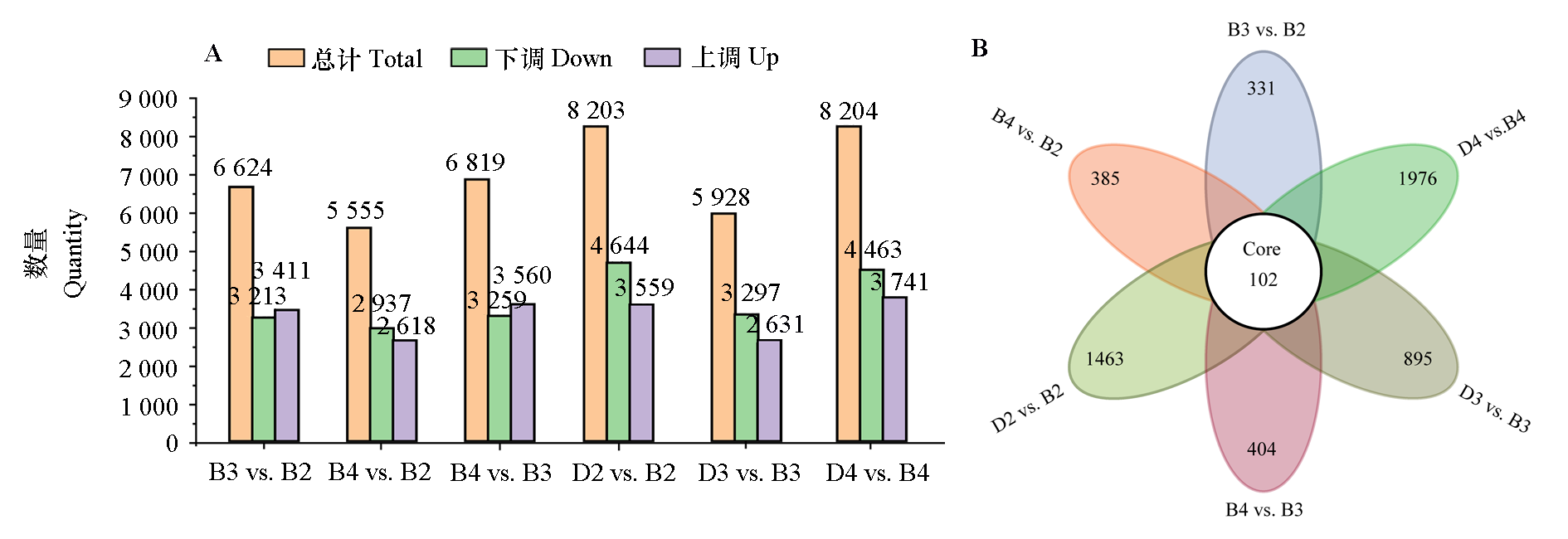
图3 不同倍性与时期‘凤丹’牡丹叶片差异表达基因(DEG)(A)和维恩图(B) B2:盛开期二倍体;B3:盛开期三倍体;B4:盛开期四倍体;D2:衰败期二倍体;D3:衰败期三倍体;D4:衰败期四倍体
Fig. 3 Differential expression genes(DEG)(A)and venn diagram(B)in the leaves Paeonia ostii‘Fengdan’under different ploidys and periods B2:Diploid during the blooming period;B3:Triploid during the blooming period;B4:Tetraploid during the blooming period;D2:Diploid during the decline period;D3:Triploid during the decline period;D4:Tetraploid during the decline period

图4 多倍体‘凤丹’牡丹叶片中生长素信号传导(A)、细胞分裂素信号传导(B)、玉米素生物合成(C)和叶绿素代谢途径(D)差异表达基因的热图 B2:盛开期二倍体;B3:盛开期三倍体;B4:盛开期四倍体;D2:衰败期二倍体;D3:衰败期三倍体;D4:衰败期四倍体。下同
Fig. 4 Heatmap of the DEG in auxin signaling transduction(A),cytokinin signaling transduction(B),zeatin biosynthesis(C)and chlorophyll metabolic pathways(D)during polyploid leaf of Paeonia ostii‘Fengdan’ B2:Diploid during the blooming period;B3:Triploid during the blooming period;B4:Tetraploid during the blooming period;D2:Diploid during the decline period;D3:Triploid during the decline period;D4:Tetraploid during the decline period. The same below

图8 qRT-PCR验证转录组数据的稳定性 不同小写字母代表有显著差异(P < 0.05)
Fig. 8 The stability of transcriptome data was verified by qRT-PCR Different lowercase letters indicate significant differences(P < 0.05)
| [1] |
|
| [2] |
|
|
程世平, 姚鹏强, 耿喜宁, 刘春洋, 谢丽华. 2022. 高温诱导牡丹产生未减数花粉. 园艺学报, 49(3):581-589.
|
|
| [3] |
|
| [4] |
|
| [5] |
|
|
康向阳. 2020. 林木三倍体育种研究进展及展望. 中国科学:生命科学, 50(2):136-143.
|
|
| [6] |
|
|
康云艳, 宋爱婷, 柴喜荣, 杨暹. 2022. 黄瓜幼苗茎段石蜡切片制作和番红染色实验方法改进. 实验技术与管理, 39(1):182-184,190.
|
|
| [7] |
|
| [8] |
|
|
李鑫. 2023. 牡丹种子发育及油脂合成相关转录因子分析及鉴定[硕士论文]. 重庆: 重庆师范大学.
|
|
| [9] |
|
| [10] |
|
| [11] |
|
|
刘春洋, 彭朝凤, 程世平, 姚鹏强, 耿喜宁, 谢丽华. 2023. 高温诱导‘凤丹’牡丹2n雌配子创制三倍体. 园艺学报, 50(7):1455-1466.
|
|
| [12] |
|
|
刘慧春, 许雯婷, 周江华, 张加强, 史小华, 朱开元. 2024. 基于牡丹涝害胁迫的转录组分析及SSR引物开发. 浙江农业学报, 36(3):544-558.
|
|
| [13] |
|
| [14] |
|
|
马鸿文, 任宇昕, 龙羿辛, 王楠, 冯祥元, 俞天泉, 华晓琴, 王君. 2024. 青黑杨杂种全同胞二倍体与三倍体长枝叶性状变异研究. 北京林业大学学报, 46(1):27-34.
|
|
| [15] |
|
|
毛常丽, 李玲, 张凤良, 李小琴, 杨湉, 吴裕. 2022. 低温胁迫下巴西橡胶树三倍体全长转录组测序分析. 热带农业科技, 45(1):1-6.
|
|
| [16] |
|
| [17] |
|
| [18] |
|
| [19] |
|
| [20] |
|
|
裴芸, 虞夏清, 赵晓坤, 张万萍, 陈劲枫. 2023. 多倍化与植物新表型关联性的研究进展. 园艺学报, 50(9):1854-1866.
|
|
| [21] |
|
| [22] |
|
| [23] |
|
|
王春夏, 尹玥, 王志平, 严瑞, 付麟岚, 孙红梅. 2019. 多倍化兰州百合和细叶百合组培苗再生和耐非生物胁迫能力. 园艺学报, 46(12):2359-2368.
|
|
| [24] |
|
| [25] |
|
| [26] |
|
| [27] |
|
|
谢洋, 邢雨蒙, 周国彦, 刘美彦, 银珊珊, 闫立英. 2023. 黄瓜二倍体及其同源四倍体果实转录组分析. 生物技术通报, 39(3):152-162.
|
|
| [28] |
|
| [29] |
|
|
杨敏, 龙宇, 王良新, 刘晓阳, 侯国艳, 蒋语嫣, 罗娅. 2022. 胞质甘油醛-3-磷酸脱氢酶GAPC2参与番茄果实成熟的调控. 四川农业大学学报, 40(2):156-162
|
|
| [30] |
|
| [31] |
|
| [32] |
|
| [33] |
|
| [34] |
|
|
赵晋锋, 杜艳伟, 余爱丽. 2020. 谷子MDH基因非生物逆境响应特性研究. 核农学报, 34(10):2152-2160.
|
|
| [35] |
|
| [36] |
|
| [1] | 何智宏, 杨国州, 李京璟, 唐小亚, 赵萍, 高斌斌, 李睿, 何丽霞. 牡丹新品种‘紫韵轮回’[J]. 园艺学报, 2025, 52(S2): 267-268. |
| [2] | 高斌斌, 何智宏, 杨国州, 李京璟, 唐小亚, 赵萍, 李睿, 何丽霞. 牡丹新品种‘茶花韵’[J]. 园艺学报, 2025, 52(S2): 269-270. |
| [3] | 邹沛姗, 郑锡荣, 胡 杏, 代色平, 倪建中. 野牡丹属新品种‘紫珊’和‘红妃’[J]. 园艺学报, 2025, 52(S1): 215-216. |
| [4] | 王心茹, 白帅帅, 孙雨乐, 武欣宇, 曾凤, 刘春英, 张玉喜, 盖树鹏, 袁延超. ‘凤丹’牡丹PoSnRK基因家族鉴定及其在花衰老中的表达分析[J]. 园艺学报, 2025, 52(9): 2363-2376. |
| [5] | 李姿燕, 陈炜曦, 李子涵, 黎茵, 梁峰铭, 曾祥利, 荐红举, 吕典秋. 基于RNA-Seq筛选调控马铃薯熟性的候选基因[J]. 园艺学报, 2025, 52(6): 1505-1518. |
| [6] | 袁娟伟, 贾利, 王涵, 严从生, 张其安, 俞飞飞, 甘德芳, 江海坤. 辣椒种质资源疫病抗性鉴定及抗病基因挖掘[J]. 园艺学报, 2025, 52(4): 857-871. |
| [7] | 智慧, 杨菁菁, 罗建让. 牡丹96个品种表型多样性分析与观赏性综合评价[J]. 园艺学报, 2025, 52(3): 635-645. |
| [8] | 李超楠, 李璐, 高丹蕾, 乔红雍, 袁涛. 原产地与引种地大花黄牡丹根系和根际土壤的AMF群落差异[J]. 园艺学报, 2025, 52(2): 467-480. |
| [9] | 李冰敏, 谭广文, 刘晓洲, 李银, 谢腾芳, 汪子涵, 周仁超. 野牡丹新品种‘霁月’[J]. 园艺学报, 2025, 52(12): 3427-3428. |
| [10] | 樊阿梅, 靳亚鑫, 何龙娇, 李琦, 张燕青, 邵祺, 宋芸, 牛颜冰, 乔永刚. 赤霉素调控连翘异型花柱生长发育的研究[J]. 园艺学报, 2025, 52(11): 3016-3030. |
| [11] | 段敏杰, 李怡斐, 王春萍, 杨小苗, 黄任中, 黄启中, 张世才. 辣椒果实类胡萝卜素调控因子转录组和靶向代谢组分析[J]. 园艺学报, 2024, 51(8): 1773-1791. |
| [12] | 李琴琴, 董山榕, 罗建让, 张延龙. 卵叶牡丹PqDFR和PqANS及启动子克隆与功能分析[J]. 园艺学报, 2024, 51(6): 1256-1272. |
| [13] | 王佩云, 李子昂, 白杨, 杨萍, 尹承芃, 李传荣, 张馨文, 宋秀华. ‘海黄’牡丹芳樟醇合酶基因PsTPS14的克隆及功能验证[J]. 园艺学报, 2024, 51(6): 1273-1283. |
| [14] | 卜文轩, 姚奕平, 黄宇, 杨星宇, 罗小宁, 张旻桓, 雷维群, 王政, 田珈宁, 陈露洁, 秦莉萍. ‘呼红’牡丹高温胁迫响应生理与转录组分析[J]. 园艺学报, 2024, 51(12): 2800-2816. |
| [15] | 吕梦雯 , 杨 勇 , 王亮生 , 李珊珊 , . 牡丹新品种‘华玉脂凝’ [J]. 园艺学报, 2023, 50(S1): 145-146. |
| 阅读次数 | ||||||
|
全文 |
|
|||||
|
摘要 |
|
|||||
版权所有 © 2012 《园艺学报》编辑部 京ICP备10030308号-2 国际联网备案号 11010802023439
编辑部地址: 北京市海淀区中关村南大街12号中国农业科学院蔬菜花卉研究所 邮编: 100081
电话: 010-82109523 E-Mail: yuanyixuebao@126.com
技术支持:北京玛格泰克科技发展有限公司