
Acta Horticulturae Sinica ›› 2023, Vol. 50 ›› Issue (11): 2365-2375.doi: 10.16420/j.issn.0513-353x.2022-0985
• Genetic & Breeding·Germplasm Resources·Molecular Biology • Previous Articles Next Articles
ZHANG Yongqi1,2, WANG Chao1,2, XU Linlin2, WU E’jiao2, LI Tianhong1, ZHAO Mizhen2, YUAN Huazhao2,*( )
)
Received:2023-03-15
Revised:2023-08-28
Online:2023-11-25
Published:2023-11-28
Contact:
YUAN Huazhao
ZHANG Yongqi, WANG Chao, XU Linlin, WU E’jiao, LI Tianhong, ZHAO Mizhen, YUAN Huazhao. Development of SSR Molecular Markers and Construction of Core Collection of Wild Diploid Strawberry[J]. Acta Horticulturae Sinica, 2023, 50(11): 2365-2375.
Add to citation manager EndNote|Ris|BibTeX
URL: https://www.ahs.ac.cn/EN/10.16420/j.issn.0513-353x.2022-0985
| 位点 Loci | 上游保守序列(5′-3′) Upstream conservative sequence | 下游保守序列(5′-3′) Downstream conservative sequence |
|---|---|---|
| SSR1 | TGCTAAGTAGAAGCCGCCA | AAAGCCATTTTACTACCTAGCC |
| SSR4 | TTAACAAGCACACGACAAAG | CATGGGTCTTGCTTATAGCC |
| SSR6 | TCCTCCATTATCTGCAGAAATC | TTCCAACCTCAAGCGTCT |
| SSR7 | CAAAGAGGGTTTCATTCATG | CGAACCCATGCACAC |
| SSR9 | AGACTTGTCTGGCCATTGTA | CACAGTACATCTCGCAAGGC |
| SSR10 | GTGACTTGATGTTGCTGTAAGT | CATACTACTGGAGATGCTC |
| SSR11 | GGTAATTTTCTTAAGTGACGCC | ATCGAACCAGCAGGCCT |
| SSR13 | CACCAATTTCACCACATATC | TTCTGTGAGTCTTGAATGTTGC |
| SSR17 | CCTCTCTTGTCCGGAACTT | CAGCAACTCCAAAGTTAAA |
| SSR18 | ATGCAATGTGTGTAAACACA | CACGTGCGTTTGTCAAC |
| SSR19 | TGTGCATTACTTGCTAATCC | CATCCTTTTGCTTTTGACCA |
| SSR20 | AGGCCCATTTCACTTCTGTC | TCACCCAACCCGGAAA |
| SSR21 | TATCTAGTAGTTGGAAAAGGA | TGTGGCATGGAAGTGG |
| SSR22 | TGCATCATGCATTCAAT | GGAAGCTGCATTTCCATAGAA |
| SSR23 | AATGTATATGTAGAATCC | GTCAATTGCCCATAGGTTAGC |
| SSR24 | TGATCGAGTTCAATTGAGTCAC | GGATCGAGGACCTAATCCC |
| SSR25 | CCCACACAAATTTAAAAGAGA | AGACTGAAATGGCCATGG |
| SSR26 | GATTGCTCAAAGTGCTTTAC | GCCTATAATTGCTGTCATGCTAG |
Table 1 Eighteen pairs of conserved sequence information before and after polymorphic SSR
| 位点 Loci | 上游保守序列(5′-3′) Upstream conservative sequence | 下游保守序列(5′-3′) Downstream conservative sequence |
|---|---|---|
| SSR1 | TGCTAAGTAGAAGCCGCCA | AAAGCCATTTTACTACCTAGCC |
| SSR4 | TTAACAAGCACACGACAAAG | CATGGGTCTTGCTTATAGCC |
| SSR6 | TCCTCCATTATCTGCAGAAATC | TTCCAACCTCAAGCGTCT |
| SSR7 | CAAAGAGGGTTTCATTCATG | CGAACCCATGCACAC |
| SSR9 | AGACTTGTCTGGCCATTGTA | CACAGTACATCTCGCAAGGC |
| SSR10 | GTGACTTGATGTTGCTGTAAGT | CATACTACTGGAGATGCTC |
| SSR11 | GGTAATTTTCTTAAGTGACGCC | ATCGAACCAGCAGGCCT |
| SSR13 | CACCAATTTCACCACATATC | TTCTGTGAGTCTTGAATGTTGC |
| SSR17 | CCTCTCTTGTCCGGAACTT | CAGCAACTCCAAAGTTAAA |
| SSR18 | ATGCAATGTGTGTAAACACA | CACGTGCGTTTGTCAAC |
| SSR19 | TGTGCATTACTTGCTAATCC | CATCCTTTTGCTTTTGACCA |
| SSR20 | AGGCCCATTTCACTTCTGTC | TCACCCAACCCGGAAA |
| SSR21 | TATCTAGTAGTTGGAAAAGGA | TGTGGCATGGAAGTGG |
| SSR22 | TGCATCATGCATTCAAT | GGAAGCTGCATTTCCATAGAA |
| SSR23 | AATGTATATGTAGAATCC | GTCAATTGCCCATAGGTTAGC |
| SSR24 | TGATCGAGTTCAATTGAGTCAC | GGATCGAGGACCTAATCCC |
| SSR25 | CCCACACAAATTTAAAAGAGA | AGACTGAAATGGCCATGG |
| SSR26 | GATTGCTCAAAGTGCTTTAC | GCCTATAATTGCTGTCATGCTAG |
| 位点Loci | Na | Ne | I | Ho | He | PIC |
|---|---|---|---|---|---|---|
| SSR1 | 27.000 | 9.435 | 2.678 | 0.188 | 0.894 | 0.887 |
| SSR4 | 13.000 | 1.479 | 0.836 | 0.006 | 0.324 | 0.315 |
| SSR6 | 27.000 | 8.005 | 2.593 | 0.443 | 0.875 | 0.867 |
| SSR7 | 24.000 | 5.728 | 2.378 | 0.188 | 0.825 | 0.816 |
| SSR9 | 36.000 | 13.120 | 2.981 | 0.216 | 0.924 | 0.920 |
| SSR10 | 29.000 | 10.574 | 2.704 | 0.352 | 0.905 | 0.898 |
| SSR11 | 16.000 | 1.751 | 1.129 | 0.028 | 0.429 | 0.419 |
| SSR13 | 7.000 | 1.687 | 0.911 | 0.051 | 0.407 | 0.387 |
| SSR17 | 34.000 | 15.419 | 3.000 | 0.477 | 0.935 | 0.932 |
| SSR18 | 18.000 | 2.448 | 1.472 | 0.119 | 0.591 | 0.569 |
| SSR19 | 11.000 | 2.886 | 1.560 | 0.119 | 0.653 | 0.632 |
| SSR20 | 49.000 | 21.158 | 3.362 | 0.432 | 0.953 | 0.951 |
| SSR21 | 23.000 | 8.990 | 2.526 | 0.153 | 0.889 | 0.879 |
| SSR22 | 21.000 | 4.063 | 2.015 | 0.222 | 0.754 | 0.739 |
| SSR23 | 25.000 | 7.088 | 2.398 | 0.222 | 0.859 | 0.846 |
| SSR24 | 30.000 | 14.681 | 2.934 | 0.307 | 0.932 | 0.928 |
| SSR25 | 34.000 | 9.835 | 2.746 | 0.347 | 0.898 | 0.891 |
| SSR26 | 20.000 | 3.827 | 2.003 | 0.199 | 0.739 | 0.726 |
| 平均Mean | 24.667 | 7.899 | 2.235 | 0.226 | 0.766 | 0.756 |
Table 2 Genetic diversity of 18 SSR in 176 strawberry materials
| 位点Loci | Na | Ne | I | Ho | He | PIC |
|---|---|---|---|---|---|---|
| SSR1 | 27.000 | 9.435 | 2.678 | 0.188 | 0.894 | 0.887 |
| SSR4 | 13.000 | 1.479 | 0.836 | 0.006 | 0.324 | 0.315 |
| SSR6 | 27.000 | 8.005 | 2.593 | 0.443 | 0.875 | 0.867 |
| SSR7 | 24.000 | 5.728 | 2.378 | 0.188 | 0.825 | 0.816 |
| SSR9 | 36.000 | 13.120 | 2.981 | 0.216 | 0.924 | 0.920 |
| SSR10 | 29.000 | 10.574 | 2.704 | 0.352 | 0.905 | 0.898 |
| SSR11 | 16.000 | 1.751 | 1.129 | 0.028 | 0.429 | 0.419 |
| SSR13 | 7.000 | 1.687 | 0.911 | 0.051 | 0.407 | 0.387 |
| SSR17 | 34.000 | 15.419 | 3.000 | 0.477 | 0.935 | 0.932 |
| SSR18 | 18.000 | 2.448 | 1.472 | 0.119 | 0.591 | 0.569 |
| SSR19 | 11.000 | 2.886 | 1.560 | 0.119 | 0.653 | 0.632 |
| SSR20 | 49.000 | 21.158 | 3.362 | 0.432 | 0.953 | 0.951 |
| SSR21 | 23.000 | 8.990 | 2.526 | 0.153 | 0.889 | 0.879 |
| SSR22 | 21.000 | 4.063 | 2.015 | 0.222 | 0.754 | 0.739 |
| SSR23 | 25.000 | 7.088 | 2.398 | 0.222 | 0.859 | 0.846 |
| SSR24 | 30.000 | 14.681 | 2.934 | 0.307 | 0.932 | 0.928 |
| SSR25 | 34.000 | 9.835 | 2.746 | 0.347 | 0.898 | 0.891 |
| SSR26 | 20.000 | 3.827 | 2.003 | 0.199 | 0.739 | 0.726 |
| 平均Mean | 24.667 | 7.899 | 2.235 | 0.226 | 0.766 | 0.756 |
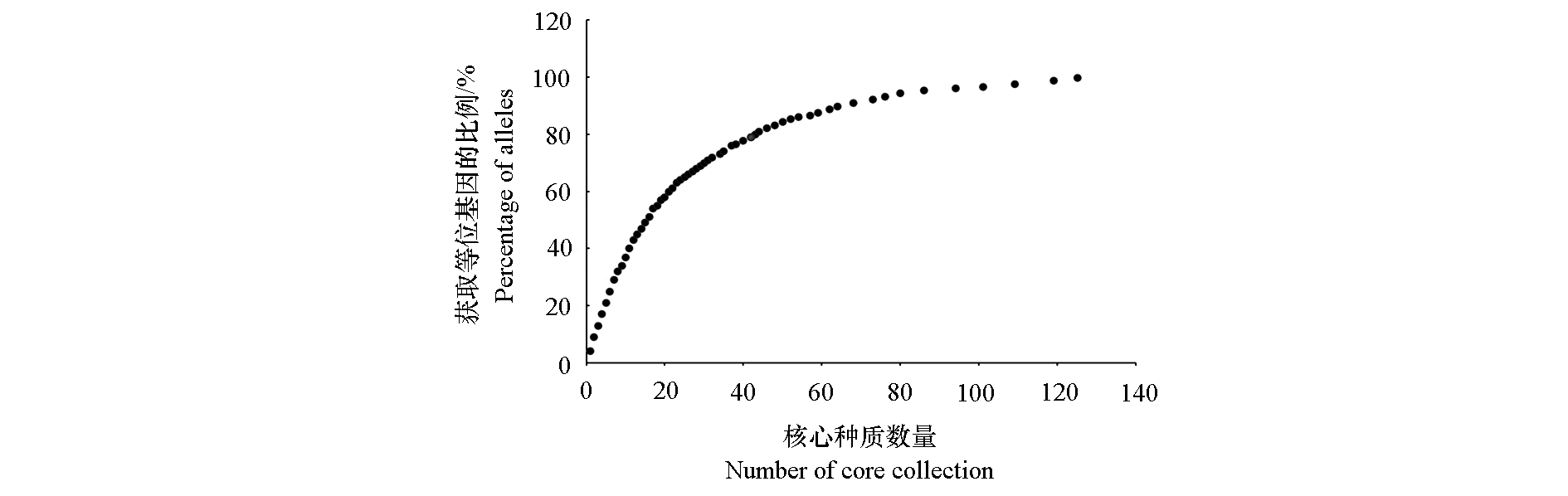
Fig. 4 Relationship between the prime number of core species and the proportion of retained alleles extracted from strawberry resources by Core Finder
| 类型 Type | 种质数Number of germplasms | N | Na | Ne | I | Ho | He | PIC |
|---|---|---|---|---|---|---|---|---|
| 原始种质 Original collection | 176.000 | 444 | 24.667 | 7.899 | 2.235 | 0.226 | 0.766 | 0.756 |
| 核心种质 Core collection | 43.000 (24.43%) | 366 (82.43%) | 20.333 (82.43%) | 10.497 (132.89%) | 2.498 (111.77%) | 0.260 (115.04%) | 0.852 (111.23%) | 0.842 (111.40%) |
| 保留种质 Reservation collection | 133.000 (75.57%) | 370 (83.33%) | 20.556 (83.33%) | 6.804 (86.14%) | 2.043 (91.41%) | 0.216 (95.58%) | 0.724 (94.52%) | 0.713 (84.65%) |
| t1 | 0.142 | 0.177 | 0.255 | 0.494 | 0.125 | 0.349 | ||
| t2 | 0.210 | 0.545 | 0.469 | 0.827 | 0.572 | 0.565 |
Table 3 Comparison of genetic diversity among wild strawberry core collection,original collection and reserved collection
| 类型 Type | 种质数Number of germplasms | N | Na | Ne | I | Ho | He | PIC |
|---|---|---|---|---|---|---|---|---|
| 原始种质 Original collection | 176.000 | 444 | 24.667 | 7.899 | 2.235 | 0.226 | 0.766 | 0.756 |
| 核心种质 Core collection | 43.000 (24.43%) | 366 (82.43%) | 20.333 (82.43%) | 10.497 (132.89%) | 2.498 (111.77%) | 0.260 (115.04%) | 0.852 (111.23%) | 0.842 (111.40%) |
| 保留种质 Reservation collection | 133.000 (75.57%) | 370 (83.33%) | 20.556 (83.33%) | 6.804 (86.14%) | 2.043 (91.41%) | 0.216 (95.58%) | 0.724 (94.52%) | 0.713 (84.65%) |
| t1 | 0.142 | 0.177 | 0.255 | 0.494 | 0.125 | 0.349 | ||
| t2 | 0.210 | 0.545 | 0.469 | 0.827 | 0.572 | 0.565 |
| [1] |
|
| [2] |
doi: 10.16420/j.issn.0513-353x.2021-0312 URL |
|
陈明堃, 陈璐, 孙维红, 马山虎, 兰思仁, 彭东辉, 刘仲健, 艾叶. 2022. 建兰种质资源遗传多样性分析及核心种质构建. 园艺学报, 49 (1):175-186.
doi: 10.16420/j.issn.0513-353x.2021-0312 URL |
|
| [3] |
doi: 10.1007/s001220051343 URL |
| [4] |
|
|
崔竣杰, 程蛟文, 曹毅, 胡开林. 2022. 基于SSR标记和表型性状构建苦瓜核心种质的研究. 中国蔬菜,(2):25-32.
|
|
| [5] |
doi: 10.1016/j.hpj.2019.11.004 |
| [6] |
|
| [7] |
|
|
贺润丽, 韩毅丽, 段静静, 刘计权, 王莉花. 2019. 款冬转录组SSR标记开发及其多态性研究. 中草药, 50 (20):5026-5032.
|
|
| [8] |
|
|
黄小凤, 韦阳连, 袁叶, 余金昌, 袁志永, 王伟山. 2022. 基于SNP分子标记的221份荔枝品种(品系)的遗传多样性分析及核心种质库构建. 植物资源与环境学报, 31 (4):74-84.
|
|
| [9] |
doi: 10.16420/j.issn.0513-353x.2020-0497 URL |
|
蒋爽, 张学英, 安海山, 徐芳杰, 章加应. 2021. 枇杷全基因组SSR标记开发及其多态性研究. 园艺学报, 48 (5):1013-1022.
doi: 10.16420/j.issn.0513-353x.2020-0497 URL |
|
| [10] |
doi: 10.16420/j.issn.0513-353x.2020-0608 URL |
|
江锡兵, 章平生, 徐阳, 吴聪连, 张东北, 龚榜初, 吴开云, 赖俊声. 2021. 栗杂交F1代SSR标记遗传多样性分析. 园艺学报, 48 (5):897-907.
doi: 10.16420/j.issn.0513-353x.2020-0608 URL |
|
| [11] |
doi: 10.1016/j.hpj.2021.05.001 URL |
| [12] |
doi: 10.1007/s11105-015-0966-7 URL |
| [13] |
|
|
雷家军, 薛莉, 代汉萍, 邓明琴. 2015. 世界草莓属(Fragaria)植物的种类与分布//草莓研究进展(Ⅳ). 北京: 中国农业出版社:364-375.
|
|
| [14] |
|
|
刘艳晶, 孙贵连, 周琴, 邹雪梅, 黄小龙. 2022. 森林草莓FvLysM基因家族的生物信息学分析. 分子植物育种, 20 (9):2864-2875.
|
|
| [15] |
|
|
刘遵春, 刘大亮, 崔美, 李敏, 焦其庆, 高利平, 陈学森. 2012. 整合农艺性状和分子标记数据构建新疆野苹果核心种质. 园艺学报, 39 (6):1045-1054.
|
|
| [16] |
|
| [17] |
|
|
缪黎明, 王神云, 邹明华, 李建斌, 孔李俊, 余小林. 2016. 园艺作物核心种质构建的研究进展. 植物遗传资源学报, 17 (5):791-800.
doi: 10.13430/j.cnki.jpgr.2016.05.001 |
|
| [18] |
doi: 10.1007/s10528-012-9509-1 URL |
| [19] |
|
|
覃阳, 全绍文, 周丽, 杨洁萍, 牛建新. 2019. 基于转录组测序的核桃(Juglans regia L.)SSR标记开发. 分子植物育种, 17 (20):6736-6742.
|
|
| [20] |
|
|
沈志军, 马瑞娟, 俞明亮, 蔡志翔, 许建兰. 2013. 国家果树种质南京桃资源圃初级核心种质构建. 园艺学报, 40 (1):125-134.
|
|
| [21] |
doi: 10.1038/ng.740 |
| [22] |
|
|
宋琳琳, 潘佳颖, 李梦思, 周晓君. 2022. 大叶铁线莲(Clematis heracleifolia DC.)多态性SSR引物开发. 分子植物育种, 20 (2):448-454.
|
|
| [23] |
|
|
孙永强, 陈建华, 张剑, 董胜君, 刘权钢, 刘青柏. 2022. 基于表型性状的西伯利亚杏核心种质构建. 沈阳农业大学学报, 53 (1):43-54.
|
|
| [24] |
doi: 10.1007/s001220051342 URL |
| [25] |
|
|
汪磊, 王姣梅, 汪魏, 王玲, 王力军, 严兴初, 谭美莲. 2021. 基于表型多样性构建向日葵核心种质. 中国油料作物学报, 43 (6):1052-1060.
doi: 10.19802/j.issn.1007-9084.2020255 |
|
| [26] |
|
|
曾钦朦, 曾亚军, 陈胜群, 侯娜. 2022. 贵州核桃核心种质筛选与构建研究. 西北植物学报, 42 (5):865-873.
|
|
| [27] |
|
|
张春雨, 陈学森, 张艳敏, 苑兆和, 刘遵春, 王延龄, 林群. 2009. 采用分子标记构建新疆野苹果核心种质的方法. 中国农业科学, 42 (2):597-604.
|
|
| [28] |
|
|
张利达, 唐克轩. 2010. 植物EST-SSR标记开发及其应用. 基因组学与应用生物学, 29 (3):534-541.
|
|
| [29] |
|
| [1] | ZHAO Xia, LI Gang, LIU Lifeng, HU Panpan, SONG Yanhong, and ZHOU Houcheng. A New Strawberry Cultivar‘Huafeng 1’ [J]. Acta Horticulturae Sinica, 2023, 50(S1): 35-36. |
| [2] | LIU Jinying, KONG Lingxi, WANG Weihao, QIN Guozheng, WANG Yuying. Advances in Aroma Compounds Biosynthesis of Strawberry Fruit [J]. Acta Horticulturae Sinica, 2023, 50(9): 1959-1970. |
| [3] | NIE Xinghua, ZHANG Yu, LIU Song, YANG Jiabin, HAO Yaqiong, LIU Yang, QIN Ling, XING Yu. Study on Genetic Characteristics and Taxonomic Status of Wild Chinese Chestnut Based on Genome Re-sequencing [J]. Acta Horticulturae Sinica, 2023, 50(8): 1622-1636. |
| [4] | MEI Yulin, XU Jinjian, JIANG Mengyue, YU Junhai, ZONG Yu, CHEN Wenrong, LIAO Fanglei, GUO Weidong. Identification of F1 Hybrids and Analysis of Fruit Shape Difference Between Fingered Citron and Citron [J]. Acta Horticulturae Sinica, 2023, 50(7): 1402-1418. |
| [5] | SUN Zeshuo, JIANG Dongyue, LIU Xinhong, SHEN Xin, LI Yingang, QU Yufei, LI Yonghua. Cluster Analysis and Construction of DNA Fingerprinting of 42 Oriental Cultivars of Flowering Cherry Based on SSR Markers [J]. Acta Horticulturae Sinica, 2023, 50(3): 657-668. |
| [6] | WANG Rui, HONG Wenjuan, LUO Hua, ZHAO Lina, CHEN Ying, WANG Jun. Construction of SSR Fingerprints of Pomegranate Cultivars and Male Parent Identification of Hybrids [J]. Acta Horticulturae Sinica, 2023, 50(2): 265-278. |
| [7] | WANG Mengmeng, SUN Deling, CHEN Rui, YANG Yingxia, ZHANG Guan, LÜ Mingjie, WANG Qian, XIE Tianyu, NIU Guobao, SHAN Xiaozheng, TAN Jin, YAO Xingwei. Construction and Evaluation of Cauliflower Core Collection [J]. Acta Horticulturae Sinica, 2023, 50(2): 421-431. |
| [8] | XIE Qian, ZHANG Shiyan, JIANG Lai, DING Mingyue, LIU Lingling, WU Rujian, CHEN Qingxi. Analysis of Canarium album Transcriptome SSR Information and Molecular Marker Development and Application [J]. Acta Horticulturae Sinica, 2023, 50(11): 2350-2364. |
| [9] | SU Qun, WANG Hongyan, LIU Jun, LI Chunniu, BU Zhaoyang, LIN Yuling, LU Jiashi, LAI Zhongxiong. Construction of Core Collection of Nymphaea Based on SSR Fluorescent Markers [J]. Acta Horticulturae Sinica, 2023, 50(10): 2128-2138. |
| [10] | LIU Yiping, NI Menghui, WU Fangfang, LIU Hongli, HE Dan, KONG Dezheng. Association Analysis of Organ Traits with SSR Markers in Lotus(Nelumbo nucifera) [J]. Acta Horticulturae Sinica, 2023, 50(1): 103-115. |
| [11] | HE Chengyong, ZHAO Xiaoli, XU Tengfei, GAO Dehang, LI Shifang, WANG Hongqing. Genome Sequence Analysis of Strawberry Virus 1 from Shandong Province,China [J]. Acta Horticulturae Sinica, 2023, 50(1): 153-160. |
| [12] | YANG Lei, LI Li, DONG Hui, FENG Jia, ZHANG Jianjun, FAN Jingfang, YANG Qiuye, and YANG Li, . A New Strawberry Cultivar‘Shimei 11’ [J]. Acta Horticulturae Sinica, 2022, 49(S2): 79-80. |
| [13] | ZHAO Xia, LI Gang, LIU Lifeng, SONG Yanhong, and ZHOU Houcheng. A New Strawberry Cultivar‘Huashuo 1’ [J]. Acta Horticulturae Sinica, 2022, 49(S2): 81-82. |
| [14] | DONG Jing, CHANG Linlin, WANG Guixia, ZHONG Chuanfei, WEI Yongqing, SUN Jian, SUN Rui, ZHANG Hongli, GAO Yongshun, XU Liping, TAO Pang, LUO Zhiwei, and ZHANG Yuntao, . A New Ever-bearing Strawberry Cultivar‘Jinghong’ [J]. Acta Horticulturae Sinica, 2022, 49(S1): 61-62. |
| [15] | LI Chao, YANG Ying, CHEN Wei, ZHENG Heyun, LIAO Xinfu, SUN Yuping. Construction of DNA Fingerprinting and Clustering Analysis with SSR Markers for the Muskmelon of Xizhoumi Series [J]. Acta Horticulturae Sinica, 2022, 49(3): 622-632. |
| Viewed | ||||||
|
Full text |
|
|||||
|
Abstract |
|
|||||
Copyright © 2012 Acta Horticulturae Sinica 京ICP备10030308号-2 国际联网备案号 11010802023439
Tel: 010-82109523 E-Mail: yuanyixuebao@126.com
Support by: Beijing Magtech Co.Ltd