
Acta Horticulturae Sinica ›› 2025, Vol. 52 ›› Issue (11): 2913-2930.doi: 10.16420/j.issn.0513-353x.2025-0011
• Genetic & Breeding · Germplasm Resources · Molecular Biology • Previous Articles Next Articles
ZHENG Conghui1,2, XU Zhenhua1,4,*( ), ZHANG Manman2,3, WANG Yuzhong1,2, LI Xiangjun1,2, DU Kejiu3, SUN Ran1,2, and LI Ya1,2
), ZHANG Manman2,3, WANG Yuzhong1,2, LI Xiangjun1,2, DU Kejiu3, SUN Ran1,2, and LI Ya1,2
Received:2025-06-20
Revised:2025-08-02
Online:2025-11-27
Published:2025-11-27
Contact:
XU Zhenhua
ZHENG Conghui, XU Zhenhua, ZHANG Manman, WANG Yuzhong, LI Xiangjun, DU Kejiu, SUN Ran, and LI Ya. Integrated Transcriptomic and Metabolomic Analysis of Flavonoid Biosynthesis in the Leaves of Ailanthus altissima‘Chaoyang’[J]. Acta Horticulturae Sinica, 2025, 52(11): 2913-2930.
Add to citation manager EndNote|Ris|BibTeX
URL: https://www.ahs.ac.cn/EN/10.16420/j.issn.0513-353x.2025-0011
| 材料 Material | 叶位 Leaf position | 样品组代号 Sample group | 颜色 Color | 颜色代码 Color code |
|---|---|---|---|---|
| 朝阳椿 Chaoyang | 1 | Y1 | 中度红橙Moderate reddish orange | N34D |
| 2 | Y2 | 浅绿黄Light greenish yellow | 6D | |
| 3 | Y3 | 亮黄绿Brilliant yellow green | 149B | |
| 普通臭椿 Common Ailanthus altissima | 1 | G1 | 中度橄榄棕色Moderate olive brown | 199A |
| 2 | G2 | 中绿Moderate green | 131C | |
| 3 | G3 | 深绿Dark green | 136A |
Table 1 Summary of leaf color
| 材料 Material | 叶位 Leaf position | 样品组代号 Sample group | 颜色 Color | 颜色代码 Color code |
|---|---|---|---|---|
| 朝阳椿 Chaoyang | 1 | Y1 | 中度红橙Moderate reddish orange | N34D |
| 2 | Y2 | 浅绿黄Light greenish yellow | 6D | |
| 3 | Y3 | 亮黄绿Brilliant yellow green | 149B | |
| 普通臭椿 Common Ailanthus altissima | 1 | G1 | 中度橄榄棕色Moderate olive brown | 199A |
| 2 | G2 | 中绿Moderate green | 131C | |
| 3 | G3 | 深绿Dark green | 136A |
| 类别 Type | 对照组—试验组 Control group— test group | 差异代谢物数 Number of differential metabolites | 差异基因数 Number of differentially expressed genes | ||
|---|---|---|---|---|---|
| 下调 Down-regulated | 上调 Up-regulated | 下调 Down-regulated | 上调 Up-regulated | ||
| 朝阳椿 Chaoyang | Y1-Y2 | 50 | 63 | 4 266 | 5 617 |
| Y1-Y3 | 69 | 67 | 12 219 | 11 431 | |
| Y2-Y3 | 39 | 35 | 6 763 | 4 828 | |
| 普通臭椿 Common Ailanthus altissima | G1-G2 | 42 | 117 | 7 968 | 8 265 |
| G1-G3 | 55 | 112 | 13 997 | 10 841 | |
| G2-G3 | 56 | 58 | 7 932 | 6 036 | |
| 普通臭椿和‘朝阳椿’ Common A. altissima and Chaoyang | G1-Y1 | 51 | 102 | 4 969 | 4 558 |
| G2-Y2 | 55 | 57 | 6 720 | 8 249 | |
| G3-Y3 | 62 | 55 | 7 970 | 10 096 | |
Table 2 Statistical table of differential metabolites and differentially expressed genes
| 类别 Type | 对照组—试验组 Control group— test group | 差异代谢物数 Number of differential metabolites | 差异基因数 Number of differentially expressed genes | ||
|---|---|---|---|---|---|
| 下调 Down-regulated | 上调 Up-regulated | 下调 Down-regulated | 上调 Up-regulated | ||
| 朝阳椿 Chaoyang | Y1-Y2 | 50 | 63 | 4 266 | 5 617 |
| Y1-Y3 | 69 | 67 | 12 219 | 11 431 | |
| Y2-Y3 | 39 | 35 | 6 763 | 4 828 | |
| 普通臭椿 Common Ailanthus altissima | G1-G2 | 42 | 117 | 7 968 | 8 265 |
| G1-G3 | 55 | 112 | 13 997 | 10 841 | |
| G2-G3 | 56 | 58 | 7 932 | 6 036 | |
| 普通臭椿和‘朝阳椿’ Common A. altissima and Chaoyang | G1-Y1 | 51 | 102 | 4 969 | 4 558 |
| G2-Y2 | 55 | 57 | 6 720 | 8 249 | |
| G3-Y3 | 62 | 55 | 7 970 | 10 096 | |
| 数据库 Database | 基因数 Number of genes | 百分比/% Percentage |
|---|---|---|
| KEGG | 68 246 | 41.33 |
| Nr | 93 228 | 56.45 |
| SwissProt | 66 292 | 40.14 |
| TrEMBL | 92 691 | 56.13 |
| KOG | 55 423 | 33.56 |
| GO | 77 384 | 46.86 |
| Pfam | 60 460 | 36.61 |
| 至少在一个数据库中有注释的基因 Annotated in at least one database | 94 616 | 57.30 |
| 基因总数 Total unigenes | 165 137 | 100.00 |
Table 3 Results of gene annotation
| 数据库 Database | 基因数 Number of genes | 百分比/% Percentage |
|---|---|---|
| KEGG | 68 246 | 41.33 |
| Nr | 93 228 | 56.45 |
| SwissProt | 66 292 | 40.14 |
| TrEMBL | 92 691 | 56.13 |
| KOG | 55 423 | 33.56 |
| GO | 77 384 | 46.86 |
| Pfam | 60 460 | 36.61 |
| 至少在一个数据库中有注释的基因 Annotated in at least one database | 94 616 | 57.30 |
| 基因总数 Total unigenes | 165 137 | 100.00 |
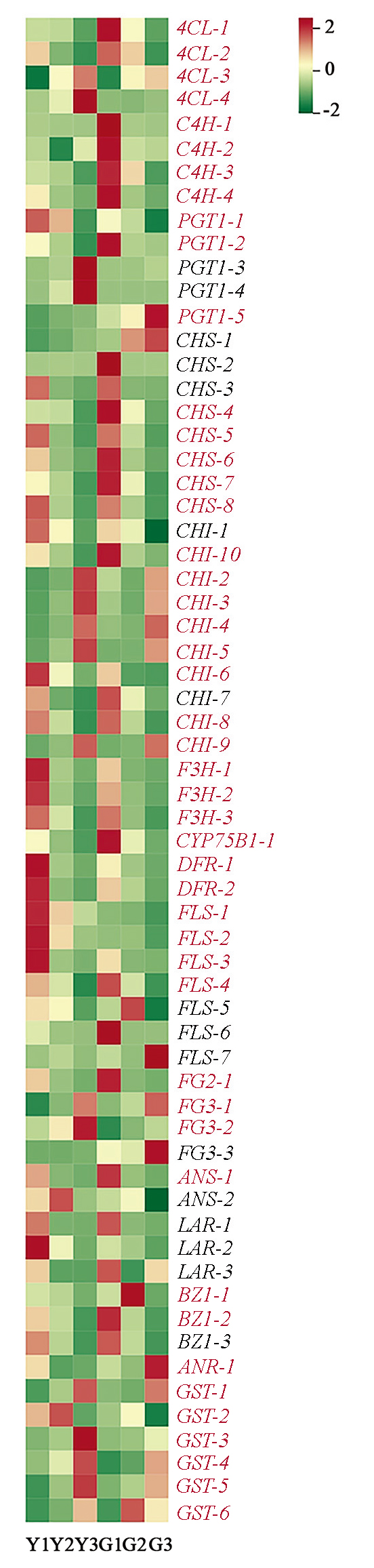
Fig. 6 Heat map of differentially expressed flavonoid related structual genes Genes in red letters represent the common genes of two types of Ailanthus altissima

Fig. 8 Correlation network of differential metabolites of flavonoids(anthocyanidins)and differentially expressed structural genes Circles represent differentially expressed structual genes,rhombuses represent differential metabolites,orange solid lines indicate a positive correlation,and green solid lines indicate a negative correlation. * indicate isomers. The same below
| 类目 Category | 代谢物 Metabolites | 结构基因 Structual genes | 转录因子基因 TF encoding genes |
|---|---|---|---|
| 花青素 Anthocyanidins | 矢车菊素-3-O-芸香糖苷 Cyanidin-3-O-rutinoside; 飞燕草素-3-O-葡萄糖苷* Delphinidin-3-O-glucoside | CHS-5;CHS-8;F3H-1;F3H-2;F3H-3;DFR-2;DFR-1;CHI-6 | MYB-4;bHLH-2;MYB-6;MYB-5;bHLH-3 |
| 查尔酮Chalcones | 柚皮素查尔酮 Naringenin chalcone;根皮苷 Phlorizin | CHS5;CHS8;PGT1-1 | - |
| 黄酮醇 Flavonols | 紫云英苷* Astragalin*;异槲皮苷* Isoquercitrin; 芦丁* Rutin;白麻苷 Baimaside | FLS-1;FLS-2;FG2-1;FG3-2 | - |
Table 4 Key differential metabolites and key differentially expressed genes of flavonoids(anthocyanidins)
| 类目 Category | 代谢物 Metabolites | 结构基因 Structual genes | 转录因子基因 TF encoding genes |
|---|---|---|---|
| 花青素 Anthocyanidins | 矢车菊素-3-O-芸香糖苷 Cyanidin-3-O-rutinoside; 飞燕草素-3-O-葡萄糖苷* Delphinidin-3-O-glucoside | CHS-5;CHS-8;F3H-1;F3H-2;F3H-3;DFR-2;DFR-1;CHI-6 | MYB-4;bHLH-2;MYB-6;MYB-5;bHLH-3 |
| 查尔酮Chalcones | 柚皮素查尔酮 Naringenin chalcone;根皮苷 Phlorizin | CHS5;CHS8;PGT1-1 | - |
| 黄酮醇 Flavonols | 紫云英苷* Astragalin*;异槲皮苷* Isoquercitrin; 芦丁* Rutin;白麻苷 Baimaside | FLS-1;FLS-2;FG2-1;FG3-2 | - |
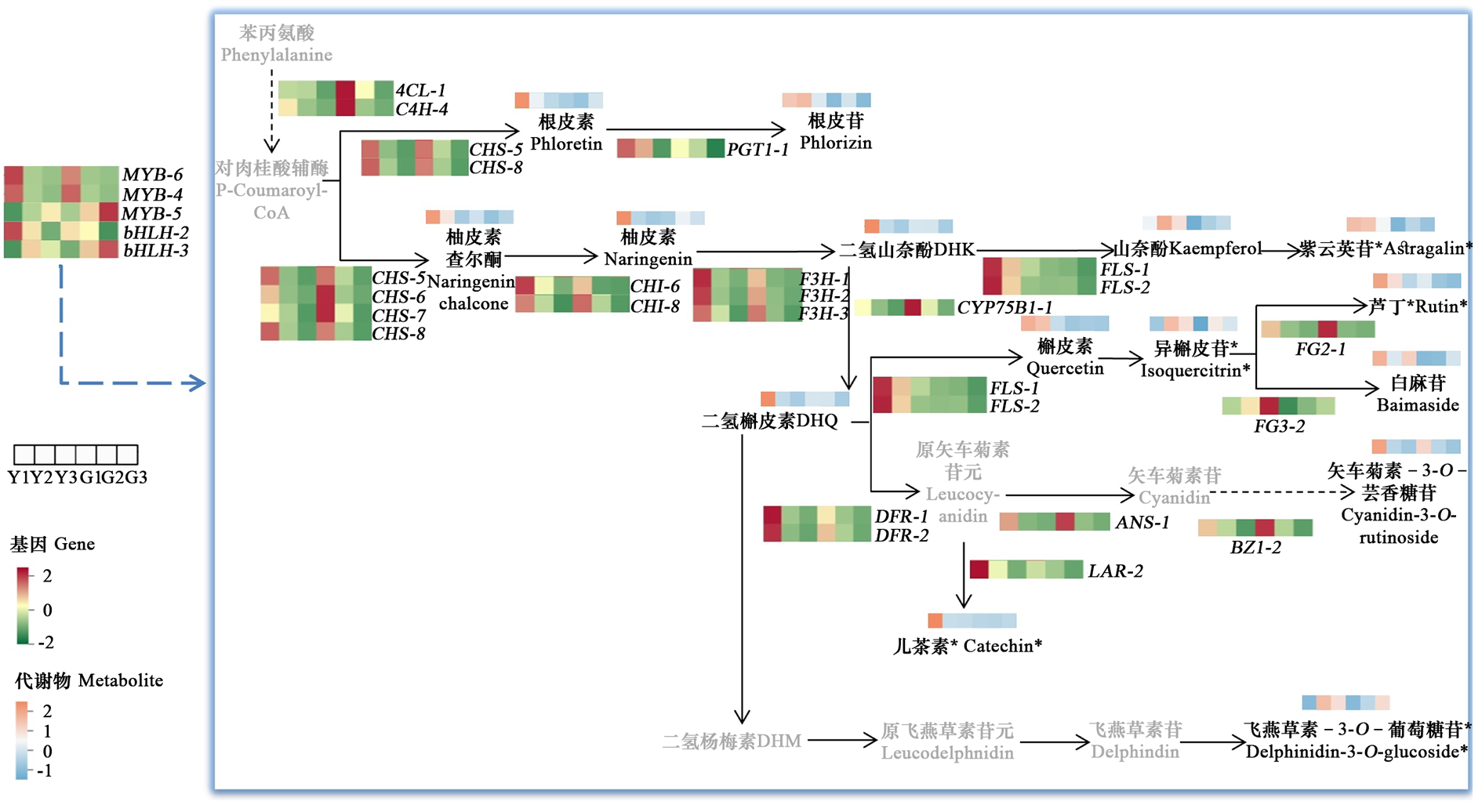
Fig. 11 Flavonoid (anthocyanin)synthesis pathway in leave of‘Chaoyang’and common Ailanthus altissima Metabolites in gray font represent the metabolites not detected in this study
| [1] |
|
| [2] |
|
|
陈梦微, 邓群仙, 龙星雨, 刘岩, 夏惠, 吕秀兰, 姚永桥. 2018. ‘脆红李’果实发育过程中花色苷含量及相关基因表达分析. 基因组学与应用生物学, 37 (7):2975-2982.
|
|
| [3] |
|
|
陈倩如, 蔡文淇, 张霞, 张大毛, 李卫东, 许璐, 于晓英, 李炎林. 2021. 檵木不同叶色形成的化学成分比较研究. 园艺学报, 48 (10):1969-1982.
|
|
| [4] |
|
|
戴思兰, 洪艳. 2016. 基于花青素苷合成和呈色机理的观赏植物花色改良分子育种. 中国农业科学, 49 (3):529-542.
|
|
| [5] |
|
| [6] |
|
| [7] |
|
| [8] |
|
|
付林江, 李厚华, 李玲, 于航, 王拉岐. 2013. 金银花花色变化原因分析. 林业科学, 49 (10):155-161.
|
|
| [9] |
|
|
葛诗蓓, 张学宁, 韩文炎, 李青云, 李鑫. 2023. 植物类黄酮的生物合成及其抗逆作用机制研究进展. 园艺学报, 50 (1):209-224.
|
|
| [10] |
|
|
葛雨萱, 王亮生, 徐彦军, 刘政安, 李崇晖, 贾妮. 2008. 蜡梅的花色和花色素组成及其在开花过程中的变化. 园艺学报, 35 (9):1331-1338.
|
|
| [11] |
|
|
郭鸿飞, 张延龙, 牛立新, 罗建让. 2015. 8种中国野生百合花色素成分分析. 西北农林科技大学学报(自然科学版), 43 (3):98-104.
|
|
| [12] |
|
| [13] |
|
|
韩雅茹, 马亚平, 陈丽华, 赵思明, 宋丽华. 2022. 转录组和代谢组联合解析气温升高和干旱互作下灵武长枣果皮花青苷代谢机制. 果树学报, 39 (5):811-825.
|
|
| [14] |
|
| [15] |
|
|
胡梦蝶, 李佳伟, 崔顺立, 侯名语, 杨鑫雷, 刘立峰, 蒋晓霞, 穆国俊. 2021. 花生花斑种皮花青素合成的转录组-代谢组联合分析. 植物遗传资源学报, 22 (6):1732-1745.
|
|
| [16] |
|
|
胡晓丹. 2021. 鲜食玉米籽粒花青素合成规律及其对糖脂代谢的调节作用[博士论文]. 广州: 华南理工大学.
|
|
| [17] |
|
| [18] |
|
|
金欣欣, 苏俏, 宋亚辉, 杨永庆, 李玉荣, 王瑾. 2024. 花生种皮类黄酮物质的代谢组与转录组分析. 作物学报, 50 (12):2950-2961.
|
|
| [19] |
|
| [20] |
|
|
李佳伟, 马钰聪, 杨鑫雷, 王梅, 崔顺立, 侯名语, 刘立峰, 胡梦蝶, 蒋晓霞, 穆国俊. 2022. 花生种皮色素合成相关通路的转录组—代谢组学联合分析. 植物遗传资源学报, 23 (1):240-254.
|
|
| [21] |
|
|
李文萍. 2014. 富含原花青素、叶酸和游离氨基酸的茶树优异种质资源的研究[硕士论文]. 福州: 福建农林大学.
|
|
| [22] |
|
|
李晓龙. 2023. ‘聊红’椿遗传转化体系的建立及果色调控相关miRNA的分离鉴定[硕士论文]. 聊城: 聊城大学.
|
|
| [23] |
|
| [24] |
|
| [25] |
|
|
李颖颖, 黄绵佳, 吴岚芳. 2012. 乙烯利对红掌佛焰苞花青素和总黄酮含量的影响. 中国农学通报, 28 (28):198-202.
|
|
| [26] |
|
| [27] |
|
|
陆小雨, 陈竹, 唐菲, 傅松玲, 任杰. 2020. 转录组与代谢组联合解析红花槭叶片中花青素苷变化机制. 林业科学, 56 (1):38-54.
|
|
| [28] |
|
| [29] |
|
|
牛义岭, 姜秀明, 许向阳. 2016. 植物转录因子MYB基因家族的研究进展. 分子植物育种,(8):2050-2059.
|
|
| [30] |
|
|
邱乾栋, 赵培宝, 郭尚敬, 于守超, 张秀省, 高祥斌. 2023. 观赏臭椿新品种‘聊红椿’. 园艺学报, 50 (7):1607-1608.
|
|
| [31] |
|
| [32] |
|
|
王超敏, 何美丹, 王文治, 袁潜华, 张树珍, 沈林波. 2024. 甘蔗条点病毒荧光定量PCR检测方法的建立及应用. 生物技术通报, 40 (6):126-133.
|
|
| [33] |
|
|
王明玉, 姜卫兵, 韩健, 王小青, 张斌斌, 马瑞娟. 2014. 不同红叶性状桃叶片花色素苷种类鉴定及呈色规律研究. 西北植物学报, 34 (7):1364-1370.
|
|
| [34] |
|
| [35] |
|
|
许海峰, 王中堂, 陈新, 刘志国, 王利虎, 刘平, 刘孟军, 张琼. 2022. 冬枣果皮着色相关类黄酮靶向代谢组学及潜在MYB 转录因子分析. 园艺学报, 49 (8):1761-1771.
|
|
| [36] |
|
|
荀守华. 2012. 彩叶乔木新品种——朝阳椿. 农业知识,(3):13.
|
|
| [37] |
|
| [38] |
|
|
余敏. 2020. 猕猴桃花青苷着色——MYB调节基因的鉴定及其功能解析[博士论文]. 武汉: 中国科学院武汉植物园.
|
|
| [39] |
|
| [40] |
|
|
郑聪慧, 徐振华, 王玉忠, 张蔓蔓, 杜克久, 李向军, 刘春鹏. 2023. 四种臭椿不同叶位叶片色素组成及分布变化. 中国农学通报, 39 (32):56-65.
|
|
| [41] |
|
|
钟培星. 2012. 芍药花瓣类黄酮成分分析及其对花色的影响[硕士论文]. 北京: 北京林业大学.
|
|
| [42] |
|
|
朱满兰. 2012. 睡莲花瓣类黄酮成分分析及其花色形成的化学机制[硕士论文]. 南京: 南京农业大学.
|
| [1] | SUN Dongyu, WEI Dong, YANG Yinyan, HU Caizhu, HU Zhiqun, ZHOU Donghui, and ZHOU Biyan. Physiological Response and Transcriptome Analysis of Wax Apple Flowering to Foliar Application of Chlorpyrifos [J]. Acta Horticulturae Sinica, 2025, 52(7): 1718-1732. |
| [2] | YU Cannan, GUO Anfang, LU Chun, MIAO Hongjun, HOU Xiaoyu, YUAN Tianyi, LIU Guoyuan, WEI Hui, ZHANG Jian, and YU Chunmei. Identification and Development of SNP Markers for Lagerstroemia indica Leaf Color Based on Transcriptome and Genome Analysis [J]. Acta Horticulturae Sinica, 2025, 52(7): 1803-1816. |
| [3] | YANG Chunmei, YU Yang, DING Yuge, XIA Jing, ZHOU Ling, and PENG Lei. Joint Transcription-Metabolism Analysis of Starch and Sucrose Metabolic Pathways in the Transformation of Axillary Buds into Flower Buds of ‘Guifei’Mango [J]. Acta Horticulturae Sinica, 2025, 52(6): 1412-1426. |
| [4] | GUO Xinmiao, TIAN Luyao, CAO Jiaqi, XIE Yan, GAO Yanxia, and SUN Zhichao. Preliminary Research on the Molecular Mechanism of Color Loss in White Mulberry Fruit [J]. Acta Horticulturae Sinica, 2025, 52(6): 1451-1462. |
| [5] | LI Ziyan, CHEN Weixi, LI Zihan, LI Yin, LIANG Fengming, ZENG Xiangli, JIAN Hongju, and LÜ Dianqiu. Screening Candidate Genes Controlling Potato Maturation Time Based on RNA-Seq [J]. Acta Horticulturae Sinica, 2025, 52(6): 1505-1518. |
| [6] | ZHANG Yue, FENG Yiliao, WANG Bei, REN Wenjing, JIANG Chunyu, ZHAO Xinyu, WANG Caihong, YANG Limei, ZHUANG Mu, LÜ Honghao, WANG Yong, ZHANG Yangyong, JI Jialei. Preliminary Transcriptome Analysis of Folate Synthesis and Metabolism in Cabbage [J]. Acta Horticulturae Sinica, 2025, 52(5): 1301-1316. |
| [7] | YUAN Juanwei, JIA Li, WANG Han, YAN Congsheng, ZHANG Qi’an, YU Feifei, GAN Defang, JIANG Haikun. Screening of Resistant Germplasm and Identifying Candidate Gene for Phytophthora capsici Resistance in Pepper Germplasm Resources [J]. Acta Horticulturae Sinica, 2025, 52(4): 857-871. |
| [8] | ZHANG Yiying, ZHANG Yu, CHU Yunxia, CHEN Hairong, DENG Shan, LI Aiai, ZHAO Hong, LIU Kun, HAN Ruixi, REN Li. Color Characteristics of Broccoli and Cauliflower Leaves Based on the Colorimeter Parameters [J]. Acta Horticulturae Sinica, 2025, 52(4): 872-882. |
| [9] | TANG Lingli, HE Yuhua, WANG Qingtao, AN Lulu, XU Yongyang, ZHANG Jian, KONG Weihu, HU Keyun, ZHAO Guangwei. Hormonal and Transcriptome Analysis of Ripe Fruits of Non-Climacteric and Climacteric Melon [J]. Acta Horticulturae Sinica, 2025, 52(4): 883-896. |
| [10] | SHAO Yifan, ZHU Baoqing, WANG Tongxin, LIAO Jianhe, WU Fanhua, YANG Siyi, FENG Jianhang, YU Xudong. Research of Genetic Regulation of Bombax ceiba Prickle Based on Hormone,Transcriptome and Metabolome [J]. Acta Horticulturae Sinica, 2025, 52(4): 933-946. |
| [11] | LIN Ruxue, WANG Fei, ZHANG Yanjie, MA Li, LIU Xiaofeng, LI Shuran, LIU Yalong, WANG Chen, JIANG Shuling, OU Chunqing. Transcriptome Screening and Functional Analysis of Dwarf Related Genes in Pear [J]. Acta Horticulturae Sinica, 2025, 52(3): 545-560. |
| [12] | FENG Kezheng, WANG Kang, WANG Yongkang, ZHAO Longlong, LIAO Na, TANG Xiaoyan, WANG Wenjie, WANG Chenggang, YUAN Lingyun, HOU Jinfeng, WU Jianqiang, and CHEN Guohu. Identification and Functional Analysis of Key Gene for Male Sterility in Wucai [J]. Acta Horticulturae Sinica, 2025, 52(11): 2876-2888. |
| [13] | FAN Amei, JIN Yaxin, HE Longjiao, LI Qi, ZHANG Yanqing, SHAO Qi, SONG Yun, NIU Yanbing, and QIAO Yonggang. Effects of Gibberellin on the Growth and Development of Forsythia suspensa Heterostyly [J]. Acta Horticulturae Sinica, 2025, 52(11): 3016-3030. |
| [14] | LI Hui, KAN Jialiang, LI Xiaogang, WANG Zhonghua, LIN Jing, YANG Qingsong. Effect of Gibberellin on Wizened Flower Buds Formation of Pyrus pyrifolia Cultivar‘Sucui 1’During the Growing Season [J]. Acta Horticulturae Sinica, 2025, 52(10): 2713-2726. |
| [15] | LI Jie, WU Chao, JIA Xiangqian, WANG Juan. Screening of Ziziphus jujuba‘Hupingzao’Fruit Coloring Substances and Their Related Genes [J]. Acta Horticulturae Sinica, 2024, 51(8): 1728-1742. |
| Viewed | ||||||
|
Full text |
|
|||||
|
Abstract |
|
|||||
Copyright © 2012 Acta Horticulturae Sinica 京ICP备10030308号-2 国际联网备案号 11010802023439
Tel: 010-82109523 E-Mail: yuanyixuebao@126.com
Support by: Beijing Magtech Co.Ltd