
园艺学报 ›› 2021, Vol. 48 ›› Issue (6): 1183-1196.doi: 10.16420/j.issn.0513-353x.2020-0510
收稿日期:2020-12-28
修回日期:2021-04-06
出版日期:2021-06-25
发布日期:2021-07-08
通讯作者:
李贵生
E-mail:gui-sheng@jsu.edu.cn
基金资助:Received:2020-12-28
Revised:2021-04-06
Online:2021-06-25
Published:2021-07-08
Contact:
LI Guisheng
E-mail:gui-sheng@jsu.edu.cn
摘要:
转录组比较发现,果肉黄色的‘金艳'猕猴桃成熟果实中优势表达基因的主要功能涉及代谢,而果肉含花青苷呈红色的‘红阳'中的优势表达基因一般与发育相关。猕猴桃果皮和外层果肉部分及内层果肉和果心部分的优势表达基因分别与光合作用和营养储备有关。猕猴桃有相对较多的R2R3类型的MYB基因,几乎所有类型的MYB基因都在果实中表达。MYB-A72位于花青苷合成相关MYB基因分支,其转录本只在‘红阳'中发现,在内层果肉和果心部分的表达水平很高。表达模式聚类分析表明:bHLH-B36和WD40-A126可能参与形成MYB-bHLH-WD40调控复合体,而CHI2、FLS1和CYP98A1可能是受其调控的靶基因。
中图分类号:
李贵生. 猕猴桃‘金艳'和‘红阳'果实转录组的比较分析[J]. 园艺学报, 2021, 48(6): 1183-1196.
LI Guisheng. Comparative Analysis of‘Jinyan'and‘Hongyang'Kiwifruit Transcriptomes[J]. Acta Horticulturae Sinica, 2021, 48(6): 1183-1196.
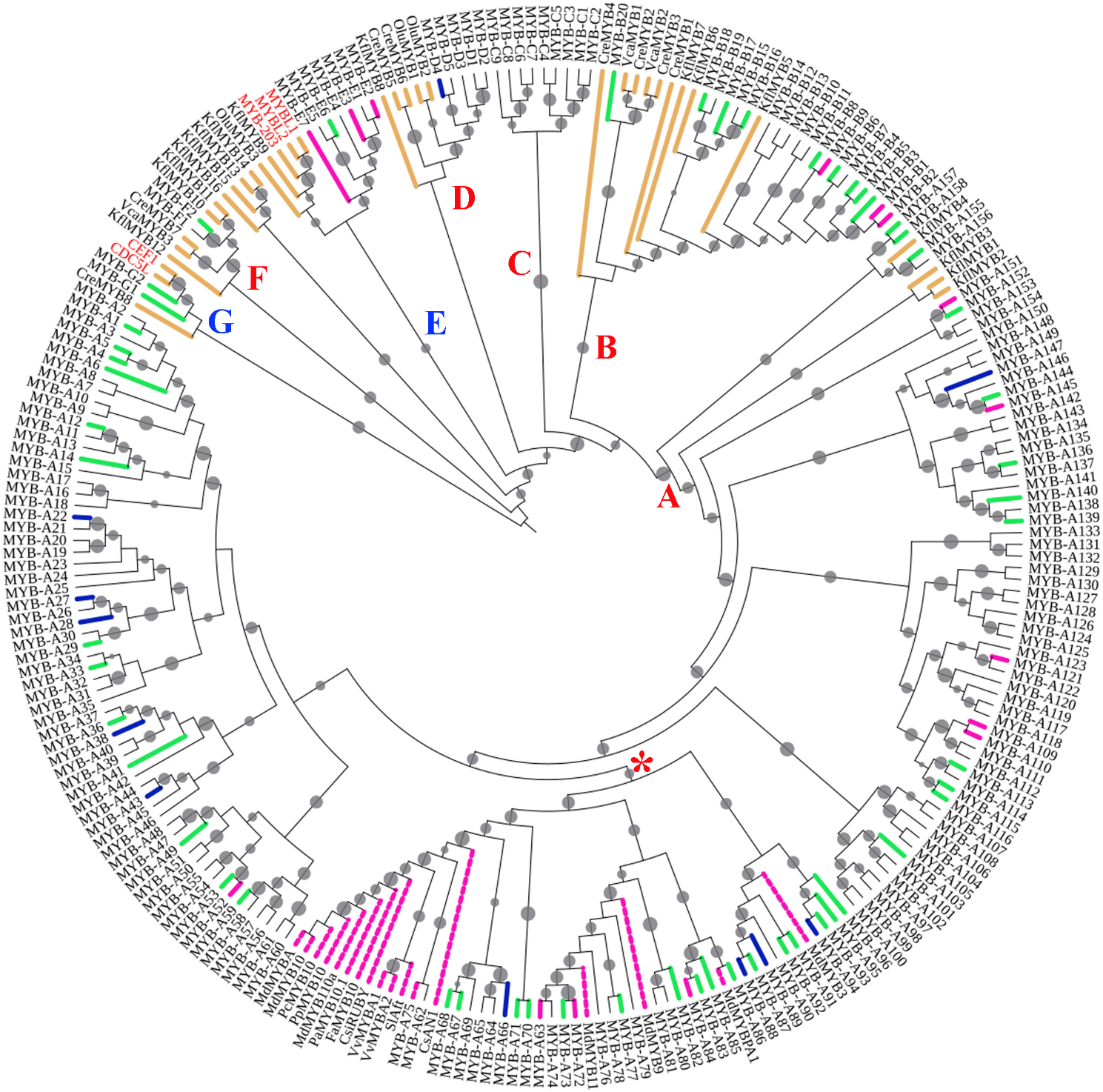
图1 猕猴桃MYB基因系统发育分析 黄色表示藻类,粉红色虚线表示已研究过的与花青苷合成相关的基因,粉红色实线表示‘红阳'果实中高表达的基因,蓝色实线是低表达基因,绿色表示差异不显著的基因。真菌和动物的基因的名字为红色。分支旁红色字母A、B、C、D、F表示R2R3类型的各个分支,蓝色字母E、G表示其他类型的分支。红色星号表示与花青苷合成有关的簇。分支上的实心圆圈表示大于50%的支持率,越大越高。
Fig. 1 The phylogenetic analysis of kiwifruit MYB genes Algae genes were indicated by yellow,MYB genes studied and related to anthocyanin synthesis were indicated by dashed pink,genes strongly expressed in‘Hongyang'fruit were indicated by pink,and genes with low expression were indicated by blue solid lines,while genes lacking significant expression difference were indicated by green lines. Fungus and animal gene names were in red. Red letters such as A,B,C,D,and F along branches indicated individual clades of the R2R3 type,while blue letters such as E and G indicated clades of other types. Red asterisks indicated the cluster involved in anthocyanin synthesis. Filled circles on branches indicated bootstrap values above 50%,with a larger circle indicating a higher value.

图2 花青苷合成相关MYB基因的多序列比对 比对底部的灰色和黑色长杠表示两个MYB模块,而第1个方框表示结合bHLH的区域,第2个方框表示花青苷合成相关MYB蛋白的特有基序。
Fig. 2 Multi-alignments of MYB genes involved in anthocyanin synthesis Two MYB domains were indicated by the grey and the black long bar below the alignment,while the first box indicated the bHLH binding region,and the second box the characteristic motif of MYB proteins related to anthocyanin synthesis.

图3 花青苷合成相关基因表达水平的聚类热图 “*”表示基因的表达水平在‘红阳'和‘金艳'之间有显著差异,在MYB基因中表示为红色,在其他基因中表示为绿色。C1 ~ C4表示4个主要的簇。
Fig. 3 The clustering heatmap of genes related to anthocyanin synthesis “*”indicated genes significantly differently expressed between‘Hongyang'and‘Jinyan',with it highlighted in red for MYB genes,and in green for other genes. C1-C4 indicated four major clusters.
| [1] |
Ampomah-Dwamena C, Thrimawithana A H, Dejnoprat S, Lewis D, Espley R V, Allan A C. 2019. A kiwifruit(Actinidia deliciosa)R2R3-MYB transcription factor modulates chlorophyll and carotenoid accumulation. New Phytol, 221 (1):309-325.
doi: 10.1111/nph.15362 |
| [2] | Bi Siqi, An Jianping, Wang Xiaofei, Hao Yujin, Rui Lin, Li Tong, Han Yuepeng, You Chunxiang. 2019. Ethylene response factor MdERF 3 promotes anthocyanin and proanthocyanidin accumulation in apple. Acta Horticulturae Sinica, 46 (12):2277-2285. (in Chinese) |
| 毕思琦, 安建平, 王小非, 郝玉金, 芮麟, 李彤, 韩月彭, 由春香. 2019. 苹果乙烯响应因子MdERF3促进花青苷和原花青苷积累. 园艺学报, 46 (12):2277-2285. | |
| [3] |
Bovy A,de Vos R, Kemper M, Schijlen E, Almenar Pertejo M, Muir S, Collins G, Robinson S, Verhoeyen M, Hughes S, Santos-Buelga C, van Tunen A. 2002. High-flavonol tomatoes resulting from the heterologous expression of the maize transcription factor genes LC and C1. Plant Cell, 14 (10):2509-2526.
doi: 10.1105/tpc.004218 URL |
| [4] |
Butelli E, Licciardello C, Zhang Y, Liu J, Mackay S, Bailey P, Reforgiato-Recupero G, Martin C. 2012. Retrotransposons control fruit-specific,cold-dependent accumulation of anthocyanins in blood oranges. Plant Cell, 24 (3):1242-1255.
doi: 10.1105/tpc.111.095232 URL |
| [5] |
Butelli E, Titta L, Giorgio M, Mock H P, Matros A, Peterek S, Schijlen E G, Hall R D, Bovy A G, Luo J, Martin C. 2008. Enrichment of tomato fruit with health-promoting anthocyanins by expression of select transcription factors. Nat Biotechnol, 26 (11):1301-1308.
doi: 10.1038/nbt.1506 URL |
| [6] | Cao Yuwei, Xu Leifeng, Yang Panpan, Xu Hua, He Guoren, Tang Yuchao, Ren Junfang, Ming Jun. 2019. Differential expression of three R2R3-MYBs genes regulating anthocyanin pigmentation patterns in Lilium spp. Acta Horticulturae Sinica, 46 (5):955-963. (in Chinese) |
| 曹雨薇, 徐雷锋, 杨盼盼, 徐华, 何国仁, 唐玉超, 任君芳, 明军. 2019. 百合花青素苷呈色类型中3种R2R3-MYBs基因的差异表达. 园艺学报, 46 (5):955-963. | |
| [7] |
Chiu L W, Zhou X, Burke S, Wu X, Prior R L, Li L. 2010. The purple cauliflower arises from activation of a MYB transcription factor. Plant Physiol, 154 (3):1470-1480.
doi: 10.1104/pp.110.164160 pmid: 20855520 |
| [8] |
Colanero S, Perata P, Gonzali S. 2020. What's behind purple tomatoes? insight into the mechanisms of anthocyanin synthesis in tomato fruits. Plant Physiol, 182 (4):1841-1853.
doi: 10.1104/pp.19.01530 pmid: 31980573 |
| [9] |
Dubos C, Stracke R, Grotewold E, Weisshaar B, Martin C, Lepiniec L. 2010. MYB transcription factors in Arabidopsis. Trends Plant Sci, 15 (10):573-581.
doi: 10.1016/j.tplants.2010.06.005 URL |
| [10] |
Espley R V, Brendolise C, Chagne D, Kutty-Amma S, Green S, Volz R, Putterill J, Schouten H J, Gardiner S E, Hellens R P, Allan A C. 2009. Multiple repeats of a promoter segment causes transcription factor autoregulation in red apples. Plant Cell, 21 (1):168-183.
doi: 10.1105/tpc.108.059329 URL |
| [11] |
Fu Z Z, Shang H Q, Jiang H, Gao J, Dong X Y, Wang H J, Li Y M, Wang L M, Zhang J, Shu Q Y, Chao Y C, Xu M L, Wang R, Wang L S, Zhang H C. 2020. Systematic identification of the light-quality responding anthocyanin synthesis-related transcripts in petunia petals. Horticultural Plant Journal, 6 (6):428-438.
doi: 10.1016/j.hpj.2020.11.006 URL |
| [12] | Gambi F, Pilkington S M, McAtee P A, Donati I, Schaffer R J, Montefiori M, Spinelli F, Burdon J. 2018. Fruit of three kiwifruit(Actinidia chinensis)cultivars differ in their degreening response to temperature after harvest. Postharvest Biology & Technology, 141:16-23. |
| [13] |
Gates D J, Strickler S R, Mueller L A, Olson B J, Smith S D. 2016. Diversification of R2R3-MYB transcription factors in the tomato family Solanaceae. Journal Mol Evol, 83 (1-2):26-37.
doi: 10.1007/s00239-016-9756-6 URL |
| [14] |
Gonzalez A, Zhao M, Leavitt J M, Lloyd A M. 2008. Regulation of the anthocyanin biosynthetic pathway by the TTG1/bHLH/Myb transcriptional complex inArabidopsis seedlings. Plant Journal, 53 (5):814-827.
doi: 10.1111/tpj.2008.53.issue-5 URL |
| [15] |
Ho L C. 1996. The mechanism of assimilate partitioning and carbohydrate compartmentation in fruit in relation to the quality and yield of tomato. Journal Exp Bot, 47:1239-1243.
doi: 10.1093/jxb/47.Special_Issue.1239 URL |
| [16] |
Holton T A, Cornish E C. 1995. Genetics and biochemistry of anthocyanin biosynthesis. Plant Cell, 7 (7):1071-1083.
doi: 10.2307/3870058 URL |
| [17] |
Hsu C C, Chen Y Y, Tsai W C, Chen W H, Chen H H. 2015. Three R2R3-MYB transcription factors regulate distinct floral pigmentation patterning in Phalaenopsis spp. Plant Physiol, 168 (1):175-191.
doi: 10.1104/pp.114.254599 URL |
| [18] |
Hsu C C, Su C J, Jeng M F, Chen W H, Chen H H. 2019. A HORT1 retrotransposon insertion in the PeMYB11 promoter causes Harlequin/Black flowers in Phalaenopsis orchids. Plant Physiol, 180 (3):1535-1548.
doi: 10.1104/pp.19.00205 URL |
| [19] | Huang S, Ding J, Deng D, Tang W, Sun H, Liu D, Zhang L, Niu X, Zhang X, Meng M, Yu J, Liu J, Han Y, Shi W, Zhang D, Cao S, Wei Z, Cui Y, Xia Y, Zeng H, Bao K, Lin L, Min Y, Zhang H, Miao M, Tang X, Zhu Y, Sui Y, Li G, Yue J, Sun J, Liu F, Zhou L, Lei L, Zheng X, Liu M, Huang L, Song J, Xu C, Li J, Ye K, Zhong S, Lu B R, He G, Xiao F, Wang H L, Zheng H, Fei Z, Liu Y. 2013. Draft genome of the kiwifruit Actinidia chinensis. Nat Commun, 4:10.1038/ncomms3640. |
| [20] |
Inagaki Y, Hisatomi Y, Suzuki T, Kasahara K, Iida S. 1994. Isolation of a Suppressor-mutator/Enhancer-like transposable element, Tpn1,from Japanese morning glory bearing variegated flowers. Plant Cell, 6 (3):375-383.
pmid: 8180498 |
| [21] |
Itoh Y, Higeta D, Suzuki A, Yoshida H, Ozeki Y. 2002. Excision of transposable elements from the chalcone isomerase and dihydroflavonol 4-reductase genes may contribute to the variegation of the yellow-flowered carnation(Dianthus caryophyllus). Plant Cell Physiol, 43 (5):578-585.
doi: 10.1093/pcp/pcf065 URL |
| [22] |
Johnson E T, Ryu S, Yi H, Shin B, Cheong H, Choi G. 2001. Alteration of a single amino acid changes the substrate specificity of dihydroflavonol 4-reductase. Plant Journal, 25 (3):325-333.
pmid: 11208024 |
| [23] |
Krga I, Milenkovic D. 2019. Anthocyanins:from sources and bioavailability to cardiovascular-health benefits and molecular mechanisms of action. Journal Agric Food Chem, 67 (7):1771-1783.
doi: 10.1021/acs.jafc.8b06737 URL |
| [24] | Li B, Xia Y, Wang Y, Qin G, Tian S. 2017a. Characterization of genes encoding key enzymes involved in anthocyanin metabolism of kiwifruit during storage period. Front Plant Sci, 8:10.3389/fpls.2017.00341. |
| [25] | Li M, Verboven P, Buchsbaum A, Cantre D, Nicola B, Heyes J, Mowat A, East A. 2015. Characterizing kiwifruit(Actinidia sp.)near skin cellular structures using optical coherence tomography. Postharvest Biology & Technology, 110:247-256. |
| [26] |
Li W, Ding Z, Ruan M, Yu X, Peng M, Liu Y. 2017b. Kiwifruit R2R3-MYB transcription factors and contribution of the novel AcMYB75 to red kiwifruit anthocyanin biosynthesis. Sci Rep, 7 (1):10.1038/s41598-017-16905-1.
doi: 10.1038/s41598-017-00036-8 URL |
| [27] |
Li W, Liu Y, Zeng S, Xiao G, Wang G, Wang Y, Peng M, Huang H. 2015. Gene expression profiling of development and anthocyanin accumulation in kiwifruit(Actinidia chinensis)based on transcriptome sequencing. PLoS ONE, 10 (8):e0136439.
doi: 10.1371/journal.pone.0136439 URL |
| [28] |
Li Y, Cui W, Wang R, Lin M, Zhong Y, Sun L, Qi X, Fang J. 2019. MicroRNA858-mediated regulation of anthocyanin biosynthesis in kiwifruit (Actinidia arguta)based on small RNA sequencing. PLoS ONE, 14 (5):e0217480.
doi: 10.1371/journal.pone.0217480 URL |
| [29] |
Li Y, Fang J, Qi X, Lin M, Zhong Y, Sun L. 2018. A key structural gene, AaLDOX,is involved in anthocyanin biosynthesis in all red-fleshed kiwifruit(Actinidia arguta)based on transcriptome analysis. Gene, 648:31-41.
doi: 10.1016/j.gene.2018.01.022 URL |
| [30] | Liu Y, Qi Y, Zhang A, Wu H, Liu Z, Ren X. 2019. Molecular cloning and functional characterization of AcGST1,an anthocyanin-related glutathione S-transferase gene in kiwifruit (Actinidia chinensis). Plant Mol Biol, 100 (4-5):451-465. |
| [31] | Liu Y, Zhou B, Qi Y, Chen X, Liu C, Liu Z, Ren X. 2017. Expression differences of pigment structural genes and transcription factors explain flesh coloration in three contrasting kiwifruit cultivars. Front Plant Sci, 8:10.3389/fpls.2017.01507. |
| [32] | Lu Jing, Ma Qijun, Liu Xiao, Kang Hui, Liu Yajing, Hao Yujin, You Chunxiang. 2019. Studies on the regulation of anthocyanin accumulation by apple sucrose transporter MdSUT2. Acta Horticulturae Sinica, 46 (1):1-10. (in Chinese) |
| 路静, 马齐军, 刘晓, 康慧, 刘亚静, 郝玉金, 由春香. 2019. 苹果蔗糖转运蛋白MdSUT2调控花青苷积累的研究. 园艺学报, 46 (1):1-10. | |
| [33] |
Man Y P, Wang Y C, Li Z Z, Jiang Z W, Yang H L, Gong J J, He S S, Wu S Q, Yang Z Q, Zheng J, Wang Z Y. 2015. High-temperature inhibition of biosynthesis and transportation of anthocyanins results in the poor red coloration in red-fleshed Actinidia chinensis. Physiol Plant, 153 (4):565-583.
doi: 10.1111/ppl.2015.153.issue-4 URL |
| [34] |
McClure M A, Smith C, Elton P. 1996. Parameterization studies for the SAM and HMMER methods of hidden Markov model generation. Proc Int Conf Intell Syst Mol Biol, 4:155-164.
pmid: 8877515 |
| [35] |
Mehrtens F, Kranz H, Bednarek P, Weisshaar B. 2005. The Arabidopsis transcription factor MYB12 is a flavonol-specific regulator of phenylpropanoid biosynthesis. Plant Physiol, 138 (2):1083-1096.
pmid: 15923334 |
| [36] |
Meng J X, Yan Gao Y, Han M L, Liu P Y, Yang C, Shen T, Li H H. 2020. In vitro anthocyanin induction and metabolite analysis in Malus spectabilis leaves under low nitrogen conditions. Horticultural Plant Journal, 6 (5):284-292.
doi: 10.1016/j.hpj.2020.06.004 URL |
| [37] |
Montefiori M, Espley R V, Stevenson D, Cooney J, Datson P M, Saiz A, Atkinson R G, Hellens R P, Allan A C. 2010. Identification and characterisation of F3GT1 and F3GGT1,two glycosyltransferases responsible for anthocyanin biosynthesis in red-fleshed kiwifruit(Actinidia chinensis). Plant Journal, 65 (1):106-118.
doi: 10.1111/j.1365-313X.2010.04409.x URL |
| [38] |
Povero G, Gonzali S, Bassolino L, Mazzucato A, Perata P. 2011. Transcriptional analysis in high-anthocyanin tomatoes reveals synergistic effect of Aft and atv genes. Journal Plant Physiol, 168 (3):270-279.
doi: 10.1016/j.jplph.2010.07.022 URL |
| [39] |
Price M N, Dehal P S, Arkin A P. 2009. FastTree:computing large minimum evolution trees with profiles instead of a distance matrix. Mol Biol Evol, 26 (7):1641-1650.
doi: 10.1093/molbev/msp077 URL |
| [40] |
Ruiz-Sanchez E, Sosa V. 2015. Origin and evolution of fleshy fruit in woody bamboos. Mol Phylogenet Evol, 91:123-134.
doi: 10.1016/j.ympev.2015.05.020 pmid: 26048705 |
| [41] | Shinozaki Y, Nicolas P, Fernandez-Pozo N, Ma Q, Evanich D J, Shi Y, Xu Y, Zheng Y, Snyder S I, Martin L B B, Ruiz-May E, Thannhauser T W, Chen K, Domozych D S, Catala C, Fei Z, Mueller L A, Giovannoni J J, Rose J K C. 2018. High-resolution spatiotemporal transcriptome mapping of tomato fruit development and ripening. Nat Commun, 9 (1):10.1038/s41467-017-02782-9. |
| [42] | Song Yang, Liu Hongdi, Wang Haibo, Zhang Hongjun, Liu Fengzhi. 2019. Molecular cloning and functional characterization of anthocyanin synthesis related genes VcTTG1of blueberry. Acta Horticulturae Sinica, 46 (7):1270-1278. (in Chinese) |
| 宋杨, 刘红弟, 王海波, 张红军, 刘凤之. 2019. 越橘花青苷合成相关基因 VcTTG1的克隆与功能鉴定. 园艺学报, 46 (7):1270-1278. | |
| [43] |
Stracke R, Ishihara H, Huep G, Barsch A, Mehrtens F, Niehaus K, Weisshaar B. 2007. Differential regulation of closely related R2R3-MYB transcription factors controls flavonol accumulation in different parts of the Arabidopsis thaliana seedling. Plant Journal, 50 (4):660-677.
pmid: 17419845 |
| [44] |
Sun C, Deng L, Du M, Zhao J, Chen Q, Huang T, Jiang H, Li C, Li C. 2020. A transcriptional network promotes anthocyanin biosynthesis in tomato flesh. Mol Plant, 13 (1):42-58.
doi: 10.1016/j.molp.2019.10.010 URL |
| [45] |
Thompson J D, Higgins D G, Gibson T J. 1994. CLUSTAL W:improving the sensitivity of progressive multiple sequence alignment through sequence weighting,position-specific gap penalties and weight matrix choice. Nucleic Acids Res, 22 (22):4673-4680.
pmid: 7984417 |
| [46] |
Wang L, Tang W, Hu Y, Zhang Y, Sun J, Guo X, Lu H, Yang Y, Fang C, Niu X, Yue J, Fei Z, Liu Y. 2019. A MYB/bHLH complex regulates tissue-specific anthocyanin biosynthesis in the inner pericarp of red-centered kiwifruit Actinidia chinensis cv. Hongyang. Plant Journal, 99 (2):359-378.
doi: 10.1111/tpj.2019.99.issue-2 URL |
| [47] |
Xu Z S, Yang Q Q, Feng K, Xiong A S. 2019. Changing carrot color:insertions in DcMYB7 alter the regulation of anthocyanin biosynthesis and modification. Plant Physiol, 181 (1):195-207.
doi: 10.1104/pp.19.00523 URL |
| [48] | Yin Cui-bo. 2008. Preliminary studies on biological characters and the rule of fruit growth and development of Hongyang kiwifruit[M. D. Dissertation]. Chengdu: Sichuan Agricultural University. (in Chinese) |
| 尹翠波. 2008. 红阳猕猴桃生物学特性及果实生长发育规律的初步研究[硕士论文]. 成都: 四川农业大学. | |
| [49] |
Yu M, Man Y, Wang Y. 2019. Light- and temperature-induced expression of an R2R3-MYB gene regulates anthocyanin biosynthesis in red-fleshed kiwifruit. Int Journal Mol Sci, 20 (20):10.3390/ijms20205228.
doi: 10.3390/ijms20010010 URL |
| [50] | Zhong Caihong, Zhang Peng, Han Fei, Li Dawei. 2015. Studies on characterization of fruit development of interspecific hybrid cultivar‘Jinyan'. Journal of Fruit Science, 32 (6):1152-1160. (in Chinese) |
| 钟彩虹, 张鹏, 韩飞, 李大卫. 2015. 猕猴桃种间杂交新品种‘金艳'的果实发育特征. 果树学报, 32 (6):1152-1160. |
| [1] | 叶子茂, 申晚霞, 刘梦雨, 王 彤, 张晓楠, 余 歆, 刘小丰, 赵晓春, . R2R3-MYB转录因子CitMYB21对柑橘类黄酮生物合成的影响[J]. 园艺学报, 2023, 50(2): 250-264. |
| [2] | 于婷婷, 李 欢, 宁源生, 宋建飞, 彭璐琳, 贾竣淇, 张玮玮, 杨洪强. 苹果GRAS全基因组鉴定及其对生长素的响应分析[J]. 园艺学报, 2023, 50(2): 397-409. |
| [3] | 翟含含, 翟宇杰, 田义, 张叶, 杨丽, 温陟良, 陈海江. 桃SAUR家族基因分析及PpSAUR5功能鉴定[J]. 园艺学报, 2023, 50(1): 1-14. |
| [4] | 袁馨, 徐云鹤, 张雨培, 单楠, 陈楚英, 万春鹏, 开文斌, 翟夏琬, 陈金印, 甘增宇. 猕猴桃后熟过程中ABA响应结合因子AcAREB1调控AcGH3.1的表达[J]. 园艺学报, 2023, 50(1): 53-64. |
| [5] | 宋 放, 陈 奇, 袁炎良, 陈 沙, 尹海军, 蒋迎春, . 黄肉猕猴桃新品种‘先沃1号’[J]. 园艺学报, 2022, 49(S2): 47-48. |
| [6] | 齐永杰, 高正辉, 马 娜, 王清明, 柯凡君, 陈 钱, 徐义流, . 黄肉抗溃疡病猕猴桃新品种‘皖农金果’[J]. 园艺学报, 2022, 49(S2): 49-50. |
| [7] | 张慧琴, 楼国荣, 陆玲鸿, 古咸彬, 宋根华, 谢 鸣. 黄肉中华猕猴桃新品种‘金义’[J]. 园艺学报, 2022, 49(S2): 51-52. |
| [8] | 王沙, 张心慧, 赵玉洁, 李变变, 招雪晴, 沈雨, 董建梅, 苑兆和. 石榴花青苷合成相关基因PgMYB111的克隆与功能分析[J]. 园艺学报, 2022, 49(9): 1883-1894. |
| [9] | 李茂福, 杨媛, 王华, 范又维, 孙佩, 金万梅. 月季中R2R3-MYB基因RhMYB113c调控花青素苷合成[J]. 园艺学报, 2022, 49(9): 1957-1966. |
| [10] | 高彦龙, 吴玉霞, 张仲兴, 王双成, 张瑞, 张德, 王延秀. 苹果ELO家族基因鉴定及其在低温胁迫下的表达分析[J]. 园艺学报, 2022, 49(8): 1621-1636. |
| [11] | 刘金明, 郭彩华, 袁星, 亢超, 全绍文, 牛建新. 梨Dof家族基因鉴定及其在宿存与脱落萼片中的表达分析[J]. 园艺学报, 2022, 49(8): 1637-1649. |
| [12] | 许海峰, 王中堂, 陈新, 刘志国, 王利虎, 刘平, 刘孟军, 张琼. 冬枣果皮着色相关类黄酮靶向代谢组学及潜在MYB转录因子分析[J]. 园艺学报, 2022, 49(8): 1761-1771. |
| [13] | 钱婕妤, 蒋玲莉, 郑钢, 陈佳红, 赖吴浩, 许梦晗, 付建新, 张超. 百日草花青素苷合成相关MYB转录因子筛选及ZeMYB9功能研究[J]. 园艺学报, 2022, 49(7): 1505-1518. |
| [14] | 吕正鑫, 贺艳群, 贾东峰, 黄春辉, 钟敏, 廖光联, 朱壹, 袁开昌, 刘传浩, 徐小彪. 猕猴桃种质资源表型性状遗传多样性分析[J]. 园艺学报, 2022, 49(7): 1571-1581. |
| [15] | 马维峰, 李艳梅, 马宗桓, 陈佰鸿, 毛娟. 苹果POD家族基因的鉴定与MdPOD15的功能分析[J]. 园艺学报, 2022, 49(6): 1181-1199. |
| 阅读次数 | ||||||
|
全文 |
|
|||||
|
摘要 |
|
|||||
版权所有 © 2012 《园艺学报》编辑部 京ICP备10030308号-2 国际联网备案号 11010802023439
编辑部地址: 北京市海淀区中关村南大街12号中国农业科学院蔬菜花卉研究所 邮编: 100081
电话: 010-82109523 E-Mail: yuanyixuebao@126.com
技术支持:北京玛格泰克科技发展有限公司
