
Acta Horticulturae Sinica ›› 2021, Vol. 48 ›› Issue (11): 2227-2238.doi: 10.16420/j.issn.0513-353x.2020-0952
• Original article • Previous Articles Next Articles
WANG Shuwen, YANG Aiyi, WANG Huasen( ), XU Yunmin(
), XU Yunmin( )
)
Received:2021-07-30
Revised:2021-10-13
Published:2021-12-02
Contact:
WANG Huasen,XU Yunmin
E-mail:wanghs@zafu.edu.cn;xuyunmin@zafu.edu.cn
CLC Number:
WANG Shuwen, YANG Aiyi, WANG Huasen, XU Yunmin. Identification and Expression Analysis of miR156/157-SPL Pathway Genes in Cucumber[J]. Acta Horticulturae Sinica, 2021, 48(11): 2227-2238.
Add to citation manager EndNote|Ris|BibTeX
URL: https://www.ahs.ac.cn/EN/10.16420/j.issn.0513-353x.2020-0952
| 基因名称 Gene name | 正向引物(5′-3′) Forward primer | 反向引物(5′-3′) Reverse primer |
|---|---|---|
| Csa-SPL1 | CCCTGTCACGAAGTATTTTGTCTC | TGTTTATCAGGAAAGTGGGTTGG |
| Csa-SPL2 | GCAGCATCAACCGACCGACTATCA | ACCAAAGGACGACCAATGGGAAT |
| Csa-SPL3 | CCACCAAACTACCTTATCAAGCG | GAAAGCACGAGGAATGTGAACTA |
| Csa-SPL4 | TAACGCCTCCATTTCTTTGCTCC | TAGATGGGGATTGATTTCCAGGATT |
| Csa-SPL5 | GCGACTGGAAAAGCTGATGGAA | ATCCCCTGAAAACCCTAATTCTGC |
| Csa-SPL6 | GAAGAACGAATCATGGGCCAACT | TCCGAAATCATCCGTCCCTCC |
| Csa-SPL7 | CAGTCCCATCTGAGTGTTCCAGT | CTTCCTCCATTTCAAGACCCA |
| Csa-SPL8 | GAAGGAAGGAGTAATGCGATGGA | GAACCGTGGCAGTGAAAAGAG |
| Csa-SPL9 | TCCAGAAACCGAACGACGACT | GCATGTGAAACTTGGTGATTGAGAC |
| Csa-SPL10 | AGAGCCAAGGGGAAAGCACAA | CTTCCTGGAGGAGCCATCTGAAT |
| Csa-SPL11 | ATCACCTACTACTCTTGGTCGTTGT | CTTAGCCTTGGTTGCGAAGA |
| Csa-SPL12 | TATGTTGCCCGGTTCGTTGT | CCGTTTCCGAGCACTTTCTG |
| Csa-SPL13 | CTAACTCATCGCAACTGATTCAAAG | TGGTTAAGGCAGTCTAGTGACATTCT |
| Csa-SPL14 | CTCAGAAGATCAAGGTTGGGAGG | ACCACCCCGAGCACTGGTTA |
| Csa-MIR156A | GTATTGAAAATTAGAGTAAGGGGAAG | CTTCAAGCATGAACCCTAACAT |
| Csa-MIR156B | GAGAGAAAAGCACAAAAGACCAAGA | CTAAGTAAAGGTATGAACTTTCAACTT |
| 基因名称 Gene name | 正向引物(5′-3′) Forward primer | 反向引物(5′-3′) Reverse primer |
| Csa-MIR156C | GTTTATCGTTCTTGAATTTGGTTAG | ATACACTTTATAGCAACTGCCTCTG |
| Csa-MIR156D | TGAATGATGGTGAGTGTGTTAGGAG | CACGCACCCGCAAAGGTAT |
| Csa-MIR156E | AAAGGTGATTAAAATTGAGGGGAA | TGGCATATTATTATACCAAAAGCCT |
| Csa-MIR156F | CATACGGAAGGTAATCTCAAGGA | AATATCTAGCATAAGCGGCCATG |
| Csa-MIR156G | TCTCACGGTCTATTTCTAACACG | CACAAAGCTTCGAGCATTGATAT |
| Csa-MIR157A | GGGTTTAGAAATTTGGAGAGAGACA | TATTAAAAGTGAAGGGATGGGATATA |
| Csa-MIR157B | ATTTATCATGCACAAGGGAGAACTT | AATTGCTATTAGTGGTACCGATTGA |
| Csa-MIR157C | CCAATACGGTGATAGCTATATGTTGT | TAACAAAGTGGTGTTTACGACTCTA |
| Csa-Tub | CACTACACCGTTGGAAAGGAAA | CAAAAGGAGGGAGCCGAGA |
| Csa-U6 | GGGGACATCCGATAAAATT | TGTGCGTGTCATCCTTGC |
| Csa-miR156 | GCGGCGGTGACAGAAGAGAG | GTGCAGGGTCCGAGGT |
| Csa-miR156 RT | GTCGTATCCAGTGCAGGGTCCGAGGTATTCGCAC | TGGATACGACGTGCTC |
Table 1 Gene detection primers
| 基因名称 Gene name | 正向引物(5′-3′) Forward primer | 反向引物(5′-3′) Reverse primer |
|---|---|---|
| Csa-SPL1 | CCCTGTCACGAAGTATTTTGTCTC | TGTTTATCAGGAAAGTGGGTTGG |
| Csa-SPL2 | GCAGCATCAACCGACCGACTATCA | ACCAAAGGACGACCAATGGGAAT |
| Csa-SPL3 | CCACCAAACTACCTTATCAAGCG | GAAAGCACGAGGAATGTGAACTA |
| Csa-SPL4 | TAACGCCTCCATTTCTTTGCTCC | TAGATGGGGATTGATTTCCAGGATT |
| Csa-SPL5 | GCGACTGGAAAAGCTGATGGAA | ATCCCCTGAAAACCCTAATTCTGC |
| Csa-SPL6 | GAAGAACGAATCATGGGCCAACT | TCCGAAATCATCCGTCCCTCC |
| Csa-SPL7 | CAGTCCCATCTGAGTGTTCCAGT | CTTCCTCCATTTCAAGACCCA |
| Csa-SPL8 | GAAGGAAGGAGTAATGCGATGGA | GAACCGTGGCAGTGAAAAGAG |
| Csa-SPL9 | TCCAGAAACCGAACGACGACT | GCATGTGAAACTTGGTGATTGAGAC |
| Csa-SPL10 | AGAGCCAAGGGGAAAGCACAA | CTTCCTGGAGGAGCCATCTGAAT |
| Csa-SPL11 | ATCACCTACTACTCTTGGTCGTTGT | CTTAGCCTTGGTTGCGAAGA |
| Csa-SPL12 | TATGTTGCCCGGTTCGTTGT | CCGTTTCCGAGCACTTTCTG |
| Csa-SPL13 | CTAACTCATCGCAACTGATTCAAAG | TGGTTAAGGCAGTCTAGTGACATTCT |
| Csa-SPL14 | CTCAGAAGATCAAGGTTGGGAGG | ACCACCCCGAGCACTGGTTA |
| Csa-MIR156A | GTATTGAAAATTAGAGTAAGGGGAAG | CTTCAAGCATGAACCCTAACAT |
| Csa-MIR156B | GAGAGAAAAGCACAAAAGACCAAGA | CTAAGTAAAGGTATGAACTTTCAACTT |
| 基因名称 Gene name | 正向引物(5′-3′) Forward primer | 反向引物(5′-3′) Reverse primer |
| Csa-MIR156C | GTTTATCGTTCTTGAATTTGGTTAG | ATACACTTTATAGCAACTGCCTCTG |
| Csa-MIR156D | TGAATGATGGTGAGTGTGTTAGGAG | CACGCACCCGCAAAGGTAT |
| Csa-MIR156E | AAAGGTGATTAAAATTGAGGGGAA | TGGCATATTATTATACCAAAAGCCT |
| Csa-MIR156F | CATACGGAAGGTAATCTCAAGGA | AATATCTAGCATAAGCGGCCATG |
| Csa-MIR156G | TCTCACGGTCTATTTCTAACACG | CACAAAGCTTCGAGCATTGATAT |
| Csa-MIR157A | GGGTTTAGAAATTTGGAGAGAGACA | TATTAAAAGTGAAGGGATGGGATATA |
| Csa-MIR157B | ATTTATCATGCACAAGGGAGAACTT | AATTGCTATTAGTGGTACCGATTGA |
| Csa-MIR157C | CCAATACGGTGATAGCTATATGTTGT | TAACAAAGTGGTGTTTACGACTCTA |
| Csa-Tub | CACTACACCGTTGGAAAGGAAA | CAAAAGGAGGGAGCCGAGA |
| Csa-U6 | GGGGACATCCGATAAAATT | TGTGCGTGTCATCCTTGC |
| Csa-miR156 | GCGGCGGTGACAGAAGAGAG | GTGCAGGGTCCGAGGT |
| Csa-miR156 RT | GTCGTATCCAGTGCAGGGTCCGAGGTATTCGCAC | TGGATACGACGTGCTC |
| 基因名称 Gene name | 基因编号 Gene ID | 长度/bp Length | 染色体 Chromosome | 位置 Location |
|---|---|---|---|---|
| Csa-SPL1 | Csa1G001450 | 379 | Chr1 | 237765..239802 (+) |
| Csa-SPL2 | Csa1G015680 | 550 | Chr1 | 2088067..2093861 (+) |
| Csa-SPL3 | Csa1G039890 | 548 | Chr1 | 3934043..3940006 (-) |
| Csa-SPL4 | Csa1G051590 | 314 | Chr1 | 6001183..6004194 (-) |
| Csa-SPL5 | Csa1G074980 | 162 | Chr1 | 7700146..7703682 (+) |
| Csa-SPL6 | Csa3G117960 | 340 | Chr3 | 6445761..6448246 (+) |
| Csa-SPL7 | Csa3G567830 | 344 | Chr3 | 22275377..22277440 (+) |
| Csa-SPL8 | Csa3G664550 | 1 013 | Chr3 | 25953081..25964518 (+) |
| Csa-SPL9 | Csa3G809420 | 382 | Chr3 | 30985575..30991777 (+) |
| Csa-SPL10 | Csa4G631590 | 202 | Chr4 | 20657444..20658877 (-) |
| Csa-SPL11 | Csa4G664590 | 1 031 | Chr4 | 23212471..23218465 (+) |
| Csa-SPL12 | Csa6G094760 | 328 | Chr6 | 6509944..6513210 (+) |
| Csa-SPL13 | Csa6G109120 | 297 | Chr6 | 7313479..7317016 (-) |
| Csa-SPL14 | Csa6G517960 | 141 | Chr6 | 27284755..27286747 (+) |
| Csa-MIR156A | 127 | Chr1 | 13072567..13072694 (+) | |
| Csa-MIR156B | 112 | Chr2 | 1701701..1701813 (+) | |
| Csa-MIR156C | 121 | Chr2 | 19844384..19844505 (+) | |
| Csa-MIR156D | 175 | Chr4 | 22021976..22022151 (+) | |
| Csa-MIR156E | 138 | Chr4 | 22620675..22620813 (-) | |
| Csa-MIR156F | 89 | Chr1 | 22345006..22345095 (+) | |
| Csa-MIR156G | 148 | Chr6 | 16464371..16464519 (-) | |
| Csa-MIR157A | 165 | Chr1 | 2522433..2522598 (+) | |
| Csa-MIR157B | 177 | Chr3 | 22790035..22790212 (-) | |
| Csa-MIR157C | 85 | Chr2 | 20893540..20893625 (-) |
Table 2 The information of miR156/157-SPL pathway genes in cucumber
| 基因名称 Gene name | 基因编号 Gene ID | 长度/bp Length | 染色体 Chromosome | 位置 Location |
|---|---|---|---|---|
| Csa-SPL1 | Csa1G001450 | 379 | Chr1 | 237765..239802 (+) |
| Csa-SPL2 | Csa1G015680 | 550 | Chr1 | 2088067..2093861 (+) |
| Csa-SPL3 | Csa1G039890 | 548 | Chr1 | 3934043..3940006 (-) |
| Csa-SPL4 | Csa1G051590 | 314 | Chr1 | 6001183..6004194 (-) |
| Csa-SPL5 | Csa1G074980 | 162 | Chr1 | 7700146..7703682 (+) |
| Csa-SPL6 | Csa3G117960 | 340 | Chr3 | 6445761..6448246 (+) |
| Csa-SPL7 | Csa3G567830 | 344 | Chr3 | 22275377..22277440 (+) |
| Csa-SPL8 | Csa3G664550 | 1 013 | Chr3 | 25953081..25964518 (+) |
| Csa-SPL9 | Csa3G809420 | 382 | Chr3 | 30985575..30991777 (+) |
| Csa-SPL10 | Csa4G631590 | 202 | Chr4 | 20657444..20658877 (-) |
| Csa-SPL11 | Csa4G664590 | 1 031 | Chr4 | 23212471..23218465 (+) |
| Csa-SPL12 | Csa6G094760 | 328 | Chr6 | 6509944..6513210 (+) |
| Csa-SPL13 | Csa6G109120 | 297 | Chr6 | 7313479..7317016 (-) |
| Csa-SPL14 | Csa6G517960 | 141 | Chr6 | 27284755..27286747 (+) |
| Csa-MIR156A | 127 | Chr1 | 13072567..13072694 (+) | |
| Csa-MIR156B | 112 | Chr2 | 1701701..1701813 (+) | |
| Csa-MIR156C | 121 | Chr2 | 19844384..19844505 (+) | |
| Csa-MIR156D | 175 | Chr4 | 22021976..22022151 (+) | |
| Csa-MIR156E | 138 | Chr4 | 22620675..22620813 (-) | |
| Csa-MIR156F | 89 | Chr1 | 22345006..22345095 (+) | |
| Csa-MIR156G | 148 | Chr6 | 16464371..16464519 (-) | |
| Csa-MIR157A | 165 | Chr1 | 2522433..2522598 (+) | |
| Csa-MIR157B | 177 | Chr3 | 22790035..22790212 (-) | |
| Csa-MIR157C | 85 | Chr2 | 20893540..20893625 (-) |
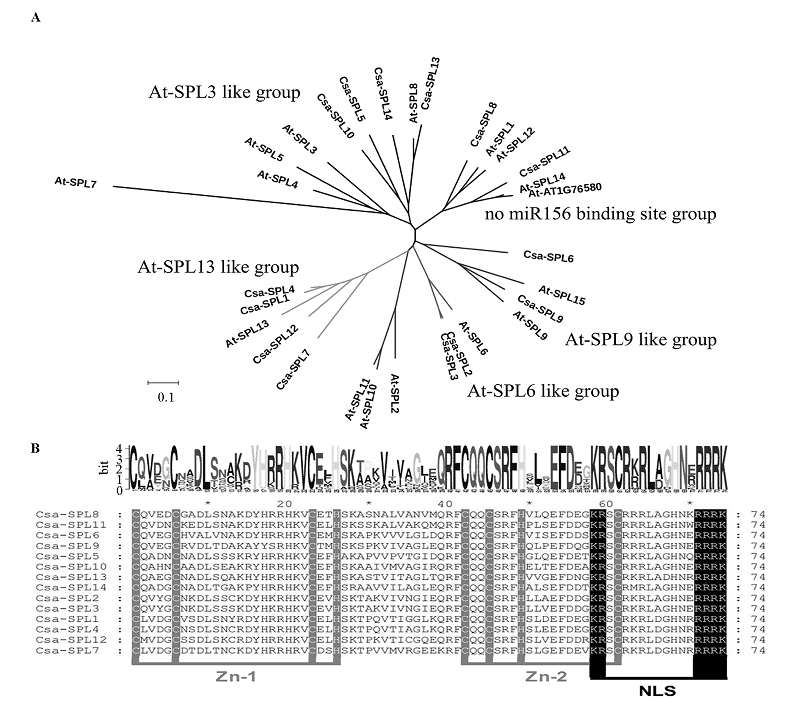
Fig. 4 NJ tree analyzing(A)and alignment of SBP domain(B)results of Csa-SPL protein sequences Zn_1 and Zn_2 indicate the Zinc finger domains,NLS indicates the nuclear localization signal.
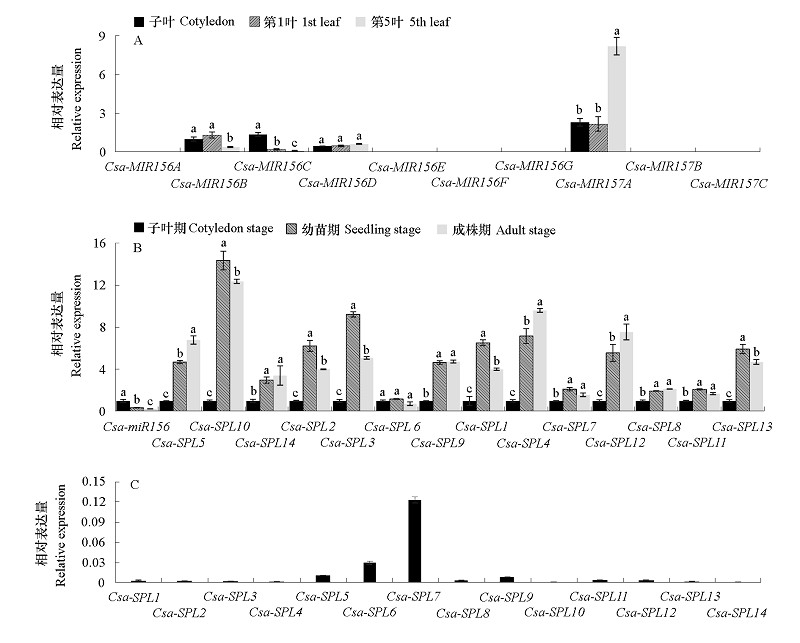
Fig. 7 Detection of miR156/157-SPLs pathway genes expression in cucumber A. Expression of Csa-MIR156/157 in different leaves;B. Expression of Csa-miR156 and Csa-SPL in different stages(The gene expression levels in cotyledon stage are normalized to 1);C. Expression of Csa-SPLs in cotyledon stage. Different letters represent significant differences at the level of 0.05.
| [1] |
Birkenbihl R P, Jach G, Saedler H, Huijser P. 2005. Functional dissection of the plant-specific SBP-domain:overlap of the DNA-binding and nuclear localization domains. Journal of Molecular Biology, 352 (3):585-596.
pmid: 16095614 |
| [2] |
Cui L, Zheng F, Wang J, Zhang C, Xiao F, Ye J, Li C, Ye Z, Zhang J. 2020. miR156a-targeted SBP-Box transcription factor SlSPL 13 regulates inflorescence morphogenesis by directly activating SFT in tomato. Plant Biotechnology Journal, 18 (8):1670-1682.
doi: 10.1111/pbi.v18.8 URL |
| [3] |
Gou J, Felippes F F, Liu C, Weigel D, Wang J. 2011. Negative regulation of anthocyanin biosynthesis in Arabidopsis by a miR156-targeted SPL transcription Factor. The Plant Cell, 23 (4):1512-1522.
doi: 10.1105/tpc.111.084525 URL |
| [4] |
He J, Xu M, Willmann M R, McCormick K, Hu T, Yang L, Starker C G, Voytas D F, Meyers B C, Poethig R S. 2018. Threshold-dependent repression of SPL gene expression by miR156/miR157 controls vegetative phase change in Arabidopsis thaliana. PLoS Genetics, 14 (4):e1007337.
doi: 10.1371/journal.pgen.1007337 URL |
| [5] |
Jiao Y, Wang Y, Xue D, Wang J, Yan M, Liu G, Dong G, Zeng D, Lu Z, Zhu X, Qian Q, Li J. 2010. Regulation of OsSPL14 by OsmiR156 defines ideal plant architecture in rice. Nature Genetics, 42 (6):541-544.
doi: 10.1038/ng.591 URL |
| [6] | Li Meng-ting. 2017. Cloning and preliminary function analysis of homologous SPL genes in cucumber(Cucumis sativus L.)[M. D. Dissertation]. Hangzhou: Hangzhou Normal University. (in Chinese) |
| 李梦婷. 2017. 黄瓜SPL同源基因的克隆及功能的初步研究[硕士论文]. 杭州: 杭州师范大学. | |
| [7] |
Liu Q, Harberd N P, Fu X. 2016a. SQUAMOSA promoter binding protein-like transcription factors: targets for improving cereal grain yield. Molecular Plant, 9 (6):765-767.
doi: 10.1016/j.molp.2016.04.008 URL |
| [8] | Liu R, Lai B, Hu B, Qin Y, Hu G, Zhao J. 2016b. Identification of MicroRNAs and their target genes related to the accumulation of anthocyanins in Litchi chinensis by high-throughput sequencing and degradome analysis. Frontiers in Plant Science, 7:2059. |
| [9] |
Luo Y, Guo Z, Li L. 2013. Evolutionary conservation of microRNA regulatory programs in plant flower development. Development Biology, 380 (2):133-144.
doi: 10.1016/j.ydbio.2013.05.009 URL |
| [10] |
Ma Y, Xue H, Zhang F, Jiang Q, Yang S, Yue P, Wang F, Zhang Y, Li L, He P, Zhang Z. 2020. The miR156/SPL module regulates apple salt stress tolerance by activating MdWRKY100 expression. Plant Biotechnology Journal, 19 (2):311-323.
doi: 10.1111/pbi.v19.2 URL |
| [11] |
Mao W, Li Z, Xia X, Li Y, Yu J. 2012. A combined approach of high-throughput sequencing and degradome analysis reveals tissue specific expression of microRNAs and their targets in cucumber. PLoS ONE, 7 (3):e33040.
doi: 10.1371/journal.pone.0033040 URL |
| [12] |
Martinez G, Forment J, Llave C, Pallas V, Gomez G. 2011. High-throughput sequencing, characterization and detection of new and conserved cucumber miRNAs. PLoS ONE, 6 (5):e19523.
doi: 10.1371/journal.pone.0019523 URL |
| [13] |
Rubio-Somoza I, Zhou C, Confraria A, Martinho C, von Born P, Baena-Gonzalez E, Wang J, Weigel D. 2014. Temporal control of leaf complexity by miRNA-regulated licensing of protein complexes. Current Biology, 24 (22):2714-2719.
doi: 10.1016/j.cub.2014.09.058 pmid: 25448000 |
| [14] |
Schwab R, Palatnik J F, Riester M, Schommer C, Schmid M, Weigel D. 2005. Specific effects of microRNAs on the plant transcriptome. Developmental Cell, 8 (4):517-527.
doi: 10.1016/j.devcel.2005.01.018 URL |
| [15] |
Silva G F F E, Silva E M, Da Silva Azevedo M, Guivin M A C, Ramiro D A, Figueiredo C R, Carrer H, Peres L E P, Nogueira F T S. 2014. microRNA156-targeted SPL/SBP box transcription factors regulate tomato ovary and fruit development. The Plant Journal, 78 (4):604-618.
doi: 10.1111/tpj.12493 URL |
| [16] |
Wang J, Czech B, Weigel D. 2009. miR156-regulated SPL transcription factors define an endogenous flowering pathway in Arabidopsis thaliana. Cell, 138 (4):738-749.
doi: 10.1016/j.cell.2009.06.014 URL |
| [17] |
Wang J, Zhou L, Shi H, Chern M, Yu H, Yi H, He M, Yin J, Zhu X, Li Y, Li W, Liu J, Wang J, Chen X, Qing H, Wang Y, Liu G, Wang W, Li P, Wu X, Zhu L, Zhou J M, Ronald P C, Li S, Li J, Chen X. 2018. A single transcription factor promotes both yield and immunity in rice. Science, 361 (6406):1026-1028.
doi: 10.1126/science.aat7675 URL |
| [18] | Wang L, Zhou C M, Mai Y X, Li L Z, Gao J, Shang G D, Lian H, Han L, Zhang T Q, Tang H B, Ren H, Wang F X, Wu L Y, Liu X L, Wang C S, Chen E W, Zhang X N, Liu C, Wang J W. 2019. A spatiotemporally regulated transcriptional complex underlies heteroblastic development of leaf hairs in Arabidopsis thaliana. EMBO Journal, 38 (8):e100063. |
| [19] |
Wang Y, Wu F, Bai J, He Y. 2014. BrpSPL9(Brassica rapa ssp. pekinensis SPL9)controls the earliness of heading time in Chinese cabbage. Plant Biotechnology Journal, 12 (3):312-321.
doi: 10.1111/pbi.2014.12.issue-3 URL |
| [20] |
Willmann M R, Poethig R S. 2007. Conservation and evolution of miRNA regulatory programs in plant development. Current Opinion in Plant Biology, 10 (5):503-511.
pmid: 17709279 |
| [21] |
Wu G, Park M Y, Conway S R, Wang J, Weigel D, Poethig R S. 2009. The sequential action of miR156 and miR172 regulates developmental timing in Arabidopsis. Cell, 138 (4):750-759.
doi: 10.1016/j.cell.2009.06.031 URL |
| [22] |
Wu G, Poethig R S. 2006. Temporal regulation of shoot development in Arabidopsis thaliana by miR156 and its target SPL3. Development, 133 (18):3539-3547.
doi: 10.1242/dev.02521 URL |
| [23] | Xu M, Hu T, Zhao J, Park M, Earley K W, Wu G, Yang L, Poethig R S. 2016. Developmental Functions of miR156-regulated SQUAMOSA PROMOTER BINDING PROTEIN-LIKE (SPL) genes in Arabidopsis thaliana. PLoS Genetics, 12 (8):e1006263. |
| [24] |
Xu Y, Qian Z, Zhou B, Wu G. 2019. Age-dependent heteroblastic development of leaf hairs in Arabidopsis. New Phytologist, 224 (2):741-748.
doi: 10.1111/nph.v224.2 URL |
| [25] |
Yamasaki K, Kigawa T, Inoue M, Tateno M, Yamasaki T, Yabuki T, Aoki M, Seki E, Matsuda T, Nunokawa E, Ishizuka Y, Terada T, Shirouzu M, Osanai T, Tanaka A, Seki M, Shinozaki K, Yokoyama S. 2004. A novel zinc-binding motif revealed by solution structures of DNA-binding domains of Arabidopsis SBP-family transcription factors. Journal of Molecular Biology, 337 (1):49-63.
doi: 10.1016/j.jmb.2004.01.015 URL |
| [26] |
Yu N, Niu Q, Ng K, Chua N. 2015. The role of miR156/SPLs modules in Arabidopsis lateral root development. The Plant Journal, 83 (4):673-685.
doi: 10.1111/tpj.12919 pmid: 26096676 |
| [27] |
Zhang H, Zhang L, Han J, Qian Z, Zhou B, Xu Y, Wu G. 2019. The nuclear localization signal is required for the function of squamosa promoter binding protein-like gene 9 to promote vegetative phase change in Arabidopsis. Plant Molecular Biology, 100 (6):571-578.
doi: 10.1007/s11103-019-00863-5 pmid: 30953277 |
| [28] |
Zhang X, Zou Z, Zhang J, Zhang Y, Han Q, Hu T, Xu X, Liu H, Li H, Ye Z. 2011. Over-expression of sly-miR156a in tomato results in multiple vegetative and reproductive trait alterations and partial phenocopy of the sft mutant. FEBS Letters, 585 (2):435-439.
doi: 10.1016/j.febslet.2010.12.036 URL |
| [29] |
Zheng C, Ye M, Sang M, Wu R. 2019. A regulatory network for miR156-SPL module in Arabidopsis thaliana. International Journal of Molecular Science, 20 (24):6166.
doi: 10.3390/ijms20246166 URL |
| [1] | WANG Xiaochen, NIE Ziye, LIU Xianju, DUAN Wei, FAN Peige, and LIANG Zhenchang, . Effects of Abscisic Acid on Monoterpene Synthesis in‘Jingxiangyu’Grape Berries [J]. Acta Horticulturae Sinica, 2023, 50(2): 237-249. |
| [2] | ZHAI Hanhan, ZHAI Yujie, TIAN Yi, ZHANG Ye, YANG Li, WEN Zhiliang, CHEN Haijiang. Genome-wide Identification of Peach SAUR Gene Family and Characterization of PpSAUR5 Gene [J]. Acta Horticulturae Sinica, 2023, 50(1): 1-14. |
| [3] | LUO Tiankuan, WU Haitao, ZHANG Shengmei, HUANG Zong’an, SUN Ji, SHUI Deju, and CHEN Xianzhi . A New Cucumber Cultivar‘Oucui 1’ [J]. Acta Horticulturae Sinica, 2022, 49(S2): 125-126. |
| [4] | WANG Hebing, XIANG Huafeng, CHEN Xinzhong, ZHANG Sheng, and ZHANG Hongcheng. A New Cucumber Hybrid‘Xinyan 095’ [J]. Acta Horticulturae Sinica, 2022, 49(S1): 79-80. |
| [5] | XU Chunmei, ZHANG Zuobiao, LIU Jinglan, WANG Xin, YANG Long, ZHAO Dan, LIU Siyu, JIA Yunhe, MENG Xuejiao, and CUI Songcen. A New Cucumber Cultivar‘Lüchun 2’ [J]. Acta Horticulturae Sinica, 2022, 49(S1): 81-82. |
| [6] | ZHANG Lidong, HUANG Hongyu, KONG Weiliang, LI Jiawang, and LI Yuhe, . A New Cucumber Cultivar of North China Type‘Jinyou 355’ [J]. Acta Horticulturae Sinica, 2022, 49(S1): 83-84. |
| [7] | WANG Huizhe, YA NG Ruihuan, DENG Qiang, CAO Mingming, and LI Shuju, . A New Cucumber Cultivar‘Jindong 369’Resistant to Scab [J]. Acta Horticulturae Sinica, 2022, 49(S1): 85-86. |
| [8] | NIE Xinmiao, LUAN Heng, FENG Gaili, WANG Chao, LI Yan, WEI Min. Effects of Silicon Nutrition and Grafting Rootstocks on Chilling Tolerance of Cucumber Seedlings [J]. Acta Horticulturae Sinica, 2022, 49(8): 1795-1804. |
| [9] | ZHANG Qiuyue, LIU Changlai, YU Xiaojing, YANG Jiading, FENG Chaonian. Screening of Reference Genes for Differentially Expressed Genes in Pyrus betulaefolia Plant Under Salt Stress by qRT-PCR [J]. Acta Horticulturae Sinica, 2022, 49(7): 1557-1570. |
| [10] | HAN Lujie, FENG Yiqing, YANG Xiuhua, ZHANG Ning, BI Huangai, AI Xizhen. Effects of Combined Application of Organic and Chemical Fertilizers on Root Zone Soil and Root Characteristics of Cucumber in Plastic Greenhouse [J]. Acta Horticulturae Sinica, 2022, 49(5): 1047-1059. |
| [11] | LI Yamei, MA Fuli, ZHANG Shanqi, HUANG Jinqiu, CHEN Mengting, ZHOU Junyong, SUN Qibao, SUN Jun. Optimization of Jujube Callus Transformation System and Application of ZjBRC1 in Regulating ZjYUCCA Expression [J]. Acta Horticulturae Sinica, 2022, 49(4): 749-757. |
| [12] | WANG Ying, AI Penghui, LI Shuailei, KANG Dongru, LI Zhongai, WANG Zicheng. Identification and Expression Analysis of Genes Related to DNA Methylation in Chrysanthemum × morifolium and C. nankingense [J]. Acta Horticulturae Sinica, 2022, 49(4): 827-840. |
| [13] | QUAN Jianhua, DUAN Yu, LUO Tian, YUAN Qiang, QI Xin, WANG Qinli. A New Cucumber Cultivar‘Yuyan 9’ [J]. Acta Horticulturae Sinica, 2022, 49(3): 703-704. |
| [14] | ZHANG Rui, ZHANG Xiayi, ZHAO Ting, WANG Shuangcheng, ZHANG Zhongxing, LIU Bo, ZHANG De, WANG Yanxiu. Transcriptome Analysis of the Molecular Mechanism of Saline-alkali Stress Response in Malus halliana Leaves [J]. Acta Horticulturae Sinica, 2022, 49(2): 237-251. |
| [15] | ZHOU Zhiming, YANG Jiabao, ZHANG Cheng, ZENG Linglu, MENG Wanqiu, SUN Li. Genome-wide Identification and Expression Analyses of Long-chain Acyl-CoA Synthetases Under Abiotic Stresses in Helianthus annuus [J]. Acta Horticulturae Sinica, 2022, 49(2): 352-364. |
| Viewed | ||||||
|
Full text |
|
|||||
|
Abstract |
|
|||||
Copyright © 2012 Acta Horticulturae Sinica 京ICP备10030308号-2 国际联网备案号 11010802023439
Tel: 010-82109523 E-Mail: yuanyixuebao@126.com
Support by: Beijing Magtech Co.Ltd