
园艺学报 ›› 2021, Vol. 48 ›› Issue (5): 947-959.doi: 10.16420/j.issn.0513-353x.2020-0800
牛西强1, 罗潇云1, 康凯程2, 黄先忠1,2,*( ), 胡能兵1, 隋益虎1, 艾昊1
), 胡能兵1, 隋益虎1, 艾昊1
收稿日期:2020-12-07
修回日期:2021-03-21
出版日期:2021-05-25
发布日期:2021-06-07
通讯作者:
黄先忠
E-mail:huangxz@ahstu.edu.cn
基金资助:
NIU Xiqaing1, LUO Xiaoyun1, KANG Kaicheng2, HUANG Xianzhong1,2,*( ), HU Nengbing1, SUI Yihu1, AI Hao1
), HU Nengbing1, SUI Yihu1, AI Hao1
Received:2020-12-07
Revised:2021-03-21
Online:2021-05-25
Published:2021-06-07
Contact:
HUANG Xianzhong
E-mail:huangxz@ahstu.edu.cn
摘要:
植物磷脂酰乙醇胺结合蛋白(Phosphatidyl ethanolamine-binding protein,PEBP)分为MFT、FT和TFL1共3个亚家族,其中FT和TFL1亚家族蛋白构成了植物的成花激素—抗成花激素系统,调控开花时间和株形结构。基于辣椒(Capsicum annuum)全基因组数据,对辣椒PEBP基因家族进行鉴定,并对基因和蛋白结构、比较进化、共线性和组织表达特征等进行了分析。结果表明,辣椒基因组包含9个PEBP成员,并且9个基因均含有4个外显子和3个内含子,定位在6条染色体上,其编码蛋白质定位在细胞质和细胞核中。与番茄PEBP同源基因的比较进化分析表明,辣椒的PEBP基因受纯化选择,其中CaFT2和CaFT4在进化中较稳定,辣椒和番茄之间存在8对共线性基因。qRT-PCR分析表明CaPEBP基因在所检测的组织中明显差异表达,如CaMFT/CaTFL1-1在果实中高表达,CaFT1在叶片中相对表达量最高,CaFT3在顶端分生组织和花中表达量最高,CaTFL1-4在根中特异表达。
中图分类号:
牛西强, 罗潇云, 康凯程, 黄先忠, 胡能兵, 隋益虎, 艾昊. 辣椒PEBP基因家族的全基因组鉴定、比较进化与组织表达分析[J]. 园艺学报, 2021, 48(5): 947-959.
NIU Xiqaing, LUO Xiaoyun, KANG Kaicheng, HUANG Xianzhong, HU Nengbing, SUI Yihu, AI Hao. Genome-wide Identification,Comparative Evolution and Expression Analysis of PEBP Gene Family from Capsicum annuum[J]. Acta Horticulturae Sinica, 2021, 48(5): 947-959.
| 基因 Gene | 上游引物序列(5′-3′) Forward primer sequence | 下游引物序列(5′-3′) Reverse primer sequence |
|---|---|---|
| CaMFT | GCTCCAAGTCCAAGTGAACC | CCGAGATCAAGATGCTGTGC |
| CaFT1 | CAATGCGCCAGACATAATTG | AGACGACGACCACCAGTACC |
| CaFT2 | GGTTCATATTGGAGGCAACG | TGCTGGGATATCTGTGACCA |
| CaFT3 | TGGTACTGATTTGAGGCCTTCT | CTCAAATTTGGGTTGCTAGGAC |
| CaFT4 | TGGTACAAATTTGAGGCCTTCT | GCATTTCCAAAGGTTACTCCTG |
| CaTFL1-1 | ACCTCCACTGGATTGTGACAG | CATTCTCTGCAGCAAACCTTC |
| CaTFL1-2 | TAGAGGGGCTTGTGAACCAC | TCTTCACCCCCAATCTCAAC |
| CaTFL1-3 | ACGGACATTCCAGGCACTAC | CAGAATCGATCCCTGGAGAG |
| CaTFL1-4 | AGATCCTGATGTTCCTGGCC | CTGAACTCCGACGTTTCTGC |
| CaUBI3 | TGTCCATCTGCTCTCTGTTG | CACCCCAAGCACAATAAGAC |
表1 辣椒PEBP基因家族基因表达分析的实时荧光定量PCR引物
Table 1 qRT-PCR primers for expression analysis of PEBP in Capsicum annuum
| 基因 Gene | 上游引物序列(5′-3′) Forward primer sequence | 下游引物序列(5′-3′) Reverse primer sequence |
|---|---|---|
| CaMFT | GCTCCAAGTCCAAGTGAACC | CCGAGATCAAGATGCTGTGC |
| CaFT1 | CAATGCGCCAGACATAATTG | AGACGACGACCACCAGTACC |
| CaFT2 | GGTTCATATTGGAGGCAACG | TGCTGGGATATCTGTGACCA |
| CaFT3 | TGGTACTGATTTGAGGCCTTCT | CTCAAATTTGGGTTGCTAGGAC |
| CaFT4 | TGGTACAAATTTGAGGCCTTCT | GCATTTCCAAAGGTTACTCCTG |
| CaTFL1-1 | ACCTCCACTGGATTGTGACAG | CATTCTCTGCAGCAAACCTTC |
| CaTFL1-2 | TAGAGGGGCTTGTGAACCAC | TCTTCACCCCCAATCTCAAC |
| CaTFL1-3 | ACGGACATTCCAGGCACTAC | CAGAATCGATCCCTGGAGAG |
| CaTFL1-4 | AGATCCTGATGTTCCTGGCC | CTGAACTCCGACGTTTCTGC |
| CaUBI3 | TGTCCATCTGCTCTCTGTTG | CACCCCAAGCACAATAAGAC |
| 基因 Gene | 基因登录号 GenBank ID | 染色体 Chr. No. | 开放阅读 框长度/bp ORF | 氨基酸数/aa Amino acid | 分子量/kD Molecular weight | 等电点 pI | 亚细胞定位 Subcellular location |
|---|---|---|---|---|---|---|---|
| CaMFT | PHT88651.1 | Chr03 | 522 | 173 | 18.98 | 9.20 | 细胞质和细胞核 Cytoplasmic and nuclear |
| CaFT1 | PHT82416.1 | Chr05 | 531 | 176 | 19.69 | 6.82 | 细胞质和细胞核 Cytoplasmic and nuclear |
| CaFT2 | PHT82541.1 | Chr05 | 528 | 175 | 19.81 | 7.76 | 细胞质和细胞核 Cytoplasmic and nuclear |
| CaFT3 | PHT65068.1 | Chr12 | 534 | 177 | 19.98 | 8.64 | 细胞质和细胞核 Cytoplasmic and nuclear |
| CaFT4 | PHT61065.1 | Sca. 2174 | 531 | 176 | 19.71 | 6.73 | 细胞质和细胞核 Cytoplasmic and nuclear |
| CaTFL1-1 | PHT94378.1 | Chr01 | 534 | 177 | 19.92 | 9.01 | 细胞质和细胞核 Cytoplasmic and nuclear |
| CaTFL1-2 | PHT87214.1 | Chr03 | 528 | 175 | 19.54 | 9.07 | 细胞质和细胞核 Cytoplasmic and nuclear |
| CaTFL1-3 | PHT80015.1 | Chr06 | 528 | 175 | 19.93 | 8.64 | 细胞质和细胞核 Cytoplasmic and nuclear |
| CaTFL1-4 | PHT71587.1 | Chr09 | 519 | 172 | 19.55 | 7.97 | 细胞质和细胞核 Cytoplasmic and nuclear |
表2 辣椒(Capsicum annuum)PEBP基因家族信息
Table 2 The information of PEBP gene family in Capsicum annuum
| 基因 Gene | 基因登录号 GenBank ID | 染色体 Chr. No. | 开放阅读 框长度/bp ORF | 氨基酸数/aa Amino acid | 分子量/kD Molecular weight | 等电点 pI | 亚细胞定位 Subcellular location |
|---|---|---|---|---|---|---|---|
| CaMFT | PHT88651.1 | Chr03 | 522 | 173 | 18.98 | 9.20 | 细胞质和细胞核 Cytoplasmic and nuclear |
| CaFT1 | PHT82416.1 | Chr05 | 531 | 176 | 19.69 | 6.82 | 细胞质和细胞核 Cytoplasmic and nuclear |
| CaFT2 | PHT82541.1 | Chr05 | 528 | 175 | 19.81 | 7.76 | 细胞质和细胞核 Cytoplasmic and nuclear |
| CaFT3 | PHT65068.1 | Chr12 | 534 | 177 | 19.98 | 8.64 | 细胞质和细胞核 Cytoplasmic and nuclear |
| CaFT4 | PHT61065.1 | Sca. 2174 | 531 | 176 | 19.71 | 6.73 | 细胞质和细胞核 Cytoplasmic and nuclear |
| CaTFL1-1 | PHT94378.1 | Chr01 | 534 | 177 | 19.92 | 9.01 | 细胞质和细胞核 Cytoplasmic and nuclear |
| CaTFL1-2 | PHT87214.1 | Chr03 | 528 | 175 | 19.54 | 9.07 | 细胞质和细胞核 Cytoplasmic and nuclear |
| CaTFL1-3 | PHT80015.1 | Chr06 | 528 | 175 | 19.93 | 8.64 | 细胞质和细胞核 Cytoplasmic and nuclear |
| CaTFL1-4 | PHT71587.1 | Chr09 | 519 | 172 | 19.55 | 7.97 | 细胞质和细胞核 Cytoplasmic and nuclear |
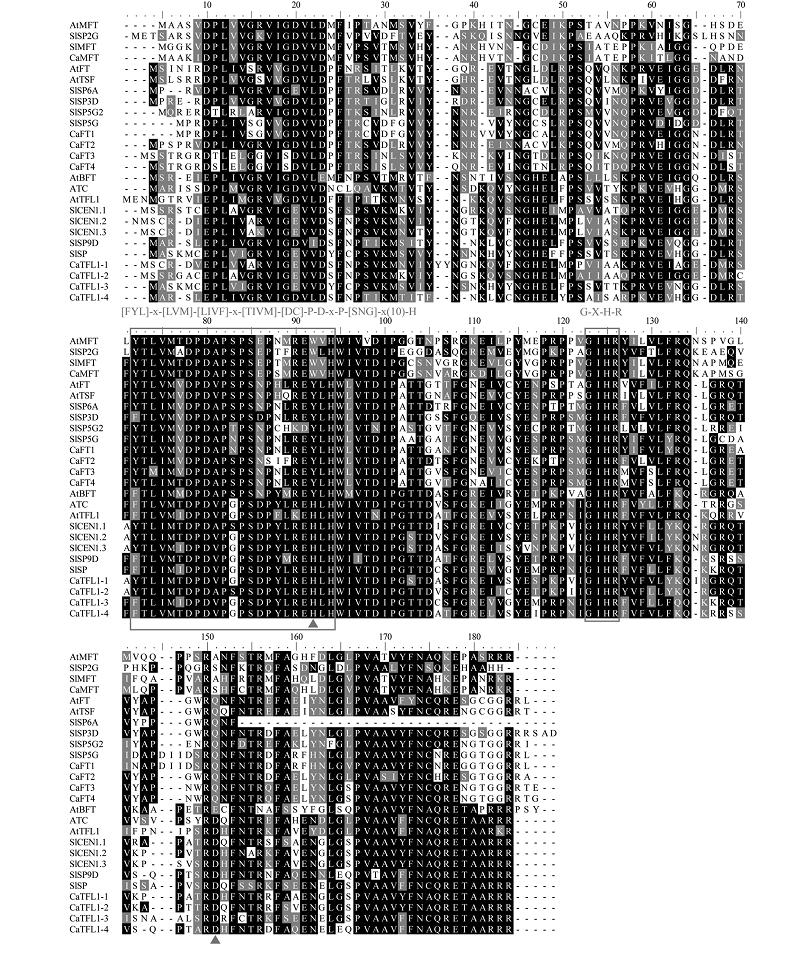
图1 拟南芥、番茄和辣椒PEBP氨基酸多重序列比对 三角形表示决定FT-like和TFL1-like功能的关键氨基酸,方框表示[FYL]-x-[LVM]-[LIVF]-x-[TIVM]-[DC]-P-D-x-P-[SNG]-x(10)-H和G-X-H-R保守基序。At:拟南芥;Sl:番茄;Ca:辣椒。
Fig. 1 Multiple amino acid sequence alignment of PEBP proteins from Arabidopsis thaliana,Solanum lycopersicum and Capsicum annuum The red triangles represent the key amino acids that determine the functions of FT-like and TFL1-like,the rectangles represent [FYL]-x-[LVM]-[LIVF]-X-[TIVM]-[DC]-P-d-x-P-[SNG]-x(10)-H and G-X-H-R conserved motif. At:Arabidopsis thaliana;Sl:Solanum lycopersicum;Ca:Capsicum annuum.
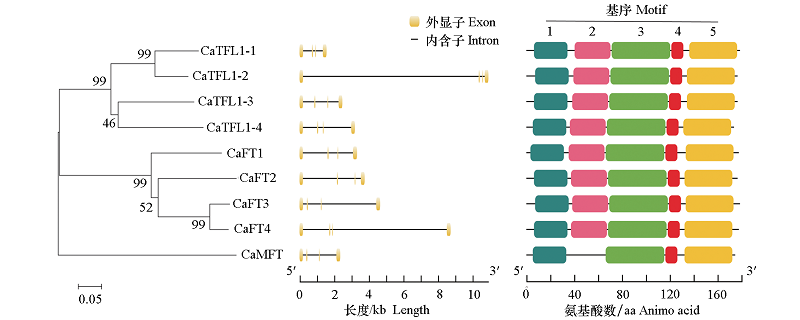
图2 CaPEBP进化树分析、辣椒PEBP基因外显子-内含子结构和motif分析 分支上的数值表示自展值;标尺表示分支长度。
Fig. 2 Phylogentic tree of CaPEBP,gene intron/exon structure and conserved motif analysis of the PEBP genes in Capsicum annuum The number in the tree represents boots value. The scale bar represents branch length.
| 基序 Motif | 长度/aa Length | 氨基酸保守序列 Amino acid conserved sequence |
|---|---|---|
| motif 1 | 29 | DPLVVGRVIGDVVDPFTPSVKMSVVYNNR |
| motif 2 | 31 | YNGHELRPSQIAAQPRVEIGGNDLRTFYTLI |
| motif 3 | 50 | MTDPDAPSPSBPYLREYLHWJVTDIPGTTDVSFGNEIVCYESPRPSIGIH |
| motif 4 | 11 | RYVFVLFRQLG |
| motif 5 | 41 | VYAPTWRDHFNTRDFAEENNLGSPVAAVYFNCQRETATRRR |
表3 辣椒PEBP蛋白氨基酸保守序列
Table 3 Conserved motifs of the PEBP proteins in Capsicum annuum
| 基序 Motif | 长度/aa Length | 氨基酸保守序列 Amino acid conserved sequence |
|---|---|---|
| motif 1 | 29 | DPLVVGRVIGDVVDPFTPSVKMSVVYNNR |
| motif 2 | 31 | YNGHELRPSQIAAQPRVEIGGNDLRTFYTLI |
| motif 3 | 50 | MTDPDAPSPSBPYLREYLHWJVTDIPGTTDVSFGNEIVCYESPRPSIGIH |
| motif 4 | 11 | RYVFVLFRQLG |
| motif 5 | 41 | VYAPTWRDHFNTRDFAEENNLGSPVAAVYFNCQRETATRRR |
| 辣椒基因 Gene ID in C. annuum | 番茄基因 Gene ID in S. lycopersicum | 基因模型 Gene model | 非同义突变频/Ka | 同义突变频率/Ks | Ka/Ks |
|---|---|---|---|---|---|
| CaMFT | SlMFT | MFT-like | 0.07 | 0.44 | 0.17 |
| CaFT1 | SlSP5G | FT-like | 0.03 | 0.39 | 0.09 |
| CaFT2 | SlSP6A | FT-like | 0.06 | 0.24 | 0.27 |
| CaFT3 | SlSP5G3 | FT-like | 0.03 | 0.29 | 0.11 |
| CaFT4 | SlSP5G1 | FT-like | 0.18 | 0.48 | 0.37 |
| CaTFL1-1 | SlCEN1.3 | TFL1-like | 0.06 | 0.56 | 0.11 |
| CaTFL1-2 | SlCEN1.1 | TFL1-like | 0.04 | 0.57 | 0.08 |
| CaTFL1-3 | SlSP | TFL1-like | 0.03 | 0.22 | 0.15 |
| SlCEN1.3 | TFL1-like | 0.26 | 1.16 | 0.22 | |
| CaTFL1-4 | SlSP9D | TFL1-like | 0.05 | 0.36 | 0.13 |
表4 辣椒和番茄PEBP同源基因的Ka/Ks
Table 4 Ka/Ks values of PEBP genes between Capsicum annuum and Solanum lycopersicum
| 辣椒基因 Gene ID in C. annuum | 番茄基因 Gene ID in S. lycopersicum | 基因模型 Gene model | 非同义突变频/Ka | 同义突变频率/Ks | Ka/Ks |
|---|---|---|---|---|---|
| CaMFT | SlMFT | MFT-like | 0.07 | 0.44 | 0.17 |
| CaFT1 | SlSP5G | FT-like | 0.03 | 0.39 | 0.09 |
| CaFT2 | SlSP6A | FT-like | 0.06 | 0.24 | 0.27 |
| CaFT3 | SlSP5G3 | FT-like | 0.03 | 0.29 | 0.11 |
| CaFT4 | SlSP5G1 | FT-like | 0.18 | 0.48 | 0.37 |
| CaTFL1-1 | SlCEN1.3 | TFL1-like | 0.06 | 0.56 | 0.11 |
| CaTFL1-2 | SlCEN1.1 | TFL1-like | 0.04 | 0.57 | 0.08 |
| CaTFL1-3 | SlSP | TFL1-like | 0.03 | 0.22 | 0.15 |
| SlCEN1.3 | TFL1-like | 0.26 | 1.16 | 0.22 | |
| CaTFL1-4 | SlSP9D | TFL1-like | 0.05 | 0.36 | 0.13 |
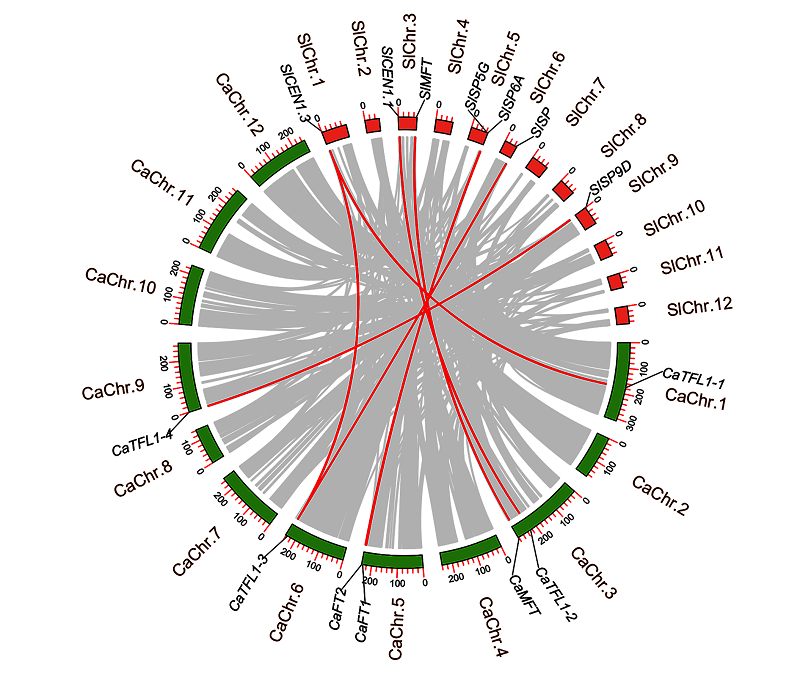
图3 番茄和辣椒的PEBP基因的共线性分析 红色线条表示种间PEBP共线性关系;灰色线条代表种间所有基因成员的共线性关系。
Fig. 3 Collinearity analysis of orthologous PEBP gene pairs betweenSolanum lycopersicum andCapsicum annuum The red lines representPEBP collinearity between species,and the gray lines are collinearity of all gene members between species.
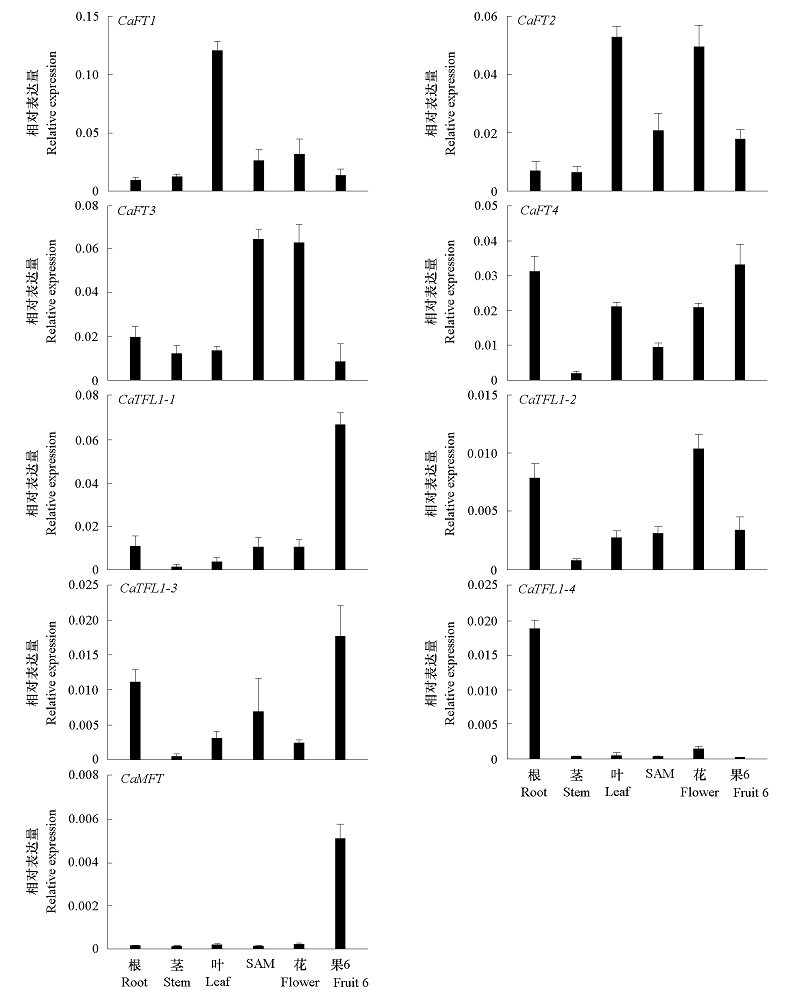
图4 辣椒根、茎、叶、顶端分生组织(SAM)、花和果(果实发育的第6时期)中PEBP基因表达的qRT-PCR分析
Fig. 4 Expression analysis of CaPEBP genes by qRT-PCR in roots,stems,leaves,shoot apical meristems(SAM),flowers and fruit(6th stage of fruit development)of pepper
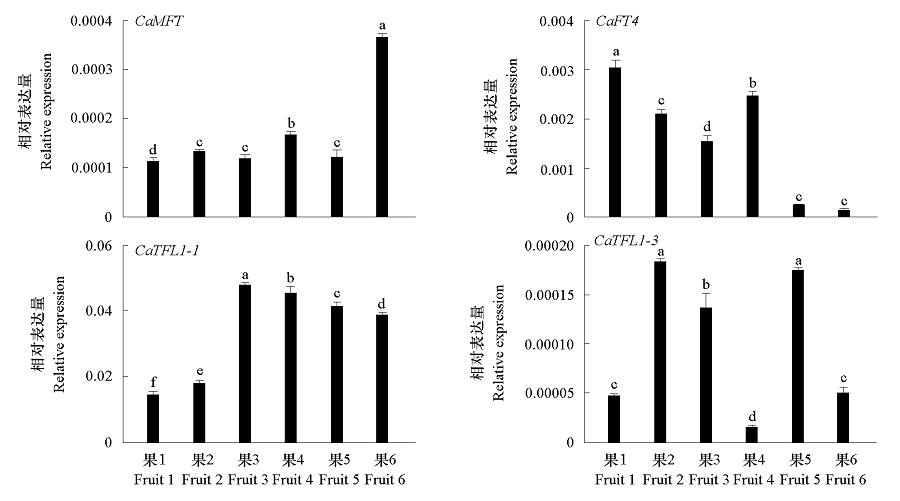
图6 辣椒果实发育(果1 ~ 果6分别表示果实发育的不同阶段)中4个CaPEBP基因的表达特征
Fig. 6 Expression profiles of four CaPEBPgenes at different developmental stages(Fruit 1-6)in fruit of Capsicum annuum
| [1] |
Abe M, Kobayashi Y, Yamamoto S, Daimon Y, Yamaguchi A, Ikeda Y, Ichinoki H, Notaguchi M, Goto K, Araki T. 2005. FD,a bZIP protein mediating signals from the floral pathway integrator FT at the shoot apex. Science, 309(5737):1052-1056.
doi: 10.1126/science.1115983 URL |
| [2] |
Ahn J H, Miller D, Winter V J, Banfield M J, Lee J H, Yoo S Y, Henz S R, Brady R L, Weigel D. 2006. A divergent external loop confers antagonistic activity on floral regulators FT and TFL1. The EMBO Journal, 25(3):605-614.
doi: 10.1038/sj.emboj.7600950 URL |
| [3] | Bernier I, Jolles P. 1984. Purification and characterisation of a basic 23 kDa cytosolic protein from bovine brain. Biochimica et Biophysica Acta, 790(2):174-181. |
| [4] | Cao K, Cui L, Zhou X, Ye L, Zou Z, Deng S. 2016. Four tomato FLOWERING LOCUS T-like proteins act antagonistically to regulate floral initiation. Frontiers in Plant Science, 6:1213. |
| [5] |
Carmel-Goren L, Liu Y S, Lifschitz E, Zamir D. 2003. The SELF-PRUNING gene family in tomato. Plant Molecular Biology, 52:1215-1222.
doi: 10.1023/B:PLAN.0000004333.96451.11 URL |
| [6] |
Carmona M J, Calonje M, Martínez-Zapater J M. 2007. The FT/TFL1 gene family in grapevine. Plant Molecular Biology, 63(5):637-650.
doi: 10.1007/s11103-006-9113-z URL |
| [7] |
Chardon F, Damerval C. 2005. Phylogenomic analysis of the PEBP gene family in cereals. Journal of Molecular Evolution, 61(5):579-590.
doi: 10.1007/s00239-004-0179-4 URL |
| [8] |
Chen C, Chen H, Zhang Y, Thomas H R, Frank M H, He Y, Xia R. 2020. TBtools:an integrative toolkit developed for interactive analyses of big biological data. Molecular Plant, 13(8):1194-1202.
doi: 10.1016/j.molp.2020.06.009 URL |
| [9] |
Elitzur T, Nahum H, Borovsky Y, Pekker I, Eshed Y, Paran I. 2009. Co-ordinated regulation of flowering time,plant architecture and growth by FASCICULATE:the pepper orthologue of SELF PRUNING. Journal of Experimental Botany, 60(3):869-880.
doi: 10.1093/jxb/ern334 URL |
| [10] |
Eshed Y, Lippman Z B. 2019. Revolutions in agriculture chart a course for targeted breeding of old and new crops. Science, 366(6466):eaax0025.
doi: 10.1126/science.aax0025 URL |
| [11] | Hanzawa Y, Money T, Bradley D. 2005. A single amino acid converts a repressor to an activator of flowering. Proceedings of the National Academy of Science of the United States of America, 102(21):7748-7753. |
| [12] |
Hedman H, Källman T, Lagercrantz U. 2009. Early evolution of the MFT-like gene family in plants. Plant Molecular Biology, 70(4):359-369.
doi: 10.1007/s11103-009-9478-x URL |
| [13] |
Ibiza V P, Blanca J, Cañizares J, Nuez F. 2012. Taxonomy and genetic diversity of domesticated Capsicum species in the Andean region. Genetic Resources and Crop Evolution, 59(6):1077-1088.
doi: 10.1007/s10722-011-9744-z URL |
| [14] |
Jaeger K E, Wigge P A. 2007. FT protein acts as a long-range signal in Arabidopsis. Current Biology, 17(12):1050-1054.
pmid: 17540569 |
| [15] | Kaneko-Suzuki M, Kurihara-Ishikawa R, Okushita-Terakawa C, Kojima C, Nagano-Fujiwara M, Ohki I, Tsuji H, Shimamoto K, Taoka K I. 2018. TFL1-Like proteins in rice antagonize rice FT-Like protein in inflorescence development by competition for complex formation with 14-3-3 and FD. Plant & Cell Physiology, 59(3):458-468. |
| [16] |
Karlgren A, Gyllenstrand N, Källman T, Sundström J F, Moore D, Lascoux M, Lagercrantz U. 2011. Evolution of the PEBP gene family in plants:functional diversification in seed plant evolution. Plant Physiology, 156(4):1967-1977.
doi: 10.1104/pp.111.176206 URL |
| [17] |
Kim S, Park J, Yeom S I, Kim Y M, Seo E, Kim K T, Kim M S, Lee J M, Cheong K, Shin H S, Kim S B, Han K, Lee J, Park M, Lee H A, Lee H Y, Lee Y, Oh S, Lee J H, Choi E, Choi E, Lee S E, Jeon J, Kim H, Choi G, Song H, Lee J, Lee S C, Kwon J K, Lee H Y, Koo N, Hong Y, Kim R W, Kang W H, Huh J H, Kang B C, Yang T J, Lee Y H, Bennetzen J L, Choi D. 2017. New reference genome sequences of hot pepper reveal the massive evolution of plant disease-resistance genes by retroduplication. Genome Biology, 18(1):210.
doi: 10.1186/s13059-017-1341-9 URL |
| [18] |
Kim S, Park M, Yeom S I, Kim Y M, Lee J M, Lee H A, Seo E, Choi J, Cheong K, Kim K T, Jung K, Lee G W, Oh H K, Bae S Y, Kim S Y, Jo Y D, Yang H B, Kang W H, Yu Y, Bark B S, Kim R Y, Choi I K, Choi B S, Lim J S, Less Y H, Choi D. 2014. Genome sequence of the hot pepper provides insights into the evolution of pungency in Capsicum species. Nature Genetics, 46(3):270-279.
doi: 10.1038/ng.2877 URL |
| [19] |
Kumar S, Stecher G, Tamura K. 2016. MEGA7:molecular evolutionary genetics analysis version 7.0 for bigger datasets. Molecular Biology and Evolution, 33(7):1870-1874.
doi: 10.1093/molbev/msw054 URL |
| [20] |
Li Chao, Zhang Yan-nan, Liu Huan-long, Huang Xian-zhong. 2015. Identification of PEBP gene family in Gossypium arboreum and Gossypium raimondii and expression analysis of the gene family in Gossypium hirsutum . Acta Agronomica Sinica, 41(3):394-404. (in Chinese)
doi: 10.3724/SP.J.1006.2015.00394 URL |
| 李超, 张彦楠, 刘焕龙, 黄先忠. 2015. 亚洲棉和雷蒙德氏棉PEBP家族基因的鉴定及该家族基因在陆地棉组织中表达分析. 作物学报, 41(3):394-404. | |
| [21] |
Li Q, Fan C, Zhang X, Wang X, Wu F, Hu R, Fu Y. 2014. Identification of a soybean MOTHER OF FT AND TFL1 homolog involved in regulation of seed germination. PLoS ONE, 9(6):e99642.
doi: 10.1371/journal.pone.0099642 URL |
| [22] |
Lifschitz E, Eshed Y. 2006. Universal florigenic signals triggered by FT homologues regulate growth and flowering cycles in perennial day-neutral tomato. Journal of Experimental Botany, 57(13):3405-3414.
doi: 10.1093/jxb/erl106 URL |
| [23] |
Livak K J, Schmittgen T D. 2001. Analysis of relative gene expression data using real-time quantitative PCR and the 2-ΔΔCT method. Methods, 25(4):402-408.
pmid: 11846609 |
| [24] |
Molinero-Rosales N, Latorre A, Jamilena M, Lozano R. 2004. SINGLE FLOWER TRUSS regulates the transition and maintenance of flowering in tomato. Planta, 218(3):427-434.
pmid: 14504922 |
| [25] |
Nakamura S, Abe F, Kawahigashi H, Nakazono K, Tagiri A, Matsumoto T, Utsugi S, Ogawa T, Handa H, Ishida H, Mori M, Kawaura K, Ogihara Y, Miura H. 2011. A wheat homolog of MOTHER OF FT AND TFL1 acts in the regulation of germination. The Plant Cell, 23(9):3215-3229.
doi: 10.1105/tpc.111.088492 pmid: 21896881 |
| [26] |
Navarro C, Abelenda J A, Cruz-Oró E, Cuéllar C A, Tamaki S, Silva J, Shimamoto k, Prat S. 2011. Control of flowering and storage organ formation in potato by FLOWERING LOCUS T. Nature, 478(7367):119-122.
doi: 10.1038/nature10431 URL |
| [27] | Qi Shiming, Liang Yan. 2020. Advances in research of the SELF-PRUNING gene family and plant architecture regulatory functions in tomato . Acta Horticulturae Sinica, 47(9):1705-1714. (in Chinese) |
| 祁世明, 梁燕. 2020. 番茄SELF-PRUNING基因家族及株形调控功能研究进展. 园艺学报, 47(9):1705-1714. | |
| [28] | Qin C, Yu C, Shen Y, Fang X, Chen L, Min J, Cheng J, Zhao S, Xu M, Luo Y, Yang Y, Wu H, Wu Z, Mao L, Zhou H, Lin H, Tang X, Zhao M, Huang Z, Zhou A, Yao X, Cui J, Li W, Chen Z, Feng Y, Niu Y, Yang X, Li W, Cai H. 2014. Whole-genome sequencing of cultivated and wild peppers provides insights into Capsicum domestication and specialization. Proceedings of the National Academy of Science of the United States of America, 111(14):5135-5140. |
| [29] |
Rozas J, Ferrer-Mata A, Sánchez-DelBarrio J C, Guirao-Rico S, Librado P, Ramos-Onsins S E, Sánchez-Gracia A. 2017. DnaSP 6:DNA sequence polymorphism analysis of large datasets. Molecular Biology and Evolution, 34(12):3299-3302.
doi: 10.1093/molbev/msx248 URL |
| [30] |
Wan H, Yuan W, Ruan M, Ye Q, Wang R, Li Z, Zhou G, Yao Z, Zhao J, Liu S, Yang Y. 2011. Identification of reference genes for reverse transcription quantitative real-time PCR normalization in pepper(Capsicum annuumL.). Biochemical and Biophysical Research Communications, 416(1-2):24-30.
doi: 10.1016/j.bbrc.2011.10.105 URL |
| [31] | Wang Y, Tang H, DeBarry J D, Tan X, Li J, Wang X, Lee T H, Jin H, Marler B, Guo H, Kissinger J C, Paterson A H. 2012. MCScanX:a toolkit for detection and evolutionary analysis of gene synteny and collinearity. Nucleic Acids Research, 40(7):49. |
| [32] |
Wang Y D, Chen G J, Lei J J, Cao B H, Chen C M. 2020. Identification and characterization of a LEA-like gene,CaMF5,specifically expressed in the anthers of male-fertileCapsicum annuum. Horticultural Plant Journal, 6(1):39-48.
doi: 10.1016/j.hpj.2019.07.004 URL |
| [33] |
Wickland D P, Hanzawa Y. 2015. TheFLOWERING LOCUS T/TERMINAL FLOWER 1 gene family:functional evolution and molecular mechanisms. Molecular Plant, 8(7):983-997.
doi: 10.1016/j.molp.2015.01.007 pmid: 25598141 |
| [34] |
Xi W, Liu C, Hou X, Yu H. 2010. MOTHER OF FT AND TFL1 regulates seed germination through a negative feedback loop modulating ABA signaling inArabidopsis. The Plant Cell, 22(6):1733-1748.
doi: 10.1105/tpc.109.073072 URL |
| [35] |
Yadav C B, Bonthala V S, Muthamilarasan M, Pandey G, Khan Y, Prasad M. 2015. Genome-wide development of transposable elements-based markers in foxtail millet and construction of an integrated database. DNA Research, 22(1):79-90.
doi: 10.1093/dnares/dsu039 URL |
| [1] | 王晓晨, 聂子页, 刘先菊, 段 伟, 范培格, 梁振昌, . 脱落酸对‘京香玉’葡萄果实单萜物质合成的影响[J]. 园艺学报, 2023, 50(2): 237-249. |
| [2] | 宋艳红, 陈亚铎, 张晓玉, 宋 盼, 刘丽锋, 李 刚, 赵 霞, 周厚成, . 森林草莓FvbHLH130转录因子调控植株提前开花[J]. 园艺学报, 2023, 50(2): 295-306. |
| [3] | 翟含含, 翟宇杰, 田义, 张叶, 杨丽, 温陟良, 陈海江. 桃SAUR家族基因分析及PpSAUR5功能鉴定[J]. 园艺学报, 2023, 50(1): 1-14. |
| [4] | 罗海林, 袁雷, 翁华, 闫佳会, 郭青云, 王文清, 马新明. 蚕豆萎蔫病毒2号青海辣椒分离物的鉴定与全基因组序列克隆[J]. 园艺学报, 2023, 50(1): 161-169. |
| [5] | 曹亚从, 张正海, 于海龙, 冯锡刚, 张宝玺, 王立浩. 螺丝椒新品种‘中椒409号’[J]. 园艺学报, 2022, 49(S2): 111-112. |
| [6] | 曹亚从, 张正海, 于海龙, 冯锡刚, 张宝玺, 王立浩. 螺丝椒抗病新品种‘中椒209号’[J]. 园艺学报, 2022, 49(S2): 113-114. |
| [7] | 王永平, 胡明文, 朱文超, 廖芳芳, 白立伟, 高 刚. 辣椒新品种‘红辣3号’[J]. 园艺学报, 2022, 49(S2): 115-116. |
| [8] | 刘术均, 惠成章, 赵丽丽, 孙永生, 刘爱群. 中早熟辣椒新品种‘美罗卡’[J]. 园艺学报, 2022, 49(S2): 117-118. |
| [9] | 王 飞, 李 宁, 尹延旭, 高升华, 徐 凯, 姚明华. 辣椒新品种‘鄂椒红元帅’[J]. 园艺学报, 2022, 49(S2): 119-120. |
| [10] | 李 宁, 尹延旭, 高升华, 徐 凯, 王 飞, 姚明华. 辣椒新品种‘锦绣红117’[J]. 园艺学报, 2022, 49(S2): 121-122. |
| [11] | 方 荣, 陈学军, 周坤华, 袁欣捷, 雷 刚, 黄月琴. 辣椒新品种‘赣椒18号’[J]. 园艺学报, 2022, 49(S1): 75-76. |
| [12] | 董桑婕, 葛诗蓓, 李岚, 贺丽群, 范飞军, 齐振宇, 喻景权, 周艳虹. 不同光质补光对辣椒幼苗生长、丛枝菌根共生和磷吸收的影响[J]. 园艺学报, 2022, 49(8): 1699-1712. |
| [13] | 李颖, 王恒明, 徐小万, 徐晓美, 黄智文. 辣椒新品种‘粤椒8号’[J]. 园艺学报, 2022, 49(8): 1837-1838. |
| [14] | 张秋悦, 刘昌来, 于晓晶, 杨甲定, 封超年. 盐胁迫条件下杜梨叶片差异表达基因qRT-PCR内参基因筛选[J]. 园艺学报, 2022, 49(7): 1557-1570. |
| [15] | 李平平, 张祥, 刘雨婷, 谢志和, 张芮豪, 赵凯, 吕俊恒, 王梓然, 文锦芬, 邹学校, 邓明华. 辣椒63份种质果皮颜色与呈色物质的关系[J]. 园艺学报, 2022, 49(7): 1589-1601. |
| 阅读次数 | ||||||
|
全文 |
|
|||||
|
摘要 |
|
|||||
版权所有 © 2012 《园艺学报》编辑部 京ICP备10030308号-2 国际联网备案号 11010802023439
编辑部地址: 北京市海淀区中关村南大街12号中国农业科学院蔬菜花卉研究所 邮编: 100081
电话: 010-82109523 E-Mail: yuanyixuebao@126.com
技术支持:北京玛格泰克科技发展有限公司