
园艺学报 ›› 2025, Vol. 52 ›› Issue (12): 3167-3179.doi: 10.16420/j.issn.0513-353x.2024-1034
陈梦如, 吴庆安, 何思佳, 蔡祥斌, 马巧利, 辜青青, 魏清江*( )
)
收稿日期:2025-02-17
修回日期:2025-10-13
出版日期:2025-12-25
发布日期:2025-12-20
通讯作者:
基金资助:
CHEN Mengru, WU Qing’an, HE Sijia, CAI Xiangbin, MA Qiaoli, GU Qingqing, WEI Qingjiang*( )
)
Received:2025-02-17
Revised:2025-10-13
Published:2025-12-25
Online:2025-12-20
摘要:
为了解甲基化修饰酶基因在柑橘基因组中的特性,在全基因组水平对枳甲基化转移酶和去甲基化酶基因进行了鉴定与分析。结果表明枳基因组中存在11个DNA甲基转移酶基因和3个去甲基化酶基因。这些基因编码蛋白的氨基酸数在665 ~ 1 962 aa之间,均属于亲水蛋白,其中13个成员定位在细胞核中。此外,PtrDME、PtrROS1、PtrDML3、PtrCMT2和PtrMET1-6中发生了3个片段重复事件,与拟南芥和水稻中同源基因都存在共线性关系。进一步分析表明,枳DNA甲基化修饰酶基因启动子区域含有较多的光响应和激素响应元件,且PtrMET1在蛋白互作网络中是核心蛋白。分析发现,PtrMET1-2和PtrCMT1在叶、根、成熟胚珠和幼嫩胚珠中的表达水平较高,PtrCMT2和PtrCMT3在成熟胚珠和幼嫩胚珠中较高。DNA甲基化抑制剂5-氮杂胞苷处理可降低枳叶片整体DNA甲基化水平,并上调甲基化修饰酶基因的表达水平,但降低了PtrDRM在茎中的表达。
陈梦如, 吴庆安, 何思佳, 蔡祥斌, 马巧利, 辜青青, 魏清江. 枳DNA甲基化修饰酶基因的鉴定与表达特征[J]. 园艺学报, 2025, 52(12): 3167-3179.
CHEN Mengru, WU Qing’an, HE Sijia, CAI Xiangbin, MA Qiaoli, GU Qingqing, WEI Qingjiang. Genomic-Wide Identification and Expression Analysis of the DNA Methylation-Modifying Enzyme Genes in Poncirus trifoliata[J]. Acta Horticulturae Sinica, 2025, 52(12): 3167-3179.
| 基因 Gene | 正向引物序列(5′-3′) Forward primer | 反向引物序列(5′-3′) Reverse primer |
|---|---|---|
| PtrActin | CATCCCTCAGCACCTTCC | CCAACCTTAGCACTTCTCC |
| Pt5g021700 | AGGTTGT·GGTGGCTTATCTGATGG | TTCTCCTGCTGGTTGCTCATACTC |
| Pt7g014310 | TGATACCTTGGTGCCTGCCTAATAC | TGCCTTCCCAATCAAGCCTTCC |
| PtUn014750 | AGATTTAATACCCTGGTGCTTACC | AAAGTTTCCTTCCCAGTCCAA |
| PtUn014790 | TTATTTCCGCCCCAGGTTT | CTCAGGTGATGCTGCCCAT |
| PtUn014840 | CTTCTGCGAACATTTATACGATCC | TCCTGCTGGTTGCTCATACTCT |
| PtUn030910 | TGCCGCCTCACATCAAGA | CCATCAGATAAGCCACCACAA |
| Pt2g001260 | AAGTAAAGAAGCAGAAGCGAAGA | TCCCATCAACTGCCTCAAAC |
| Pt5g018630 | AGACGGTTCCTACAACAGCTACTTC | CGAGGCATAATCCAGTTGACATTCC |
| Pt5g023400 | AGCGAAGAAGAAGAAGAGGAGGAAG | TTCGGATCACCATAGCAGACAGC |
| Pt9g000560 | GTTGTCAGTCTTCAGCGGAATTGG | CCCACCACCTCTTGAGAATCCTTC |
| Pt1g006140 | GACATAGACCAGGAAGGCAAGGAG | ATGACAATCACCGCAAACACGAAG |
| Pt6g006730 | TGCGACTTCTAACACTCCACCATC | GCACCCAGCCGAGTCTTACAG |
| Pt8g006600 | GACTGTTGCCAGAACTGAGTATTGC | TGATCGCCTGTGCCTGTCTTG |
| Pt4g018910 | ACAACCGATGCTAGTGCGAGAC | TCTCTGCCTGCTCACCAATTACTTC |
表1 qRT-PCR引物序列
Table 1 The sequences of qRT-PCR primers
| 基因 Gene | 正向引物序列(5′-3′) Forward primer | 反向引物序列(5′-3′) Reverse primer |
|---|---|---|
| PtrActin | CATCCCTCAGCACCTTCC | CCAACCTTAGCACTTCTCC |
| Pt5g021700 | AGGTTGT·GGTGGCTTATCTGATGG | TTCTCCTGCTGGTTGCTCATACTC |
| Pt7g014310 | TGATACCTTGGTGCCTGCCTAATAC | TGCCTTCCCAATCAAGCCTTCC |
| PtUn014750 | AGATTTAATACCCTGGTGCTTACC | AAAGTTTCCTTCCCAGTCCAA |
| PtUn014790 | TTATTTCCGCCCCAGGTTT | CTCAGGTGATGCTGCCCAT |
| PtUn014840 | CTTCTGCGAACATTTATACGATCC | TCCTGCTGGTTGCTCATACTCT |
| PtUn030910 | TGCCGCCTCACATCAAGA | CCATCAGATAAGCCACCACAA |
| Pt2g001260 | AAGTAAAGAAGCAGAAGCGAAGA | TCCCATCAACTGCCTCAAAC |
| Pt5g018630 | AGACGGTTCCTACAACAGCTACTTC | CGAGGCATAATCCAGTTGACATTCC |
| Pt5g023400 | AGCGAAGAAGAAGAAGAGGAGGAAG | TTCGGATCACCATAGCAGACAGC |
| Pt9g000560 | GTTGTCAGTCTTCAGCGGAATTGG | CCCACCACCTCTTGAGAATCCTTC |
| Pt1g006140 | GACATAGACCAGGAAGGCAAGGAG | ATGACAATCACCGCAAACACGAAG |
| Pt6g006730 | TGCGACTTCTAACACTCCACCATC | GCACCCAGCCGAGTCTTACAG |
| Pt8g006600 | GACTGTTGCCAGAACTGAGTATTGC | TGATCGCCTGTGCCTGTCTTG |
| Pt4g018910 | ACAACCGATGCTAGTGCGAGAC | TCTCTGCCTGCTCACCAATTACTTC |
| 基因名称 Gene name | 基因号 Gene ID | 编码序列长度/bp CDS Length | 氨基酸数 No. of amino acids | 分子量/D Molecular weight | 等电点 pI | 脂溶性指数 Aliphatic index | 不稳定系数 Instability index (II) | 亲水性 Gravy | 亚细胞定位 Subcellular localization |
|---|---|---|---|---|---|---|---|---|---|
| PtrMET1-1 | Pt5g021700.1 | 5 247 | 1 748 | 196 887.65 | 6.18 | 78.07 | 40.18 | -0.439 | 细胞核 Nucleus |
| PtrMET1-2 | Pt7g014310.1 | 4 677 | 1 558 | 175 472.98 | 5.85 | 74.85 | 43.70 | -0.502 | 细胞核 Nucleus |
| PtrMET1-3 | PtUn014750.1 | 2 829 | 942 | 105 994.39 | 6.27 | 77.09 | 35.16 | -0.433 | 细胞质 Cytosol |
| PtrMET1-4 | PtUn014790.1 | 4 488 | 1 495 | 168 753.73 | 6.27 | 77.00 | 42.11 | -0.469 | 细胞核 Nucleus |
| PtrMET1-5 | PtUn014840.1 | 4 305 | 434 | 161 602.75 | 5.84 | 77.97 | 37.77 | -0.392 | 细胞核 Nucleus |
| PtrMET1-6 | PtUn030910.1 | 5 088 | 1 695 | 190 981.84 | 6.21 | 77.70 | 41.75 | -0.434 | 细胞核 Nucleus |
| PtrCMT1 | Pt2g001260.2 | 2 616 | 871 | 99 098.07 | 5.06 | 77.07 | 45.44 | -0.569 | 细胞核 Nucleus |
| PtrCMT2 | Pt5g018630.1 | 1 998 | 665 | 75 200.79 | 8.04 | 77.05 | 43.81 | -0.437 | 细胞核 Nucleus |
| PtrCMT3 | Pt5g023400.1 | 2 493 | 830 | 93 924.59 | 5.60 | 78.57 | 42.02 | -0.484 | 细胞核 Nucleus |
| PtrDRM | Pt9g000560.2 | 2 094 | 697 | 78 513.03 | 5.26 | 75.35 | 50.29 | -0.546 | 细胞核 Nucleus |
| PtrDNMT2 | Pt1g006140.1 | 3 543 | 1 180 | 132 606.24 | 5.80 | 70.49 | 39.30 | -0.580 | 细胞核 Nucleus |
| PtrROS1 | Pt6g006730.1 | 5 889 | 1 962 | 218 463.80 | 6.91 | 68.59 | 48.65 | -0.733 | 细胞核 Nucleus |
| PtrDML3 | Pt8g006600.1 | 5 250 | 1 749 | 196 115.50 | 8.25 | 73.01 | 54.29 | -0.689 | 细胞核 Nucleus |
| PtrDME | Pt4g018910.1 | 5 874 | 1 957 | 219 765.43 | 6.49 | 66.69 | 55.85 | -0.763 | 细胞核 Nucleus |
表2 枳DNA甲基化修饰酶蛋白理化性质
Table 2 Physicochemical properties of DNA methylation-modifying enzyme genes in Poncirus trifoliata
| 基因名称 Gene name | 基因号 Gene ID | 编码序列长度/bp CDS Length | 氨基酸数 No. of amino acids | 分子量/D Molecular weight | 等电点 pI | 脂溶性指数 Aliphatic index | 不稳定系数 Instability index (II) | 亲水性 Gravy | 亚细胞定位 Subcellular localization |
|---|---|---|---|---|---|---|---|---|---|
| PtrMET1-1 | Pt5g021700.1 | 5 247 | 1 748 | 196 887.65 | 6.18 | 78.07 | 40.18 | -0.439 | 细胞核 Nucleus |
| PtrMET1-2 | Pt7g014310.1 | 4 677 | 1 558 | 175 472.98 | 5.85 | 74.85 | 43.70 | -0.502 | 细胞核 Nucleus |
| PtrMET1-3 | PtUn014750.1 | 2 829 | 942 | 105 994.39 | 6.27 | 77.09 | 35.16 | -0.433 | 细胞质 Cytosol |
| PtrMET1-4 | PtUn014790.1 | 4 488 | 1 495 | 168 753.73 | 6.27 | 77.00 | 42.11 | -0.469 | 细胞核 Nucleus |
| PtrMET1-5 | PtUn014840.1 | 4 305 | 434 | 161 602.75 | 5.84 | 77.97 | 37.77 | -0.392 | 细胞核 Nucleus |
| PtrMET1-6 | PtUn030910.1 | 5 088 | 1 695 | 190 981.84 | 6.21 | 77.70 | 41.75 | -0.434 | 细胞核 Nucleus |
| PtrCMT1 | Pt2g001260.2 | 2 616 | 871 | 99 098.07 | 5.06 | 77.07 | 45.44 | -0.569 | 细胞核 Nucleus |
| PtrCMT2 | Pt5g018630.1 | 1 998 | 665 | 75 200.79 | 8.04 | 77.05 | 43.81 | -0.437 | 细胞核 Nucleus |
| PtrCMT3 | Pt5g023400.1 | 2 493 | 830 | 93 924.59 | 5.60 | 78.57 | 42.02 | -0.484 | 细胞核 Nucleus |
| PtrDRM | Pt9g000560.2 | 2 094 | 697 | 78 513.03 | 5.26 | 75.35 | 50.29 | -0.546 | 细胞核 Nucleus |
| PtrDNMT2 | Pt1g006140.1 | 3 543 | 1 180 | 132 606.24 | 5.80 | 70.49 | 39.30 | -0.580 | 细胞核 Nucleus |
| PtrROS1 | Pt6g006730.1 | 5 889 | 1 962 | 218 463.80 | 6.91 | 68.59 | 48.65 | -0.733 | 细胞核 Nucleus |
| PtrDML3 | Pt8g006600.1 | 5 250 | 1 749 | 196 115.50 | 8.25 | 73.01 | 54.29 | -0.689 | 细胞核 Nucleus |
| PtrDME | Pt4g018910.1 | 5 874 | 1 957 | 219 765.43 | 6.49 | 66.69 | 55.85 | -0.763 | 细胞核 Nucleus |

图1 DNA甲基化修饰酶基因进化、染色体定位和共线性分析 A:枳(Poncirus trifoliata,Ptr)、拟南芥(Arabidopsis thaliana,At)、水稻(Oryza sativa,Os)DNA甲基化修饰酶基因蛋白统进化分析;B:枳DNA甲基化修饰酶基因染色体定位和物种内的共线性分析;C:枳、拟南芥、水稻DNA甲基化修饰酶基因共线性分析
Fig. 1 Phylogenetic analysis,chromosome localization,and syntenic analysis of DNA methylation-modifying enzyme genes A:Phylogenetic analysis of DNA methylation-modifying enzyme genes from Poncirus trifoliata(Ptr),Arabidopsis thaliana(At),and Oryza sativa(Os);B:Chromosome localization and syntenic analysis of DNA methylation-modifying enzyme genes in Poncirus trifoliata;C:Syntenic analysis of DNA methylation-modifying enzyme genes from Poncirus trifoliata,Arabidopsis thaliana,and Oryza sativa

图3 枳DNA甲基化修饰酶基因启动子顺式作用元件种类(A)、数量(B)和蛋白互作分析(C)
Fig. 3 The type(A)and number(B)of cis-acting elements in the promoter region,and protein interaction network(C)of DNA methylation-modifying enzyme genes in Poncirus trifoliata
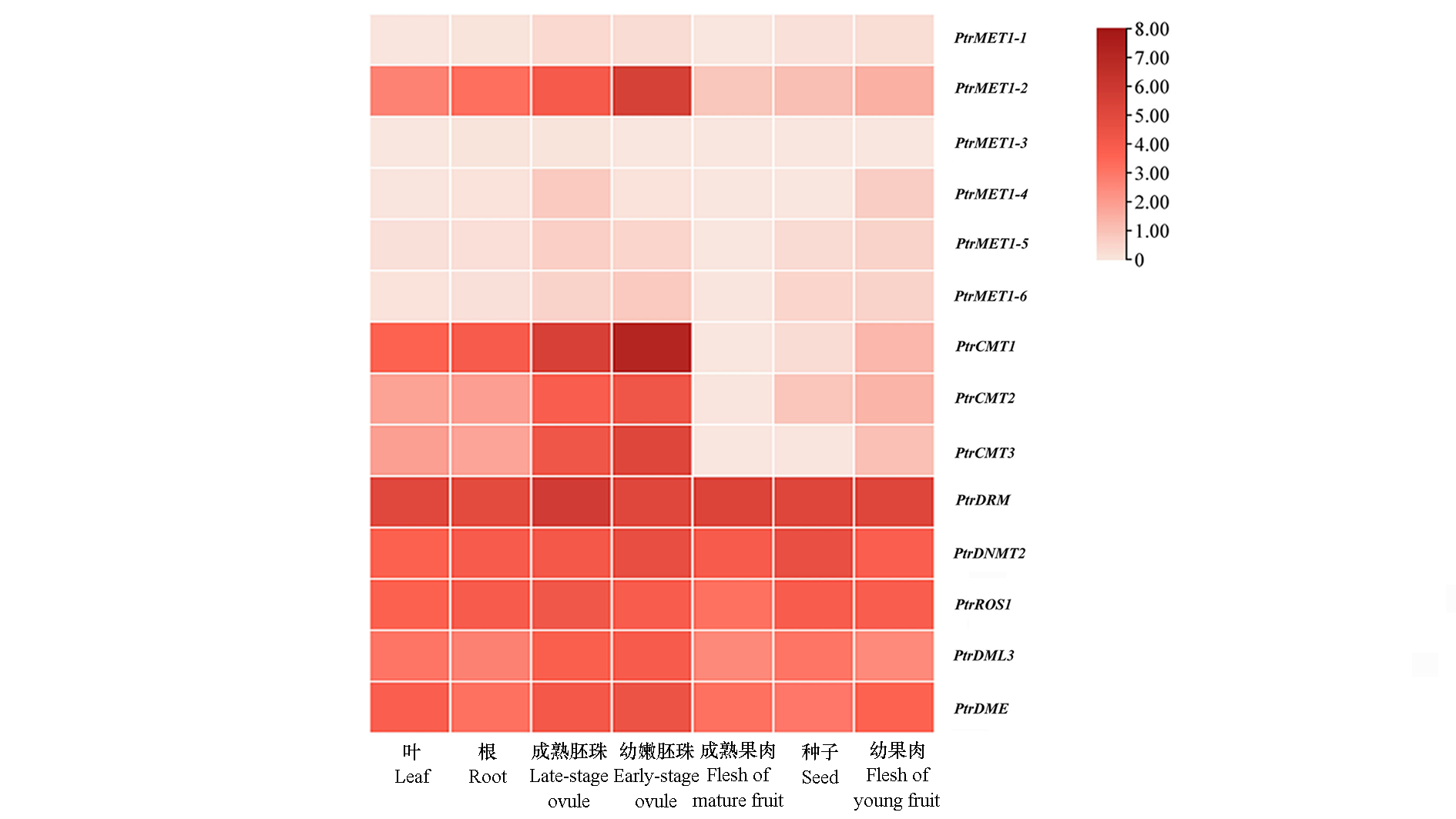
图4 枳DNA甲基化修饰酶基因组织特异性表达 数据显示值为log2FPKM
Fig. 4 Expression heatmap of DNA methylation-modifying enzyme genes in different tissues of Poncirus trifoliata The data in the heatmap shows the log2FPKM value
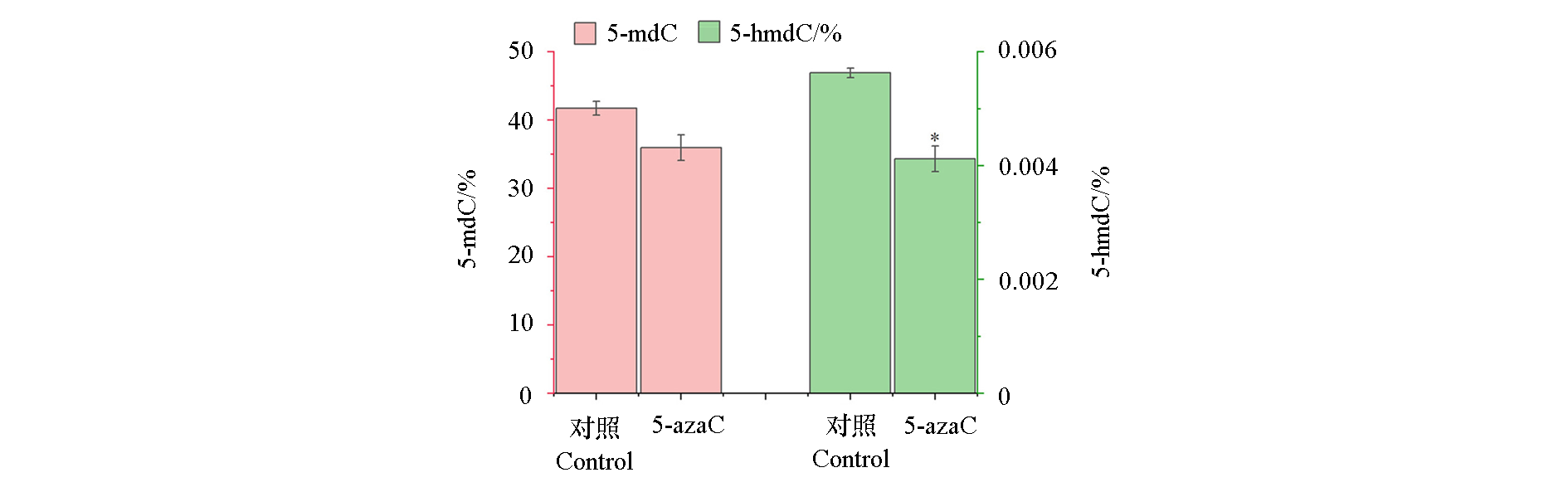
图5 5-azaC处理后枳叶片DNA甲基化水平 平均值差异采用t检测,*代表处理和对照之间差异显著(P < 0.05)
Fig. 5 DNA methylation level in leaf of Poncirus trifoliata after 5-azaC treatment Differences between the means were evaluated using Student’s t-test,and the * indicates significant difference between the control and 5-azaC treatment(P < 0.05)
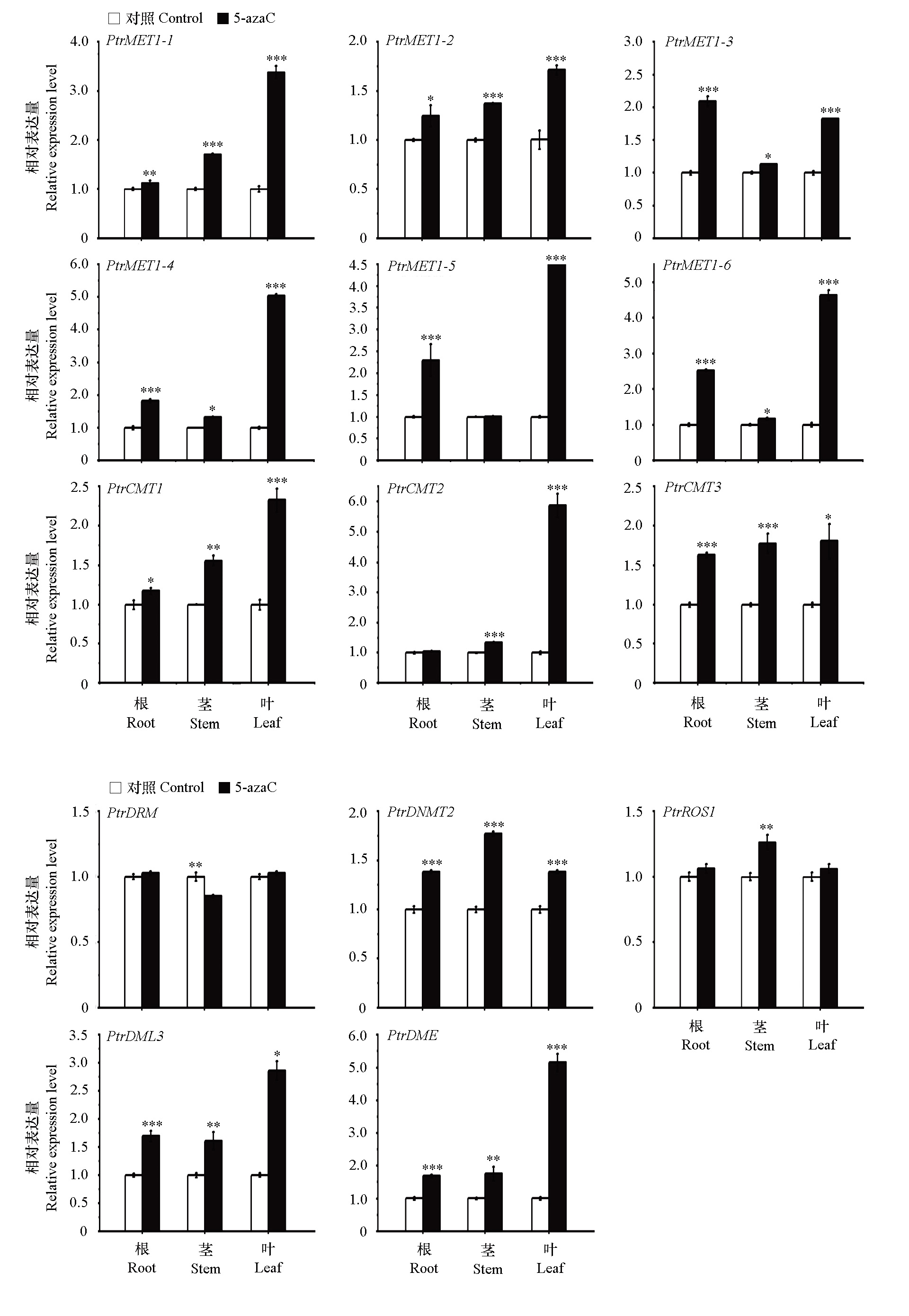
图6 5-azaC处理后枳根、茎、叶中DNA甲基化修饰酶基因的表达 平均值差异采用t检测,*,**和***分别代表在P < 0.05,P < 0.01和P < 0.001水平差异显著
Fig. 6 Expression of DNA methylation-modifying enzyme genes in root,stem,and leaf of Poncirus trifoliata after 5-azaC treatment Differences between the means were evaluated using Student’s t-test,and the *,**,and *** indicate significant difference between the CK and 5-azaC treatment at P < 0.05,P < 0.01,and P < 0.001,respectively
| [35] |
|
| [36] |
|
|
徐记迪. 2015. 柑橘全基因组DNA甲基化分析及调控作用研究[博士论文]. 武汉: 华中农业大学.
|
|
| [37] |
|
| [38] |
|
|
杨惠栋, 彭震宇, 彭良志, 袁梦, 淳长品, 袁高鹏. 2023. 不同砧木对纽荷尔脐橙嫁接苗生长和生理指标的影响. 江西农业大学学报, 45(3):575-583.
|
|
| [39] |
|
| [40] |
|
|
杨志宁, 陆许可, 范亚朋, 孙玉平, 喻欣, 王亮, 高永健, 古丽加依那什·哈不力, 古丽沙西·拿斯依, 吾木尔·阿不都, 黄慧, 张梦豪, 王立东, 陈筱, 肖磊, 张鑫蕊, 王帅, 陈修贵, 王俊娟, 郭丽雪, 高文伟, 叶武威. 2025. 棉花GhDMT7基因对5-氮杂胞嘧啶核苷的响应机制. 中国农业科技导报, 27(8):28-35.
|
|
| [41] |
|
| [1] |
|
| [2] |
|
| [3] |
|
| [4] |
|
| [5] |
|
| [6] |
|
| [7] |
|
| [8] |
|
| [9] |
|
|
陈婷欣, 符敏, 李娜, 杨蕾蕾, 李凌飞, 钟春梅. 2024. 铁甲秋海棠DNA甲基转移酶全基因组鉴定及表达分析. 植物学报, 59(5):726-737.
|
|
| [10] |
|
| [11] |
|
|
邓秀新. 2022. 中国柑橘育种60年回顾与展望. 园艺学报, 49(10):2063-2074.
|
|
| [12] |
|
| [13] |
|
| [14] |
|
| [15] |
|
| [16] |
|
| [17] |
|
| [18] |
|
| [19] |
|
| [20] |
|
| [21] |
|
| [22] |
|
| [23] |
|
| [24] |
|
| [25] |
|
| [26] |
|
| [27] |
|
| [28] |
|
| [42] |
|
|
张盼, 余永旭, 曹领改, 朱广兵, 吴伟, 郭玉双, 尹国英, 贾蒙骜. 2023. m6A甲基化修饰响应植物生物胁迫和非生物胁迫的研究进展. 园艺学报, 50(9):1841-1853.
|
|
| [43] |
|
| [44] |
|
| [45] |
|
|
朱世平, 王福生, 陈娇, 余歆, 余洪, 罗国涛, 胡洲, 冯锦英, 赵晓春, 洪棋斌. 2020. 柑橘不同类型砧木的种子和苗期性状. 中国农业科学, 53(3):585-599.
|
|
| [46] |
|
|
庄泽岳. 2023. 辣椒DRM基因家族的分析及CaDRM1在花器官发育中的功能研究[硕士论文]. 广州: 仲恺农业工程学院.
|
|
| [29] |
|
|
马青龄, 梁贝贝, 杨莉, 胡威, 刘勇, 刘德春. 2022. 枳WOX基因家族全基因组鉴定及表达分析. 果树学报, 39(5):712-729.
|
|
| [30] |
|
| [31] |
|
| [32] |
|
|
祁春节, 顾雨檬, 曾彦. 2021. 我国柑橘产业经济研究进展. 华中农业大学学报, 40(1):58-69.
|
|
| [33] |
|
| [34] |
|
| [1] | 唐嘉怡, 缪绸雨, 尹豪杰, 吴明月, 曹向敏, 张惠敏, 金燕, 卢晓鹏, 朱亦赤, 李大志, 盛玲. 外源草酸对采后血橙果实品质和花色苷代谢的影响[J]. 园艺学报, 2025, 52(9): 2425-2438. |
| [2] | 窦雪婷, 朱熙, 张宁, 司怀军. 马铃薯低温胁迫相关基因StHY5的功能分析[J]. 园艺学报, 2025, 52(7): 1745-1757. |
| [3] | 周天, 吕帅丽, 韩晗, 李雨佳, 刘海强, 刘永忠. 外源独脚金内酯对枳幼苗茎增粗生长的影响[J]. 园艺学报, 2025, 52(7): 1843-1854. |
| [4] | 王毅, 梁振昌. 基于群体转录组的葡萄远缘杂交遗传机制分析[J]. 园艺学报, 2025, 52(4): 835-845. |
| [5] | 唐伶俐, 贺玉花, 王庆涛, 安璐璐, 徐永阳, 张健, 孔维虎, 户克云, 赵光伟. 非呼吸跃变和呼吸跃变型甜瓜成熟果实的激素与转录组分析[J]. 园艺学报, 2025, 52(4): 883-896. |
| [6] | 汪进萱, 倪莹, 孟昕, 冷卓, 马波, 冷平生, 吴静, 胡增辉. 紫丁香SoCHR35参与罗勒烯合成的功能解析[J]. 园艺学报, 2025, 52(10): 2655-2665. |
| [7] | 李慧, 阚家亮, 李晓刚, 王中华, 蔺经, 杨青松. 赤霉素对‘苏翠1号’梨花芽松散的影响[J]. 园艺学报, 2025, 52(10): 2713-2726. |
| [8] | 周小林, 林俊轩, 李彩霞, 孙美华, 白龙强, 侯雷平, 苗妍秀. 外源谷胱甘肽对高温胁迫下叶用莴苣光合特性的影响[J]. 园艺学报, 2025, 52(10): 2761-2773. |
| [9] | 贺丹丹, 何宏泰, 王文庭, 周文美, 刘燕敏, 刘骕骦. 甜瓜GolS家族基因鉴定及其响应低温胁迫的表达分析[J]. 园艺学报, 2025, 52(1): 136-148. |
| [10] | 史兴秀, 冯贝贝, 闫鹏, 耿文娟, 朱玛孜拉·沙尔山木汗. ‘王林’苹果早期水心病发生过程中的相关生理变化[J]. 园艺学报, 2025, 52(1): 171-184. |
| [11] | 任思源, 陈森, 龙治坚, 王博雅, 唐登国, 王正前, 杨斌, 胡尚连, 曹颖. 花魔芋球茎发育期氮分配变化和相关基因表达分析[J]. 园艺学报, 2024, 51(9): 2019-2030. |
| [12] | 周平, 颜少宾, 郭瑞, 金光. 桃镁离子转运蛋白MGT基因家族鉴定与表达分析[J]. 园艺学报, 2024, 51(3): 463-478. |
| [13] | 闫超凡, 孙雪梅, 钟启文, 邵登魁, 邓昌蓉, 文军琴. 番茄20S蛋白酶体基因家族鉴定及生物信息学分析[J]. 园艺学报, 2024, 51(2): 266-280. |
| [14] | 崔一琼, 李菊, 刘晓奇, 王俊文, 唐中祺, 武玥, 肖雪梅, 郁继华. 水分亏缺对设施基质栽培番茄果实蔗糖和淀粉代谢的影响[J]. 园艺学报, 2024, 51(11): 2607-2619. |
| [15] | 韩世文, 刘涛, 王丽萍, 李楠洋, 王素娜, 王星. 黄瓜SRS基因家族鉴定及胁迫响应表达分析[J]. 园艺学报, 2024, 51(10): 2281-2296. |
| 阅读次数 | ||||||
|
全文 |
|
|||||
|
摘要 |
|
|||||
版权所有 © 2012 《园艺学报》编辑部 京ICP备10030308号-2 国际联网备案号 11010802023439
编辑部地址: 北京市海淀区中关村南大街12号中国农业科学院蔬菜花卉研究所 邮编: 100081
电话: 010-82109523 E-Mail: yuanyixuebao@126.com
技术支持:北京玛格泰克科技发展有限公司