
Acta Horticulturae Sinica ›› 2021, Vol. 48 ›› Issue (6): 1208-1216.doi: 10.16420/j.issn.0513-353x.2020-0750
• Research Notes • Previous Articles Next Articles
YANG Fei1, ZHANG Aihong1, WANG Xiuzhi2, LI Xiwang1, DI Dianping1,*, MIAO Hongqin1, ZHANG Shangqing3, SUN Xiangrui1, SUN Qian1
Received:2020-11-18
Revised:2021-03-10
Online:2021-06-25
Published:2021-07-08
Contact:
DI Dianping
CLC Number:
YANG Fei, ZHANG Aihong, WANG Xiuzhi, LI Xiwang, DI Dianping, MIAO Hongqin, ZHANG Shangqing, SUN Xiangrui, SUN Qian. Identification of Tomato spotted wilt orthotospovirus in Chifeng,Inner Mongolia[J]. Acta Horticulturae Sinica, 2021, 48(6): 1208-1216.
Add to citation manager EndNote|Ris|BibTeX
URL: https://www.ahs.ac.cn/EN/10.16420/j.issn.0513-353x.2020-0750
| 序号 No. | Genbank登录号 Genbank Accession No. | 地区 Area | 寄主 Host |
|---|---|---|---|
| 1 | MN365037 | 中国云南Yunnan China | 花生Arachis hypogaea |
| 2 | MF688996 | 中国云南Yunnan China | 桔梗Platycodon grandiflorus |
| 3 | KY495612 | 中国云南Yunnan China | 刺天茄Solanum indicum |
| 4 | KU976396 | 中国江苏Jiangsu China | 莴苣Lactuca sativa |
| 5 | MG878875 | 中国黑龙江Helongjiang China | 番茄Solanum lycopersicum |
| 6 | MF422034 | 中国云南Yunnan China | 旱金莲Tropaeolum majus |
| 7 | KU356854 | 中国云南Yunnan China | 芹菜Apium graveliens |
| 8 | MG593199 | 美国USA | 菊苣Cichorium intybus |
| 9 | KU179603 | 美国USA | 球兰Hoya carnosa |
| 10 | KC261949 | 韩国Korea | 辣椒Capsicum annuum |
| 11 | KU179561 | 美国USA | 番茄Solanum lycopersicum |
| 12 | KU179533 | 美国USA | 辣椒Capsicum annuum |
| 13 | MG983521 | 意大利Italy | 辣椒Capsicum annuum |
| 14 | KC261958 | 韩国Korea | 辣椒Capsicum annuum |
| 15 | KC261961 | 韩国Korea | 山莴苣Lactuca indica |
| 16 | KC261970 | 韩国Korea | 番茄Solanum lycopersicum |
| 17 | KM365066 | 澳大利亚Australia | 番茄Solanum lycopersicum |
| 18 | AB088385 | 日本Janpan | — |
| 19 | MG602673 | 南非South Africa | 菊花Dendranthema morifolium |
| 20 | AJ418781 | 德国Germany | 金钱草Lysimachia christinaeHance |
| 21 | KJ649612 | 匈牙利Hungary | 辣椒Capsicum annuum |
| 22 | MH367502 | 土耳其Turkey | 番茄Solanum lycopersicum |
Table 1 Sequence information of TSWV isolates used in constructing phylogenetic tree
| 序号 No. | Genbank登录号 Genbank Accession No. | 地区 Area | 寄主 Host |
|---|---|---|---|
| 1 | MN365037 | 中国云南Yunnan China | 花生Arachis hypogaea |
| 2 | MF688996 | 中国云南Yunnan China | 桔梗Platycodon grandiflorus |
| 3 | KY495612 | 中国云南Yunnan China | 刺天茄Solanum indicum |
| 4 | KU976396 | 中国江苏Jiangsu China | 莴苣Lactuca sativa |
| 5 | MG878875 | 中国黑龙江Helongjiang China | 番茄Solanum lycopersicum |
| 6 | MF422034 | 中国云南Yunnan China | 旱金莲Tropaeolum majus |
| 7 | KU356854 | 中国云南Yunnan China | 芹菜Apium graveliens |
| 8 | MG593199 | 美国USA | 菊苣Cichorium intybus |
| 9 | KU179603 | 美国USA | 球兰Hoya carnosa |
| 10 | KC261949 | 韩国Korea | 辣椒Capsicum annuum |
| 11 | KU179561 | 美国USA | 番茄Solanum lycopersicum |
| 12 | KU179533 | 美国USA | 辣椒Capsicum annuum |
| 13 | MG983521 | 意大利Italy | 辣椒Capsicum annuum |
| 14 | KC261958 | 韩国Korea | 辣椒Capsicum annuum |
| 15 | KC261961 | 韩国Korea | 山莴苣Lactuca indica |
| 16 | KC261970 | 韩国Korea | 番茄Solanum lycopersicum |
| 17 | KM365066 | 澳大利亚Australia | 番茄Solanum lycopersicum |
| 18 | AB088385 | 日本Janpan | — |
| 19 | MG602673 | 南非South Africa | 菊花Dendranthema morifolium |
| 20 | AJ418781 | 德国Germany | 金钱草Lysimachia christinaeHance |
| 21 | KJ649612 | 匈牙利Hungary | 辣椒Capsicum annuum |
| 22 | MH367502 | 土耳其Turkey | 番茄Solanum lycopersicum |

Fig. 1 Symptoms of TSWV in tomato fruit,leaves and plants A:Shrinkage;B:Uneven coloration;C:Yellow halo;D:Dappled;E:Bright yellow leaves with brown spots;F:Symptoms of tomato bitten by thrips;G,H:Symptoms of tomato plant.
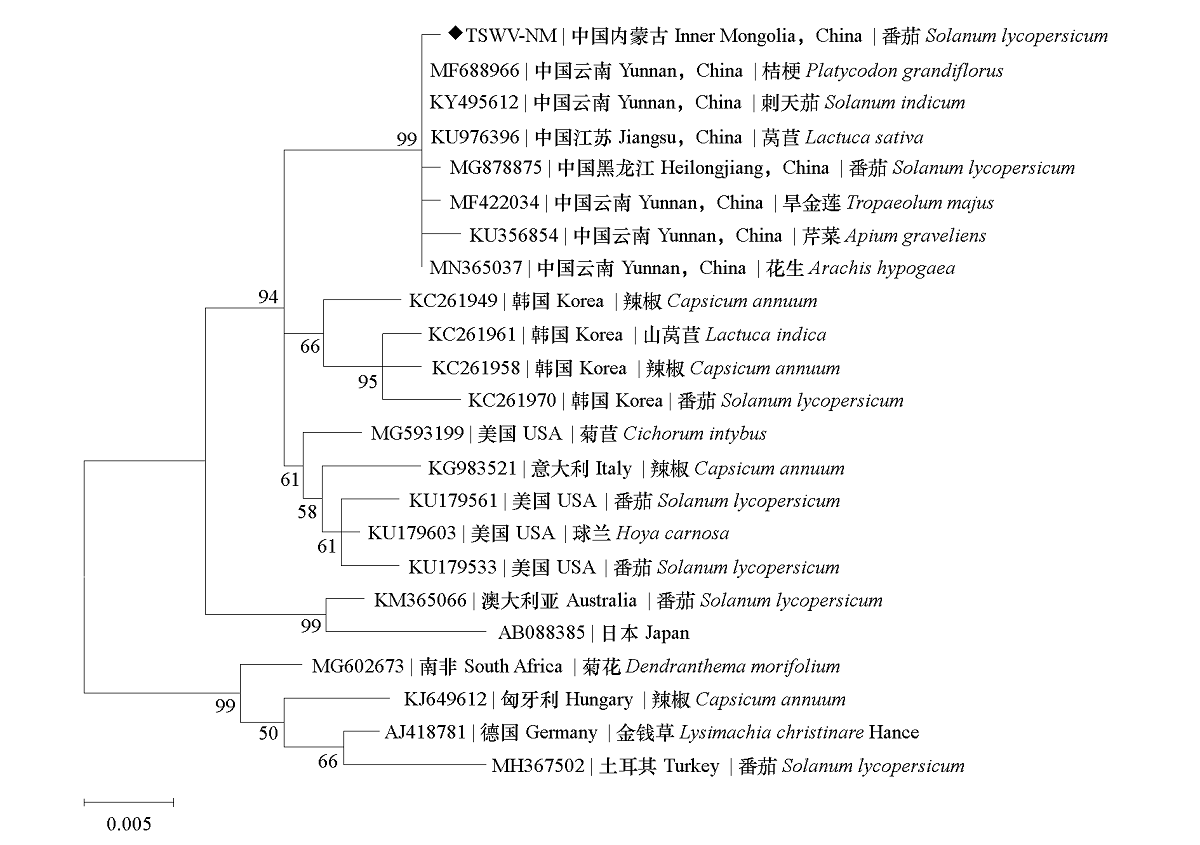
Fig. 4 Phylogenetic tree based on 914 nt fragment containing N gene sequence of different isolates of TSWV The bootstrap confidence values for each branch are shown above or below the branch. The value of the distance scale represents the nucleotide substitution rate, and 0.005 represents 5 different nucleotides per 1 000 nucleotides. ◆ Isolate of TSWV sequenced in this study.

Fig. 5 Symptoms of pepper induced by sap inoculation of infected tomato fruit juice(28 d) Control:Plants inoculated with phosphate buffer;TSWV:Plants inoculated with diseased tomato fruit juice.
| [1] | Ashfaq M, Ahamd A. 2017. First report of tomato spotted wilt virus in hot pepper in Pakistan. Journal of Plant Pathology, 99 (1):287-304. |
| [2] | Brittlebank C C. 1919. Tomato diseases. Journal of Agriculture, 17:231-235. |
| [3] | Cao Jin-qiang, Xie Xue-wen, Chai A-li, Li Bao-ju. 2016. Identification and control of tomato spotted wilt virus in Ningxia. China Vegetables,(4):87-89. (in Chinese) |
| 曹金强, 谢学文, 柴阿丽, 李宝聚. 2016. 李宝聚博士诊病手记(九十三)番茄斑萎病毒病在宁夏的发现与防治. 中国蔬菜,(4):87-89. | |
| [4] | Chen Dong-liang, Li Ming-yuan, Li Xue-mei, Wang Ling-ling, Huang Cong-lin. 2018. Identifcation of tomato spotted wilt virus from Lettuce in Beijing. China Vegetables,(1):65-69. (in Chinese) |
| 陈东亮, 李明远, 李雪梅, 王玲玲, 黄丛林. 2018. 北京地区莴苣中番茄斑萎病毒的鉴定. 中国蔬菜,(1):65-69. | |
| [5] |
Cho S Y, Kim S M, Kim S, Lee B C. 2020. First report of tomato spotted wilt virus infecting Arachis hypogaea in Korea. Journal of Plant Pathology, 102 (1):271.
doi: 10.1007/s42161-019-00410-7 URL |
| [6] | Choi S K, Choi G S. 2015. First report of tomato spotted wilt virus in Solanum tuberosum in Korea. Plant Disease, 99 (11):1657-1658. |
| [7] |
Crowley N C. 1957. Studies on the seed transmission of plant virus diseases. Australian Journal of Biological Sciences, 10 (4):449-464.
doi: 10.1071/BI9570449 URL |
| [8] |
Delić D, Balech B, Radulović M, Đurić Z, Lolić B, Santamaria M, Đurić G. 2017. Molecular identification of tomato spotted wilt virus on pepper and tobacco in Republic of Srpska (Bosnia and Herzegovina). European Journal of Plant Pathology, 150:785-789.
doi: 10.1007/s10658-017-1313-7 URL |
| [9] | Feng Li-xia, Wu Mu-tao, Yu Cui, Wu Xing-hai. 2017. Detection and identification of tomato spotted wilt virus in lettuce seeds from American. China Plant Protection, 37 (1):9-12. (in Chinese) |
| 冯黎霞, 武目涛, 于翠, 吴兴海. 2017. 由美国进境生菜种子上番茄斑萎病毒的检测鉴定. 中国植保导刊, 37 (1):9-12. | |
| [10] |
Goldbach R, Peters D. 1994. Possible causes of the emergence of tospovirus diseases. Seminars in Virology, 5:113-120.
doi: 10.1006/smvy.1994.1012 URL |
| [11] |
Kohnić A, Radulović M, Delić D. 2019. First report of tomato spotted wilt virus on Chrysanthemum in Bosnia and Herzegovina. Journal of Plant Pathology, 101:421.
doi: 10.1007/s42161-018-0195-7 URL |
| [12] | Li Fei, Wu Qing-jun, Xu Bao-yun, Xie Wen, Wang Shao-li, Zhang You-jun. 2012. Tomato spotted wilt virus was identified in Beijing. Plant Protection, 38 (6):186-188. (in Chinese) |
| 李飞, 吴青君, 徐宝云, 谢文, 王少丽, 张友军. 2012. 北京地区发现番茄斑萎病毒. 植物保护, 38 (6):186-188. | |
| [13] | Li Jie, Chi Sheng-qi, Yang Qin-min, Ding Tian-bo, Chu Dong. 2017. Identification of tomato spotted wilt virus s disease in Shandong,China. Plant Protection, 43 (1):228-232. (in Chinese) |
| 李洁, 迟胜起, 杨勤民, 丁天波, 褚栋. 2017. 山东烟台地区发生番茄斑萎病毒病危害. 植物保护, 43 (1):228-232. | |
| [14] | Liu Yi-di. 2019. Gongzhuling Agricultural co-operative Industrial Park,Damiao town,Chifeng:tomatoes“transformed”into a new driving force for poverty alleviation and prosperity. CNR,Oct.18th: http://news.cnr.cn/dj/20191018/t20191018_524821159.shtml. (in Chinese) |
| 刘一荻. 2019. 赤峰大庙镇公主岭农业合作产业园:西红柿“变身”脱贫致富新动力. 央广网,10月18日: http://news.cnr.cn/dj/20191018/t20191018_524821159.shtml. | |
| [15] |
Mason G, Roggero P, Tavella L. 2003. Detection of tomato spotted wilt virus in its vector Frankliniella occidentalis by reverse transcription-polymerase chain reaction. Journal of Virological Methods, 109 (1):69-73.
doi: 10.1016/S0166-0934(03)00048-X URL |
| [16] | Ohnishi J, Katsuzaki H, Tsuda S, Sakurai T, Murai T. 2006. Frankliniella cephalica,a new vector for tomato spotted wilt virus. Plant Disease, 90 (5):685-685. |
| [17] |
Pappu H R, Jones R A C, Jain R K. 2009. Global status of Tospovirus epidemics in diverse cropping systems:successes achieved and challenges ahead. Virus Research, 141 (2):219-236.
doi: 10.1016/j.virusres.2009.01.009 URL |
| [18] |
Reitz S R. 2009. Biology and Ecology of the western flower thrips (Thysanoptera:Thripidae):the making of a pest. Florida Entomologist, 92 (1):7-13.
doi: 10.1653/024.092.0102 URL |
| [19] |
Rodríguez-Román E, Mejías A, Marys E. 2018. First detection of tomato spotted wilt virus in tomato in Venezuela. Journal of Plant Pathology, 100:363.
doi: 10.1007/s42161-018-0074-2 URL |
| [20] | Samuel G, Bald J G, Pittman H A. 1930. Investigations on‘spotted wilt'of tomatoes. Australia Council for Scientific & Industrial Research Bulletin. 44,8-11. |
| [21] |
Scholthof K B G, Adkins S, Czosnek H, Palukaitis P, Jacquot E, Hohn T, Hohn B, Saunders K, Candresse T, Ahlquist P, Hemenway C, Foster G D. 2011. Top 10 plant viruses in molecular plant pathology. Molecular Plant Pathology, 12 (9):938-954.
doi: 10.1111/j.1364-3703.2011.00752.x URL |
| [22] | Song Xiao-yu, Liu Yong, Chen Jian-bin, Shi Xiao-bin, Zhang De-yong. 2019. Systemic infection of Chinese garlic by Tomato spotted wilt tospovirus. Plant Protection, 45 (3):149-151. (in Chinese) |
| 宋晓宇, 刘勇, 陈建斌, 史晓斌, 张德咏. 2019. 番茄斑萎病毒系统侵染我国大蒜. 植物保护, 45 (3):149-151. | |
| [23] | Su Da-kun, Yuan Xuan-ze, Xie Yong-hong, Wang Shu-rong, Ding Hai. 1987. Tomato spotted wilt virus in tomato in Chengdu and Dukou. Acta Phytopathologica Sinica, 17 (4):255-256. (in Chinese) |
| 苏大昆, 袁宣泽, 谢永红, 王淑荣, 丁海. 1987. 成都、渡口两市番茄中检出番茄斑萎病毒. 植物病理学报, 17 (4):255-256. | |
| [24] | Wang Kai-na, Zhan Bin-hui, Zhou Xue-ping. 2019. Detection and partial biological characterization of Tomato spotted wilt tospovirus in Heilongjiang Province. Plant Protection, 45 (1):37-43. (in Chinese) |
| 王凯娜, 战斌慧, 周雪平. 2019. 黑龙江地区番茄斑萎病毒的鉴定及其部分生物学特征分析. 植物保护, 45 (1):37-43. | |
| [25] |
Whitfield A E, Ullman D E, German T L. 2005. Tospovirus-thrips interactions. Annual Review of Phytopathology, 43 (1):459-489.
doi: 10.1146/annurev.phyto.43.040204.140017 URL |
| [26] | Wu Shuhua, Tu Liqin, Xian Wenrong, Yan Jiahui, Gao Danna, Ji Ying, Tao Xiaorong, Zhou Yijun, Guo Qingyun, Ji Yinghua. 2020. Detection and identification of Tomato spotted wilt tospovirus on pepperfrom Qinghai province. Acta Horticulturae Sinica, 47 (7):1391-1400. (in Chinese) |
| 吴淑华, 涂丽琴, 咸文荣, 闫佳会, 高丹娜, 吉颖, 陶小荣, 周益军, 郭青云, 季英华. 2020. 青海辣椒上番茄斑萎病毒检测及鉴定. 园艺学报, 47 (7):1391-1400. | |
| [27] | Xu Jing, Liu Shu-sheng. 2007. Biological interactions and biological invasion. Science, 59 (5):9-12. (in Chinese) |
| 徐婧, 刘树生. 2007. 生物互作与生物入侵. 科学, 59 (5):9-12. | |
| [28] | Xu Ze-yong, Reddy D V R., Rajeshwari R, Zhang Zong-yi, Huang Li-xin. 1986. A new peanut virus disease caused by tomato spotted wilt virus in southern China. Chinese Journal of Virology, 2 (3):271-274. (in Chinese) |
| 许泽永, Reddy D V R, Rajeshwari R, 张宗义, 黄立新. 1986. 我国南方花生发生一种由蕃茄斑萎病毒引起的新病害. 病毒学报, 2 (3):271-274. | |
| [29] | Zhang Zhong-kai, Fang Qi, Ding Ming, Peng Lu-bo, Zhang Li-zhen. 2000. Detection of infection of tomato spotted wilt virus(TSWV)on tobacco by electro-microscopy. Journal of Chinese Electron Microscopy Society, 19 (3):339-340. (in Chinese) |
| 张仲凯, 方琦, 丁铭, 彭潞波, 张丽珍. 2000. 侵染烟草的番茄斑萎病毒(TSWV)电镜诊断鉴定. 电子显微学报, 19 (3):339-340. |
| [1] | SHI Hongli, LI La, GUO Cuimei, YU Tingting, JIAN Wei, YANG Xingyong. Isolation,Identification and Analysis of Biocontrol Ability of Biocontrol Strain TL1 Against Tomato Botrytis cinerea [J]. Acta Horticulturae Sinica, 2023, 50(1): 79-90. |
| [2] | HU Jingyu, QUE Kaijuan, MIAO Tianli, WU Shaozheng, WANG Tiantian, ZHANG Lei, DONG Xian, JI Pengzhang, DONG Jiahong. Identification of Tomato Spotted Wilt Orthotospovirus Infecting Iris tectorum [J]. Acta Horticulturae Sinica, 2023, 50(1): 170-176. |
| [3] | ZHENG Jirong, WANG Tonglin, and HU Songshen. A New Tomato Cultivar‘Hangza 603’with High Quality [J]. Acta Horticulturae Sinica, 2022, 49(S2): 103-104. |
| [4] | ZHENG Jirong and WANG Tonglin. A New Tomato Cultivar‘Hangza 601’ [J]. Acta Horticulturae Sinica, 2022, 49(S2): 105-106. |
| [5] | ZHENG Jirong and WANG Tonglin. A New Cherry Tomato Cultivar‘Hangza 503’ [J]. Acta Horticulturae Sinica, 2022, 49(S2): 107-108. |
| [6] | HUANG Tingting, LIU Shuqin, ZHANG Yongzhi, LI Ping, ZHANG Zhihuan, and SONG Libo. A New Cherry Tomato Cultivar‘Yingshahong 4’ [J]. Acta Horticulturae Sinica, 2022, 49(S2): 109-110. |
| [7] | ZHANG Qianrong, LI Dazhong, QIU Boyin, LIN Hui, MA Huifei, YE Xinru, LIU Jianting, ZHU Haisheng, and WEN Qingfang. A New Tomato Cultivar‘Minnongke 2’ [J]. Acta Horticulturae Sinica, 2022, 49(S1): 73-74. |
| [8] | HAN Shuai, WU Jie, ZHANG Heqing, XI Yadong. Identification and Sequence Analysis of Tomato Spotted Wilt Orthotospovirus Infecting Lettuce in Sichuan [J]. Acta Horticulturae Sinica, 2022, 49(9): 2007-2016. |
| [9] | CHEN Lilang, YANG Tianzhang, CAI Ruping, LIN Xiaoman, DENG Nankang, CHE Haiyan, LIN Yating, KONG Xiangyi. Molecular Detection and Identification of Viruses from Passiflora edulis in Hainan [J]. Acta Horticulturae Sinica, 2022, 49(8): 1785-1794. |
| [10] | XIA Yan, HUANG Song, WU Xueli, LIU Yiqi, WANG Miaomiao, SONG Chunhui, BAI Tuanhui, SONG Shangwei, PANG Hongguang, JIAO Jian, ZHENG Xianbo. Identification and Analysis of Apple Viruses Diseases Based on Virome Sequencing Technology [J]. Acta Horticulturae Sinica, 2022, 49(7): 1415-1428. |
| [11] | ZHANG Qiuyue, LIU Changlai, YU Xiaojing, YANG Jiading, FENG Chaonian. Screening of Reference Genes for Differentially Expressed Genes in Pyrus betulaefolia Plant Under Salt Stress by qRT-PCR [J]. Acta Horticulturae Sinica, 2022, 49(7): 1557-1570. |
| [12] | LU Tao, YU Hongjun, LI Qiang, JIANG Weijie. Effects of Leaf and Fruit Quantity Regulation on Growth,Fruit Quality and Yield of Tomato [J]. Acta Horticulturae Sinica, 2022, 49(6): 1261-1274. |
| [13] | MENG Xianmin, CUI Qingqing, DUAN Yundan, ZHUANG Tuanjie, PU Dan, DONG Chunjuan, YANG Wencai, SHANG Qingmao. Promoting Effects of Uniconazole on Grafting Formation of Tomato Seedlings and Underlying Mechanisms [J]. Acta Horticulturae Sinica, 2022, 49(6): 1275-1289. |
| [14] | GONG Mingxia, ZHAO Hu, WANG Meng, WU Xing, ZHAO Zengjing, HE Zhi, HUANG Jinmei, MENG Shengde, WANG Risheng. Identification of Viruses Infecting Peppers in Guangxi by Small RNA Deep Sequencing and RT-PCR [J]. Acta Horticulturae Sinica, 2022, 49(5): 1060-1072. |
| [15] | CUI Dongyu, LI Changqing, SUN Yanxin, WANG Jiqing, ZOU Guoyuan, YANG Jungang. Effects of Dwarf Close Planting on Growth and Yield of Tomato Under East-West Cultivation in Greenhouse [J]. Acta Horticulturae Sinica, 2022, 49(4): 875-884. |
| Viewed | ||||||
|
Full text |
|
|||||
|
Abstract |
|
|||||
Copyright © 2012 Acta Horticulturae Sinica 京ICP备10030308号-2 国际联网备案号 11010802023439
Tel: 010-82109523 E-Mail: yuanyixuebao@126.com
Support by: Beijing Magtech Co.Ltd