
Acta Horticulturae Sinica ›› 2022, Vol. 49 ›› Issue (11): 2455-2470.doi: 10.16420/j.issn.0513-353x.2021-0876
• Research Notes • Previous Articles Next Articles
WU Zhijun1,5, LI Wei2, WANG Xinghua3, ZHANG Cheng2, JIANG Xun4, LI Sheng1, LUO Liyong1,5, SUN Kang1,5,*( ), ZENG Liang1,5,*(
), ZENG Liang1,5,*( )
)
Received:2021-12-02
Revised:2022-06-13
Online:2022-11-25
Published:2022-11-25
Contact:
SUN Kang,ZENG Liang
E-mail:sunkang@swu.edu.cn;zengliangbaby@126.com
CLC Number:
WU Zhijun, LI Wei, WANG Xinghua, ZHANG Cheng, JIANG Xun, LI Sheng, LUO Liyong, SUN Kang, ZENG Liang. Genetic Relationship Analysis of the Population Germplasms of Section Thea with Unstable Ovary Locule Number Based on SLAF-seq[J]. Acta Horticulturae Sinica, 2022, 49(11): 2455-2470.
Add to citation manager EndNote|Ris|BibTeX
URL: https://www.ahs.ac.cn/EN/10.16420/j.issn.0513-353x.2021-0876
| 来源地 Source | 种质名称 Germplasm name | 张宏达分类系统 Classification of Zhang Hongda | 闵天禄分类系统 Classification of Min Tianlu | 子房 室数 Locule number | 花柱柱头数 Stigma number | 有无毛 Hairy or not | 样本号 S ample ID | 测序号 BMK ID |
|---|---|---|---|---|---|---|---|---|
| 重庆北碚区 Beibei,Chongqing | 川小种 Chuanxiaozhong | 茶系 Ser. Sinenses | 茶 C. sinenses | 3 | 3 | 有Yes | BBCX1 | aj |
| 云南大叶种 Yunnan Large Leaf | 茶系 Ser. Sinenses | 茶 C. sinenses | 3 ~ 4 | 3 ~ 4 | 有Yes | BBYNDY1 | ai | |
| 七心白 Qixinbai | 红山茶组 Sect. Camellia | 山茶组 Sect. Camellia | - | - | - | CH | ab | |
| 茶梅 C. sasanqua | 油茶组 Sect. Oleifera | 油茶组 Sect. Paracamellia | - | - | - | CM | ac | |
| 来源地 Source | 种质名称 Germplasm name | 张宏达分类系统 Classification of Zhang Hongda | 闵天禄分类系统 Classification of Min Tianlu | 子房 室数 Locule number | 花柱柱头数 Stigma number | 有无毛 Hairy or not | 样本号 Sample ID | 测序号 BMK ID |
| 油茶 C. oleifera | 油茶组 Sect. Oleifera | 油茶组 Sect. Paracamellia | - | - | - | YC | bt | |
| 重庆江津区 Jiangjin,Chongqing | 江津大茶树 Big tea tree of Jiangjin | 秃房茶系 Ser. Gymnogynae | 疑似疏齿大厂茶 Suspected C. tachangensis var. remotiserrata | 3 ~ 4 | 3 ~ 4 | 无No | JJDS1 | df |
| JJDS2 | dg | |||||||
| JJDS3 | dh | |||||||
| JJDS4 | di | |||||||
| JJDS5 | dj | |||||||
| JJDS6 | dk | |||||||
| 重庆南川区 Nanchuan,Chongqing | 南川大茶树 Big tea tree of Nanchuan | 秃房茶系 Ser. Gymnogynae | 疏齿大厂茶 C. tachangensis var. remotiserrata | 3 | 3 | 无No | NCDS26 | ax |
| NCDS94 | bg | |||||||
| 3 ~ 4 | 3 ~ 4 | 无No | NCDS8 | bb | ||||
| NCDS13 | ap | |||||||
| NCDS14 | aq | |||||||
| NCDS15 | ar | |||||||
| NCDS24 | aw | |||||||
| NCDS89 | bd | |||||||
| NCDS93 | bf | |||||||
| NCDS108 | bl | |||||||
| NCDS109 | bm | |||||||
| NCDS113 | an | |||||||
| 五室茶系 Ser. Guinquelocularis | 疏齿大厂茶 C. tachangensis var. remotiserrata | 3 ~ 5 | 3 ~ 5 | 无No | NCDS18 | at | ||
| NCDS19 | au | |||||||
| NCDS20 | av | |||||||
| NCDS50 | ay | |||||||
| NCDS52 | az | |||||||
| NCDS88 | bc | |||||||
| NCDS97 | bh | |||||||
| NCDS111 | am | |||||||
| 疑似秃房茶/五室茶系 Suspected Ser. Gymnogynae/Ser. Guinquelocularis | 疑似疏齿大厂茶 Suspected C. tachangensis var. remotiserrata | - | - | - | NCDS107 | bk | ||
| NCDS110 | ao | |||||||
| 茶系(疑似秃房茶系 与茶系杂交个体) Ser. Sinenses(suspected hybrid individuals of Ser. Gymnogynae and Ser. Sinense) | 茶(疑似疏齿大厂茶与茶杂交个体) C. sinenses(suspected hybrid individuals of C. tachangensis var. remotiserrata and C. sinenses) | 3 ~ 4 | 3 ~ 4 | 有Yes | NCDS17 | as | ||
| NCDS53 | ba | |||||||
| NCDS90 | be | |||||||
| NCDS105 | bi | |||||||
| - | - | - | NCDS106 | bj | ||||
| 川小种 Chuanxiaozhong | 茶系 Ser. Sinenses | 茶 C. sinenses | 3 | 3 | 有Yes | NCCX8 | br | |
| NCCX18 | bp | |||||||
| NCCX24 | bq | |||||||
| 3 ~ 4 | 3 ~ 4 | 有Yes | NCCX10 | bn | ||||
| NCCX12 | bo | |||||||
| 重庆綦江区 Qijiang,Chongqing | 綦江大茶树 Big tea tree of Qijiang | 秃房茶系 Ser. Gymnogynae | 疏齿大厂茶 C. tachangensis var. remotiserrata | 3 ~ 4 | 3 ~ 4 | 无No | QJDS3 | de |
| 茶系(疑似秃房茶系 与茶系杂交个体) Ser. Sinenses(suspected hybrid individuals of Ser. Gymnogynae and Ser. Sinense) | 茶(疑似疏齿大厂茶与茶杂交个体) C. sinenses(suspected hybrid individuals of C. tachangensis var. remotiserrata and C. sinenses) | 3 ~ 4 | 3 ~ 4 | 有Yes | QJDS1 | dc | ||
| QJDS2 | dd | |||||||
| 重庆酉阳县 Youyang,Chongqing | 酉阳黄叶 Youyang Huangye | 茶系 Ser. Sinenses | 茶 C. sinenses | 3 | 3 | 有Yes | YYHY | dl |
| 福建安溪县 Anxi,Fujian | 黄旦 Huangdan | 茶系 Ser. Sinenses | 茶 C. sinenses | 3 | 3 | 有Yes | HD | ae |
| 福建福安市 Fu'an,Fujian | 金冠茶 Jinguancha | 茶系 Ser. Sinenses | 茶 C. sinenses | 3 | 3 | 有Yes | JGC | ag |
| 金牡丹 Jinmudan | 茶系 Ser. Sinenses | 茶 C. sinenses | 3 | 3 | 有Yes | JMD | ah | |
| 来源地 Source | 种质名称 Germplasm name | 张宏达分类系统 Classification of Zhang Hongda | 闵天禄分类系统 Classification of Min Tianlu | 子房 室数 Locule number | 花柱柱头数 Stigma number | 有无毛 Hairy or not | 样本号 Sample ID | 测序号 BMK ID |
| 茗冠 Mingguan | 茶系 Ser. Sinenses | 茶 C. sinenses | 3 | 3 | 有Yes | MG | al | |
| 福建福鼎市 Fuding,Fujian | 福鼎大白茶 Fuding Dabaicha | 茶系 Ser. Sinenses | 茶 C. sinenses | 3 | 3 | 有Yes | FDDB | ad |
| 广西昭平县 Zhaoping,Guangxi | 突肋茶 C. costata | 秃房茶系 Ser. Gymnogynae | 秃房茶 C. gymnogyna | 3 | 3 | 无No | GXTLC | db |
| 湖南汝城县 Rucheng,Hunan | 汝城毛叶茶 C. pubescens | 茶系 Ser. Sinenses | 茶 C. sinenses | 3 | 3 | 有Yes | RCMYC | da |
| 云南昌宁县 Changning,Yunnan | 昌宁秃房茶 C. gymnogyna | 秃房茶系 Ser. Gymnogynae | 秃房茶 C. gymnogyna | 3 | 3 | 无No | CN1 | dm |
| 云南江城县 Jiangcheng,Yunnan | 白毛茶 C. pubilimba | 茶系 Ser. Sinenses | 茶 C. sinenses | 3 | 3 | 有Yes | JC06-003 | cs |
| 云南景东县 Jingdong,Yunnan | 茶 C. sinensis | 茶系 Ser. Sinenses | 茶 C. sinenses | 3 | 3 | 有Yes | JD06-009 | cw |
| 云南景谷县 Jinggu,Yunnan | 大苞茶 C. grandibracteata | 五室茶系 Ser. Guinquelocularis | 大苞茶 C. grandibracteata | 5 | 5 | 无No | JG06-041 | cp |
| 茶C. sinensis | 茶系Ser. Sinenses | 茶C. sinenses | 3 | 3 | 有Yes | JG06-049 | cy | |
| JG06-048 | cx | |||||||
| 3 ~ 4 | 3 ~ 4 | 有Yes | JG06-035 | cz | ||||
| 云南澜沧县 Lancang,Yunnan | 普洱茶 C. assamica | 茶系 Ser. Sinenses | 茶 C. sinenses | 3 | 3 | 有Yes | LC06-003 | cq |
| LC06-027 | ck | |||||||
| 云南勐海县 Menghai,Yunnan | 紫鹃 Zijuan | 茶系 Ser. Sinenses | 茶 C. sinenses | 3 | 3 | 有Yes | ZJ | bx |
| 云南墨江县 Mojiang,Yunnan | 普洱茶 C. assamica | 茶系 Ser. Sinenses | 茶 C. sinenses | 3 | 3 | 有Yes | MJ06-009 | ch |
| MJ06-018 | ci | |||||||
| 云南宁洱县 Ning'er,Yunnan | 白毛茶 C. pubilimba | 茶系 Ser. Sinenses | 茶 C. sinenses | 3 | 3 | 有Yes | PR06-002 | ct |
| 云南思茅区 Simao,Yunnan | 普洱茶 C. assamica | 茶系 Ser. Sinenses | 茶 C. sinenses | 3 | 3 | 有Yes | CY06-008 | cj |
| 白毛茶 C. pubilimba | 茶系 Ser. Sinenses | 茶 C. sinenses | 3 | 3 | 有Yes | CY06-025 | cr | |
| 茶 C. sinensis | 茶系 Ser. Sinenses | 茶 C. sinenses | 3 | 3 | 有Yes | CY06-010 | cv | |
| 云南西盟县 Ximeng,Yunnan | 大理茶 C. taliensis | 五柱茶系 Ser. Pentastyla | 大理茶 C. taliensis | 5 | 5 | 有Yes | XM06-011 | by |
| XM06-010 | bz | |||||||
| XM06-015 | cb | |||||||
| 普洱茶 C. assamica | 茶系 Ser. Sinenses | 茶 C. sinenses | 3 | 3 | 有Yes | XM06-013 | ca | |
| 云南镇远县 Zhenyuan,Yunnan | 大理茶 C. taliensis | 五柱茶系 Ser. Pentastyla | 大理茶 C. taliensis | 5 | 5 | 有Yes | ZY06-001 | cc |
| ZY06-003 | cd | |||||||
| 普洱茶 C. assamica | 茶系 Ser. Sinenses | 茶 C. sinenses | 3 | 3 | 有Yes | ZY06-033 | cf | |
| 3 ~ 4 | 3 ~ 4 | 有Yes | ZY06-019 | ce | ||||
| ZY06-037 | cg | |||||||
| 茶 C. sinensis | 茶系 Ser. Sinenses | 茶 C. sinenses | 3 | 3 | 有Yes | ZY06-023 | cu | |
| 浙江嵊州市 Shengzhou,Zhejiang | 越黄1号/龙井早黄 Yuehuang 1/ Longjing Zaohuang | 茶系 Ser. Sinenses | 茶 C. sinenses | 3 | 3 | 有Yes | YH1 | bu |
| LJZH | ak | |||||||
| 浙江安吉县 Anji,Zhejiang | 白叶1号 Baiye 1 | 茶系 Ser. Sinenses | 茶 C. sinenses | 3 | 3 | 有Yes | BY | aa |
| 浙江余姚市 Yuyao,Zhejiang | 黄金芽 Huangjinya | 茶系 Ser. Sinenses | 茶 C. sinenses | 3 | 3 | 有Yes | HJY | af |
| 浙江永嘉县 Yongjia,Zhejiang | 乌牛早 Wuniuzao | 茶系 Ser. Sinenses | 茶 C. sinenses | 3 | 3 | 有Yes | WNZ | bs |
| 浙江宁波市 Ningbo,Zhejiang | 郁金香 Yujinxiang | 茶系 Ser. Sinenses | 茶 C. sinenses | 3 | 3 | 有Yes | YJX | bv |
| 浙江缙云县 Jinyun,Zhejiang | 中黄2号 Zhonghuang 2 | 茶系 Ser. Sinenses | 茶 C. sinenses | 3 | 3 | 有Yes | ZH2 | bw |
Table 1 Germplasm basic information of plant samples
| 来源地 Source | 种质名称 Germplasm name | 张宏达分类系统 Classification of Zhang Hongda | 闵天禄分类系统 Classification of Min Tianlu | 子房 室数 Locule number | 花柱柱头数 Stigma number | 有无毛 Hairy or not | 样本号 S ample ID | 测序号 BMK ID |
|---|---|---|---|---|---|---|---|---|
| 重庆北碚区 Beibei,Chongqing | 川小种 Chuanxiaozhong | 茶系 Ser. Sinenses | 茶 C. sinenses | 3 | 3 | 有Yes | BBCX1 | aj |
| 云南大叶种 Yunnan Large Leaf | 茶系 Ser. Sinenses | 茶 C. sinenses | 3 ~ 4 | 3 ~ 4 | 有Yes | BBYNDY1 | ai | |
| 七心白 Qixinbai | 红山茶组 Sect. Camellia | 山茶组 Sect. Camellia | - | - | - | CH | ab | |
| 茶梅 C. sasanqua | 油茶组 Sect. Oleifera | 油茶组 Sect. Paracamellia | - | - | - | CM | ac | |
| 来源地 Source | 种质名称 Germplasm name | 张宏达分类系统 Classification of Zhang Hongda | 闵天禄分类系统 Classification of Min Tianlu | 子房 室数 Locule number | 花柱柱头数 Stigma number | 有无毛 Hairy or not | 样本号 Sample ID | 测序号 BMK ID |
| 油茶 C. oleifera | 油茶组 Sect. Oleifera | 油茶组 Sect. Paracamellia | - | - | - | YC | bt | |
| 重庆江津区 Jiangjin,Chongqing | 江津大茶树 Big tea tree of Jiangjin | 秃房茶系 Ser. Gymnogynae | 疑似疏齿大厂茶 Suspected C. tachangensis var. remotiserrata | 3 ~ 4 | 3 ~ 4 | 无No | JJDS1 | df |
| JJDS2 | dg | |||||||
| JJDS3 | dh | |||||||
| JJDS4 | di | |||||||
| JJDS5 | dj | |||||||
| JJDS6 | dk | |||||||
| 重庆南川区 Nanchuan,Chongqing | 南川大茶树 Big tea tree of Nanchuan | 秃房茶系 Ser. Gymnogynae | 疏齿大厂茶 C. tachangensis var. remotiserrata | 3 | 3 | 无No | NCDS26 | ax |
| NCDS94 | bg | |||||||
| 3 ~ 4 | 3 ~ 4 | 无No | NCDS8 | bb | ||||
| NCDS13 | ap | |||||||
| NCDS14 | aq | |||||||
| NCDS15 | ar | |||||||
| NCDS24 | aw | |||||||
| NCDS89 | bd | |||||||
| NCDS93 | bf | |||||||
| NCDS108 | bl | |||||||
| NCDS109 | bm | |||||||
| NCDS113 | an | |||||||
| 五室茶系 Ser. Guinquelocularis | 疏齿大厂茶 C. tachangensis var. remotiserrata | 3 ~ 5 | 3 ~ 5 | 无No | NCDS18 | at | ||
| NCDS19 | au | |||||||
| NCDS20 | av | |||||||
| NCDS50 | ay | |||||||
| NCDS52 | az | |||||||
| NCDS88 | bc | |||||||
| NCDS97 | bh | |||||||
| NCDS111 | am | |||||||
| 疑似秃房茶/五室茶系 Suspected Ser. Gymnogynae/Ser. Guinquelocularis | 疑似疏齿大厂茶 Suspected C. tachangensis var. remotiserrata | - | - | - | NCDS107 | bk | ||
| NCDS110 | ao | |||||||
| 茶系(疑似秃房茶系 与茶系杂交个体) Ser. Sinenses(suspected hybrid individuals of Ser. Gymnogynae and Ser. Sinense) | 茶(疑似疏齿大厂茶与茶杂交个体) C. sinenses(suspected hybrid individuals of C. tachangensis var. remotiserrata and C. sinenses) | 3 ~ 4 | 3 ~ 4 | 有Yes | NCDS17 | as | ||
| NCDS53 | ba | |||||||
| NCDS90 | be | |||||||
| NCDS105 | bi | |||||||
| - | - | - | NCDS106 | bj | ||||
| 川小种 Chuanxiaozhong | 茶系 Ser. Sinenses | 茶 C. sinenses | 3 | 3 | 有Yes | NCCX8 | br | |
| NCCX18 | bp | |||||||
| NCCX24 | bq | |||||||
| 3 ~ 4 | 3 ~ 4 | 有Yes | NCCX10 | bn | ||||
| NCCX12 | bo | |||||||
| 重庆綦江区 Qijiang,Chongqing | 綦江大茶树 Big tea tree of Qijiang | 秃房茶系 Ser. Gymnogynae | 疏齿大厂茶 C. tachangensis var. remotiserrata | 3 ~ 4 | 3 ~ 4 | 无No | QJDS3 | de |
| 茶系(疑似秃房茶系 与茶系杂交个体) Ser. Sinenses(suspected hybrid individuals of Ser. Gymnogynae and Ser. Sinense) | 茶(疑似疏齿大厂茶与茶杂交个体) C. sinenses(suspected hybrid individuals of C. tachangensis var. remotiserrata and C. sinenses) | 3 ~ 4 | 3 ~ 4 | 有Yes | QJDS1 | dc | ||
| QJDS2 | dd | |||||||
| 重庆酉阳县 Youyang,Chongqing | 酉阳黄叶 Youyang Huangye | 茶系 Ser. Sinenses | 茶 C. sinenses | 3 | 3 | 有Yes | YYHY | dl |
| 福建安溪县 Anxi,Fujian | 黄旦 Huangdan | 茶系 Ser. Sinenses | 茶 C. sinenses | 3 | 3 | 有Yes | HD | ae |
| 福建福安市 Fu'an,Fujian | 金冠茶 Jinguancha | 茶系 Ser. Sinenses | 茶 C. sinenses | 3 | 3 | 有Yes | JGC | ag |
| 金牡丹 Jinmudan | 茶系 Ser. Sinenses | 茶 C. sinenses | 3 | 3 | 有Yes | JMD | ah | |
| 来源地 Source | 种质名称 Germplasm name | 张宏达分类系统 Classification of Zhang Hongda | 闵天禄分类系统 Classification of Min Tianlu | 子房 室数 Locule number | 花柱柱头数 Stigma number | 有无毛 Hairy or not | 样本号 Sample ID | 测序号 BMK ID |
| 茗冠 Mingguan | 茶系 Ser. Sinenses | 茶 C. sinenses | 3 | 3 | 有Yes | MG | al | |
| 福建福鼎市 Fuding,Fujian | 福鼎大白茶 Fuding Dabaicha | 茶系 Ser. Sinenses | 茶 C. sinenses | 3 | 3 | 有Yes | FDDB | ad |
| 广西昭平县 Zhaoping,Guangxi | 突肋茶 C. costata | 秃房茶系 Ser. Gymnogynae | 秃房茶 C. gymnogyna | 3 | 3 | 无No | GXTLC | db |
| 湖南汝城县 Rucheng,Hunan | 汝城毛叶茶 C. pubescens | 茶系 Ser. Sinenses | 茶 C. sinenses | 3 | 3 | 有Yes | RCMYC | da |
| 云南昌宁县 Changning,Yunnan | 昌宁秃房茶 C. gymnogyna | 秃房茶系 Ser. Gymnogynae | 秃房茶 C. gymnogyna | 3 | 3 | 无No | CN1 | dm |
| 云南江城县 Jiangcheng,Yunnan | 白毛茶 C. pubilimba | 茶系 Ser. Sinenses | 茶 C. sinenses | 3 | 3 | 有Yes | JC06-003 | cs |
| 云南景东县 Jingdong,Yunnan | 茶 C. sinensis | 茶系 Ser. Sinenses | 茶 C. sinenses | 3 | 3 | 有Yes | JD06-009 | cw |
| 云南景谷县 Jinggu,Yunnan | 大苞茶 C. grandibracteata | 五室茶系 Ser. Guinquelocularis | 大苞茶 C. grandibracteata | 5 | 5 | 无No | JG06-041 | cp |
| 茶C. sinensis | 茶系Ser. Sinenses | 茶C. sinenses | 3 | 3 | 有Yes | JG06-049 | cy | |
| JG06-048 | cx | |||||||
| 3 ~ 4 | 3 ~ 4 | 有Yes | JG06-035 | cz | ||||
| 云南澜沧县 Lancang,Yunnan | 普洱茶 C. assamica | 茶系 Ser. Sinenses | 茶 C. sinenses | 3 | 3 | 有Yes | LC06-003 | cq |
| LC06-027 | ck | |||||||
| 云南勐海县 Menghai,Yunnan | 紫鹃 Zijuan | 茶系 Ser. Sinenses | 茶 C. sinenses | 3 | 3 | 有Yes | ZJ | bx |
| 云南墨江县 Mojiang,Yunnan | 普洱茶 C. assamica | 茶系 Ser. Sinenses | 茶 C. sinenses | 3 | 3 | 有Yes | MJ06-009 | ch |
| MJ06-018 | ci | |||||||
| 云南宁洱县 Ning'er,Yunnan | 白毛茶 C. pubilimba | 茶系 Ser. Sinenses | 茶 C. sinenses | 3 | 3 | 有Yes | PR06-002 | ct |
| 云南思茅区 Simao,Yunnan | 普洱茶 C. assamica | 茶系 Ser. Sinenses | 茶 C. sinenses | 3 | 3 | 有Yes | CY06-008 | cj |
| 白毛茶 C. pubilimba | 茶系 Ser. Sinenses | 茶 C. sinenses | 3 | 3 | 有Yes | CY06-025 | cr | |
| 茶 C. sinensis | 茶系 Ser. Sinenses | 茶 C. sinenses | 3 | 3 | 有Yes | CY06-010 | cv | |
| 云南西盟县 Ximeng,Yunnan | 大理茶 C. taliensis | 五柱茶系 Ser. Pentastyla | 大理茶 C. taliensis | 5 | 5 | 有Yes | XM06-011 | by |
| XM06-010 | bz | |||||||
| XM06-015 | cb | |||||||
| 普洱茶 C. assamica | 茶系 Ser. Sinenses | 茶 C. sinenses | 3 | 3 | 有Yes | XM06-013 | ca | |
| 云南镇远县 Zhenyuan,Yunnan | 大理茶 C. taliensis | 五柱茶系 Ser. Pentastyla | 大理茶 C. taliensis | 5 | 5 | 有Yes | ZY06-001 | cc |
| ZY06-003 | cd | |||||||
| 普洱茶 C. assamica | 茶系 Ser. Sinenses | 茶 C. sinenses | 3 | 3 | 有Yes | ZY06-033 | cf | |
| 3 ~ 4 | 3 ~ 4 | 有Yes | ZY06-019 | ce | ||||
| ZY06-037 | cg | |||||||
| 茶 C. sinensis | 茶系 Ser. Sinenses | 茶 C. sinenses | 3 | 3 | 有Yes | ZY06-023 | cu | |
| 浙江嵊州市 Shengzhou,Zhejiang | 越黄1号/龙井早黄 Yuehuang 1/ Longjing Zaohuang | 茶系 Ser. Sinenses | 茶 C. sinenses | 3 | 3 | 有Yes | YH1 | bu |
| LJZH | ak | |||||||
| 浙江安吉县 Anji,Zhejiang | 白叶1号 Baiye 1 | 茶系 Ser. Sinenses | 茶 C. sinenses | 3 | 3 | 有Yes | BY | aa |
| 浙江余姚市 Yuyao,Zhejiang | 黄金芽 Huangjinya | 茶系 Ser. Sinenses | 茶 C. sinenses | 3 | 3 | 有Yes | HJY | af |
| 浙江永嘉县 Yongjia,Zhejiang | 乌牛早 Wuniuzao | 茶系 Ser. Sinenses | 茶 C. sinenses | 3 | 3 | 有Yes | WNZ | bs |
| 浙江宁波市 Ningbo,Zhejiang | 郁金香 Yujinxiang | 茶系 Ser. Sinenses | 茶 C. sinenses | 3 | 3 | 有Yes | YJX | bv |
| 浙江缙云县 Jinyun,Zhejiang | 中黄2号 Zhonghuang 2 | 茶系 Ser. Sinenses | 茶 C. sinenses | 3 | 3 | 有Yes | ZH2 | bw |

Fig. 1 Representative germplasm types investigated in Chongqing A. The population germplasms of section Thea with unstable ovary locule number(UONG);B. Suspected hybrid individuals of UONG and Ser. Sinenses germplasms.
| 样本编号 Sample ID | SLAF 数量 SLAF number | 平均深度/× Average depth | SNP 数量 SNP number | 完整性/% Integrity | 杂合率/% Heter ratio |
|---|---|---|---|---|---|
| BY1 | 414 220 | 5.55 | 6 454 305 | 25.75 | 3.34 |
| CH | 250 085 | 9.56 | 3 929 083 | 15.67 | 6.47 |
| CM | 332 076 | 8.02 | 4 926 261 | 19.65 | 10.57 |
| FDDB | 481 959 | 8.10 | 7 351 225 | 29.32 | 4.46 |
| HD | 506 591 | 8.91 | 7 763 226 | 30.97 | 4.14 |
| HJY | 486 495 | 8.03 | 7 480 906 | 29.84 | 3.99 |
| JGC | 459 171 | 8.25 | 7 190 058 | 28.68 | 4.13 |
| JMD | 449 894 | 7.43 | 7 142 843 | 28.49 | 3.96 |
| BBYNDY1 | 428 896 | 7.95 | 7 029 547 | 28.04 | 4.55 |
| BBCX1 | 461 256 | 10.3 | 7 211 715 | 28.77 | 3.92 |
| LJZH | 474 604 | 8.54 | 7 419 029 | 29.59 | 3.75 |
| MG | 461 136 | 7.34 | 7 384 539 | 29.46 | 4.10 |
| NCDS111 | 291 238 | 8.10 | 5 453 610 | 21.75 | 3.38 |
| NCDS113 | 320 219 | 10.52 | 5 890 530 | 23.50 | 3.61 |
| NCDS110 | 324 279 | 9.68 | 5 970 167 | 23.81 | 3.53 |
| NCDS13 | 339 038 | 11.17 | 6 099 118 | 24.33 | 3.71 |
| NCDS14 | 308 380 | 12.69 | 5 565 239 | 22.20 | 3.63 |
| NCDS15 | 323 666 | 11.57 | 5 833 824 | 23.27 | 3.75 |
| NCDS17 | 448 147 | 8.76 | 7 351 167 | 29.32 | 4.48 |
| NCDS18 | 326 089 | 9.15 | 5 961 543 | 23.78 | 3.37 |
| NCDS19 | 260 316 | 9.02 | 4 803 209 | 19.16 | 2.83 |
| NCDS20 | 282 640 | 9.73 | 5 134 816 | 20.48 | 3.17 |
| NCDS24 | 293 598 | 10.38 | 5 272 901 | 21.03 | 3.32 |
| NCDS26 | 328 771 | 10.99 | 5 940 411 | 23.70 | 3.48 |
| NCDS50 | 368 320 | 13.21 | 6 541 063 | 26.09 | 3.87 |
| NCDS52 | 347 655 | 11.97 | 6 253 213 | 24.94 | 3.65 |
| NCDS53 | 444 298 | 8.03 | 7 375 079 | 29.42 | 4.49 |
| NCDS8 | 292 326 | 9.71 | 5 303 949 | 21.16 | 3.22 |
| NCDS88 | 309 393 | 9.74 | 5 542 995 | 22.11 | 3.42 |
| NCDS89 | 320 760 | 9.42 | 5 790 379 | 23.10 | 3.53 |
| NCDS90 | 575 792 | 11.92 | 8 983 398 | 35.83 | 5.31 |
| NCDS93 | 338 289 | 10.75 | 5 949 992 | 23.73 | 3.69 |
| NCDS94 | 344 422 | 10.80 | 6 151 755 | 24.54 | 3.74 |
| NCDS97 | 270 397 | 7.26 | 4 931 892 | 19.67 | 3.07 |
| NCDS105 | 596 526 | 15.27 | 9 113 088 | 36.35 | 5.41 |
| NCDS106 | 448 437 | 13.71 | 7 229 172 | 28.84 | 4.44 |
| NCDS107 | 483 678 | 21.66 | 7 990 956 | 31.88 | 4.66 |
| NCDS108 | 400 903 | 16.93 | 6 815 528 | 27.19 | 4.16 |
| NCDS109 | 451 745 | 18.89 | 7 560 007 | 30.16 | 4.49 |
| NCCX10 | 446 271 | 9.38 | 6 912 577 | 27.57 | 3.81 |
| NCCX12 | 386 912 | 7.78 | 6 075 948 | 24.24 | 3.46 |
| NCCX18 | 368 217 | 7.18 | 5 805 146 | 23.16 | 3.63 |
| NCCX24 | 329 609 | 6.76 | 5 123 490 | 20.44 | 2.94 |
| NCCX8 | 758 583 | 18.24 | 10 167 608 | 40.56 | 5.50 |
| WNZ | 577 519 | 14.82 | 8 141 922 | 32.48 | 4.69 |
| YC | 251 323 | 7.35 | 3 804 367 | 15.18 | 9.00 |
| YH1 | 539 877 | 6.78 | 7 841 706 | 31.28 | 3.70 |
| YJX | 500 139 | 6.98 | 7 392 604 | 29.49 | 3.58 |
| ZH2 | 541 289 | 6.46 | 8 008 655 | 31.95 | 3.95 |
| 样本编号 Sample ID | SLAF 数量 SLAF number | 平均深度/× Average depth | SNP 数量 SNP number | 完整性/% Integrity | 杂合率/% Heter ratio |
| ZJ | 407 185 | 11.75 | 6 355 024 | 25.35 | 4.03 |
| XM06-011 | 457 256 | 13.40 | 7 898 877 | 31.51 | 5.03 |
| XM06-010 | 470 333 | 13.51 | 8 089 389 | 32.27 | 4.89 |
| XM06-013 | 530 537 | 10.70 | 8 703 605 | 34.72 | 4.48 |
| XM06-015 | 468 836 | 14.20 | 8 169 820 | 32.59 | 4.71 |
| ZY06-001 | 661 570 | 13.65 | 10 768 142 | 42.95 | 6.52 |
| ZY06-003 | 532 811 | 11.17 | 8 338 167 | 33.26 | 5.04 |
| ZY06-019 | 527 067 | 12.63 | 8 289 233 | 33.07 | 4.64 |
| ZY06-033 | 426 746 | 12.22 | 6 319 921 | 25.21 | 3.51 |
| ZY06-037 | 602 969 | 15.19 | 9 522 082 | 37.98 | 4.68 |
| MJ06-009 | 520 429 | 13.97 | 8 348 878 | 33.30 | 4.56 |
| MJ06-018 | 530 471 | 9.67 | 8 246 077 | 32.89 | 4.20 |
| CY06-008 | 441 645 | 11.65 | 7 146 514 | 28.51 | 3.79 |
| LC06-027 | 399 991 | 11.92 | 6 634 614 | 26.47 | 3.29 |
| JG06-041 | 544 734 | 11.78 | 8 940 958 | 35.67 | 5.49 |
| LC06-003 | 471 185 | 15.77 | 7 558 053 | 30.15 | 3.58 |
| CY06-025 | 515 104 | 12.35 | 8 162 424 | 32.56 | 4.39 |
| JC06-003 | 679 013 | 23.88 | 10 828 450 | 43.19 | 6.45 |
| PR06-002 | 604 820 | 18.15 | 9 397 217 | 37.49 | 4.81 |
| ZY06-023 | 666 653 | 12.28 | 11 356 933 | 45.30 | 7.35 |
| CY06-010 | 363 660 | 10.96 | 5 800 945 | 23.14 | 3.10 |
| JD06-009 | 449 962 | 7.96 | 7 124 788 | 28.42 | 4.17 |
| JG06-048 | 509 334 | 11.65 | 8 091 180 | 32.28 | 4.21 |
| JG06-049 | 457 856 | 11.21 | 7 283 365 | 29.05 | 3.74 |
| JG06-035 | 740 212 | 9.88 | 12 615 010 | 50.32 | 10.11 |
| RCMYC | 492 005 | 9.00 | 7 729 188 | 30.83 | 3.88 |
| GXTLC | 407 528 | 8.47 | 7 115 287 | 28.38 | 4.21 |
| QJDS1 | 515 390 | 11.03 | 8 268 692 | 32.98 | 4.96 |
| QJDS2 | 476 054 | 10.06 | 7 687 007 | 30.66 | 4.69 |
| QJDS3 | 513 155 | 20.96 | 8 747 664 | 34.89 | 5.14 |
| JJDS1 | 507 692 | 20.69 | 8 569 999 | 34.19 | 5.07 |
| JJDS2 | 512 383 | 26.77 | 8 532 684 | 34.04 | 5.00 |
| JJDS3 | 571 736 | 10.75 | 8 347 775 | 33.30 | 5.36 |
| JJDS4 | 609 961 | 18.93 | 9 615 444 | 38.36 | 5.68 |
| JJDS5 | 484 151 | 17.86 | 8 182 597 | 32.64 | 4.89 |
| JJDS6 | 423 805 | 15.78 | 7 195 866 | 28.70 | 4.15 |
| YYHY | 586 477 | 14.33 | 8 926 982 | 35.61 | 4.22 |
| CN1 | 460 655 | 13.79 | 7 382 921 | 29.45 | 3.63 |
Table 2 SLAF labels and SNP markers statistics of each sample
| 样本编号 Sample ID | SLAF 数量 SLAF number | 平均深度/× Average depth | SNP 数量 SNP number | 完整性/% Integrity | 杂合率/% Heter ratio |
|---|---|---|---|---|---|
| BY1 | 414 220 | 5.55 | 6 454 305 | 25.75 | 3.34 |
| CH | 250 085 | 9.56 | 3 929 083 | 15.67 | 6.47 |
| CM | 332 076 | 8.02 | 4 926 261 | 19.65 | 10.57 |
| FDDB | 481 959 | 8.10 | 7 351 225 | 29.32 | 4.46 |
| HD | 506 591 | 8.91 | 7 763 226 | 30.97 | 4.14 |
| HJY | 486 495 | 8.03 | 7 480 906 | 29.84 | 3.99 |
| JGC | 459 171 | 8.25 | 7 190 058 | 28.68 | 4.13 |
| JMD | 449 894 | 7.43 | 7 142 843 | 28.49 | 3.96 |
| BBYNDY1 | 428 896 | 7.95 | 7 029 547 | 28.04 | 4.55 |
| BBCX1 | 461 256 | 10.3 | 7 211 715 | 28.77 | 3.92 |
| LJZH | 474 604 | 8.54 | 7 419 029 | 29.59 | 3.75 |
| MG | 461 136 | 7.34 | 7 384 539 | 29.46 | 4.10 |
| NCDS111 | 291 238 | 8.10 | 5 453 610 | 21.75 | 3.38 |
| NCDS113 | 320 219 | 10.52 | 5 890 530 | 23.50 | 3.61 |
| NCDS110 | 324 279 | 9.68 | 5 970 167 | 23.81 | 3.53 |
| NCDS13 | 339 038 | 11.17 | 6 099 118 | 24.33 | 3.71 |
| NCDS14 | 308 380 | 12.69 | 5 565 239 | 22.20 | 3.63 |
| NCDS15 | 323 666 | 11.57 | 5 833 824 | 23.27 | 3.75 |
| NCDS17 | 448 147 | 8.76 | 7 351 167 | 29.32 | 4.48 |
| NCDS18 | 326 089 | 9.15 | 5 961 543 | 23.78 | 3.37 |
| NCDS19 | 260 316 | 9.02 | 4 803 209 | 19.16 | 2.83 |
| NCDS20 | 282 640 | 9.73 | 5 134 816 | 20.48 | 3.17 |
| NCDS24 | 293 598 | 10.38 | 5 272 901 | 21.03 | 3.32 |
| NCDS26 | 328 771 | 10.99 | 5 940 411 | 23.70 | 3.48 |
| NCDS50 | 368 320 | 13.21 | 6 541 063 | 26.09 | 3.87 |
| NCDS52 | 347 655 | 11.97 | 6 253 213 | 24.94 | 3.65 |
| NCDS53 | 444 298 | 8.03 | 7 375 079 | 29.42 | 4.49 |
| NCDS8 | 292 326 | 9.71 | 5 303 949 | 21.16 | 3.22 |
| NCDS88 | 309 393 | 9.74 | 5 542 995 | 22.11 | 3.42 |
| NCDS89 | 320 760 | 9.42 | 5 790 379 | 23.10 | 3.53 |
| NCDS90 | 575 792 | 11.92 | 8 983 398 | 35.83 | 5.31 |
| NCDS93 | 338 289 | 10.75 | 5 949 992 | 23.73 | 3.69 |
| NCDS94 | 344 422 | 10.80 | 6 151 755 | 24.54 | 3.74 |
| NCDS97 | 270 397 | 7.26 | 4 931 892 | 19.67 | 3.07 |
| NCDS105 | 596 526 | 15.27 | 9 113 088 | 36.35 | 5.41 |
| NCDS106 | 448 437 | 13.71 | 7 229 172 | 28.84 | 4.44 |
| NCDS107 | 483 678 | 21.66 | 7 990 956 | 31.88 | 4.66 |
| NCDS108 | 400 903 | 16.93 | 6 815 528 | 27.19 | 4.16 |
| NCDS109 | 451 745 | 18.89 | 7 560 007 | 30.16 | 4.49 |
| NCCX10 | 446 271 | 9.38 | 6 912 577 | 27.57 | 3.81 |
| NCCX12 | 386 912 | 7.78 | 6 075 948 | 24.24 | 3.46 |
| NCCX18 | 368 217 | 7.18 | 5 805 146 | 23.16 | 3.63 |
| NCCX24 | 329 609 | 6.76 | 5 123 490 | 20.44 | 2.94 |
| NCCX8 | 758 583 | 18.24 | 10 167 608 | 40.56 | 5.50 |
| WNZ | 577 519 | 14.82 | 8 141 922 | 32.48 | 4.69 |
| YC | 251 323 | 7.35 | 3 804 367 | 15.18 | 9.00 |
| YH1 | 539 877 | 6.78 | 7 841 706 | 31.28 | 3.70 |
| YJX | 500 139 | 6.98 | 7 392 604 | 29.49 | 3.58 |
| ZH2 | 541 289 | 6.46 | 8 008 655 | 31.95 | 3.95 |
| 样本编号 Sample ID | SLAF 数量 SLAF number | 平均深度/× Average depth | SNP 数量 SNP number | 完整性/% Integrity | 杂合率/% Heter ratio |
| ZJ | 407 185 | 11.75 | 6 355 024 | 25.35 | 4.03 |
| XM06-011 | 457 256 | 13.40 | 7 898 877 | 31.51 | 5.03 |
| XM06-010 | 470 333 | 13.51 | 8 089 389 | 32.27 | 4.89 |
| XM06-013 | 530 537 | 10.70 | 8 703 605 | 34.72 | 4.48 |
| XM06-015 | 468 836 | 14.20 | 8 169 820 | 32.59 | 4.71 |
| ZY06-001 | 661 570 | 13.65 | 10 768 142 | 42.95 | 6.52 |
| ZY06-003 | 532 811 | 11.17 | 8 338 167 | 33.26 | 5.04 |
| ZY06-019 | 527 067 | 12.63 | 8 289 233 | 33.07 | 4.64 |
| ZY06-033 | 426 746 | 12.22 | 6 319 921 | 25.21 | 3.51 |
| ZY06-037 | 602 969 | 15.19 | 9 522 082 | 37.98 | 4.68 |
| MJ06-009 | 520 429 | 13.97 | 8 348 878 | 33.30 | 4.56 |
| MJ06-018 | 530 471 | 9.67 | 8 246 077 | 32.89 | 4.20 |
| CY06-008 | 441 645 | 11.65 | 7 146 514 | 28.51 | 3.79 |
| LC06-027 | 399 991 | 11.92 | 6 634 614 | 26.47 | 3.29 |
| JG06-041 | 544 734 | 11.78 | 8 940 958 | 35.67 | 5.49 |
| LC06-003 | 471 185 | 15.77 | 7 558 053 | 30.15 | 3.58 |
| CY06-025 | 515 104 | 12.35 | 8 162 424 | 32.56 | 4.39 |
| JC06-003 | 679 013 | 23.88 | 10 828 450 | 43.19 | 6.45 |
| PR06-002 | 604 820 | 18.15 | 9 397 217 | 37.49 | 4.81 |
| ZY06-023 | 666 653 | 12.28 | 11 356 933 | 45.30 | 7.35 |
| CY06-010 | 363 660 | 10.96 | 5 800 945 | 23.14 | 3.10 |
| JD06-009 | 449 962 | 7.96 | 7 124 788 | 28.42 | 4.17 |
| JG06-048 | 509 334 | 11.65 | 8 091 180 | 32.28 | 4.21 |
| JG06-049 | 457 856 | 11.21 | 7 283 365 | 29.05 | 3.74 |
| JG06-035 | 740 212 | 9.88 | 12 615 010 | 50.32 | 10.11 |
| RCMYC | 492 005 | 9.00 | 7 729 188 | 30.83 | 3.88 |
| GXTLC | 407 528 | 8.47 | 7 115 287 | 28.38 | 4.21 |
| QJDS1 | 515 390 | 11.03 | 8 268 692 | 32.98 | 4.96 |
| QJDS2 | 476 054 | 10.06 | 7 687 007 | 30.66 | 4.69 |
| QJDS3 | 513 155 | 20.96 | 8 747 664 | 34.89 | 5.14 |
| JJDS1 | 507 692 | 20.69 | 8 569 999 | 34.19 | 5.07 |
| JJDS2 | 512 383 | 26.77 | 8 532 684 | 34.04 | 5.00 |
| JJDS3 | 571 736 | 10.75 | 8 347 775 | 33.30 | 5.36 |
| JJDS4 | 609 961 | 18.93 | 9 615 444 | 38.36 | 5.68 |
| JJDS5 | 484 151 | 17.86 | 8 182 597 | 32.64 | 4.89 |
| JJDS6 | 423 805 | 15.78 | 7 195 866 | 28.70 | 4.15 |
| YYHY | 586 477 | 14.33 | 8 926 982 | 35.61 | 4.22 |
| CN1 | 460 655 | 13.79 | 7 382 921 | 29.45 | 3.63 |
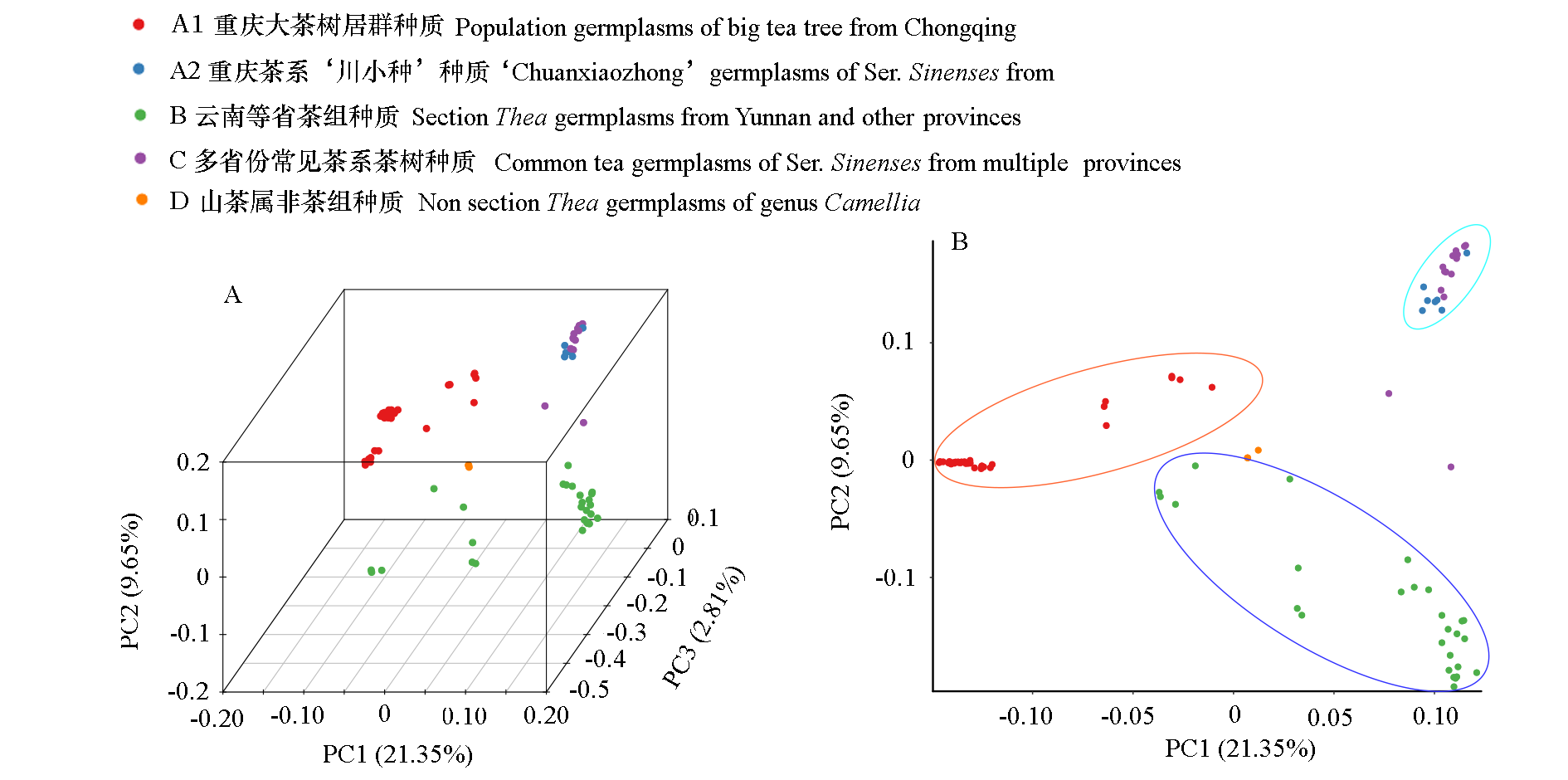
Fig. 6 PCA three-dimensional cluster diagram(A)and two-dimensional cluster diagram(B)of each sample A point represents a sample and a color represents a group.
| 样本群Sample group | A1 | A2 | B | C | D |
|---|---|---|---|---|---|
| A1 | |||||
| A2 | 0.396658 | ||||
| B | 0.311787 | 0.218657 | |||
| C | 0.401042 | 0.084062 | 0.219729 | ||
| D | 0.379444 | 0.416460 | 0.272662 | 0.396544 |
Table 3 Pairwise comparison of genetic differentiation coefficient among sample groups
| 样本群Sample group | A1 | A2 | B | C | D |
|---|---|---|---|---|---|
| A1 | |||||
| A2 | 0.396658 | ||||
| B | 0.311787 | 0.218657 | |||
| C | 0.401042 | 0.084062 | 0.219729 | ||
| D | 0.379444 | 0.416460 | 0.272662 | 0.396544 |
| [1] |
Alexander D H, Novembre J, Lange K. 2009. Fast model-based estimation of ancestry in unrelated individuals. Genome Research, 19:1655-1664.
doi: 10.1101/gr.094052.109 pmid: 19648217 |
| [2] | Chen Jiedan, Ma Chunlei, Chen Liang. 2019. Research on tea germplasm resources in China for 40 years. China Tea, 41:1-5,46. (in Chinese) |
| 陈杰丹, 马春雷, 陈亮. 2019. 我国茶树种质资源研究40年. 中国茶叶, 41:1-5,46. | |
| [3] | Chen Liang, Yu Fulian, Tong Qiqing. 2000. Discussions on phylogenetic classification and evolution of Sect. Thea. Journal of Tea Science, 20 (2):89-94. (in Chinese) |
| 陈亮, 虞富莲, 童启庆. 2000. 关于茶组植物分类与演化的讨论. 茶叶科学, 20 (2):89-94. | |
| [4] |
Clark S E, Running M P, Meyerowitz E M. 1995. CLAVATA3 is a specific regulator of shoot and floral meristem development affecting the same processes as CLAVATA1. Development, 121:2057-2067.
doi: 10.1242/dev.121.7.2057 URL |
| [5] |
Excoffier L, Lischer H E L. 2010. Arlequin suite ver 3.5:a new series of programs to perform population genetics analyses under Linux and Windows. Molecular Ecology Resources, 10:564-567.
doi: 10.1111/j.1755-0998.2010.02847.x pmid: 21565059 |
| [6] | Gong Wanzhuo, Zhang Zecen. 2006. A review on the classification of section Thea. Tea In Fujian,(2):9-12. (in Chinese) |
| 龚万灼, 张泽岑. 2006. 茶组植物分类研究综述. 福建茶叶,(2):9-12. | |
| [7] | Guo Yan, Qiao Dahe, Yang Chun, Li Yan, Chen Zhengwu,Chen Juan. 2019. Genetic diversity of old tea plant resources in Jiuan city of Guizhou Province,using Genome-Wide SNP. Journal of Plant Genetic Resources, 20:26-36. (in Chinese) |
| 郭燕, 乔大河, 杨春, 李燕, 陈正武, 陈娟. 2019. 基于全基因组SNP的贵州久安古茶树遗传关系分析. 植物遗传资源学报, 20:26-36. | |
| [8] | Hicks A. 2009. Current status and future development of global tea production and tea products. Tempo Social, 12:251-264. |
| [9] |
Holsinger K E, Weir B S. 2009. Genetics in geographically structured populations:defining,estimating and interpreting Fst. Nature Reviews Genetics, 10:639-650.
doi: 10.1038/nrg2611 pmid: 19687804 |
| [10] |
Kumar S, Stecher G, Li M, Knyaz C, Tamura K. 2018. MEGA X:molecular evolutionary genetics analysis across computing platforms. Molecular Biology and Evolution, 35:1547-1549.
doi: 10.1093/molbev/msy096 URL |
| [11] |
Li H, Durbin R. 2009. Fast and accurate short read alignment with Burrows-Wheeler transform. Bioinformatics, 25:1754-1760.
doi: 10.1093/bioinformatics/btp324 pmid: 19451168 |
| [12] |
Li H, Handsaker B, Wysoker A, Fennell T, Ruan J, Homer N, Marth G, Abecasis G, Durbin R. 2009. The sequence alignment/map format and SAMtools. Bioinformatics, 25:2078-2079.
doi: 10.1093/bioinformatics/btp352 pmid: 19505943 |
| [13] | Li Huajun, Wei Xu, Tan Shuli, Xu Rongbo, Liu Qinjin. 2018. Three novel wild tea in Chongqing and their status in botanical classification. Journal of Tea, 44 (1):1-5. (in Chinese) |
| 李华钧, 魏旭, 谭树立, 徐荣波, 刘勤晋. 2018. 重庆大茶树“三新种”及其分类学意义. 茶叶, 44 (1):1-5. | |
| [14] |
Li R, Li Y, Kristiansen K, Wang J. 2008. SOAP:short oligonucleotide alignment program. Bioinformatics, 24 (5):713-714.
doi: 10.1093/bioinformatics/btn025 URL |
| [15] |
Li S, Pan Y, Wen C, Li Y, Liu X, Zhang X, Behera T K, Xing G, Weng Y. 2016. Integrated analysis in bi-parental and natural populations reveals CsCLAVATA3(CsCLV3)underlying carpel number variations in cucumber. Theoretical and Applied Genetics, 129:1007-1022.
doi: 10.1007/s00122-016-2679-1 URL |
| [16] | Li Xinghui, Shi Zhaopeng, Tang Heping, Li Milu. 2002. Research on genetic relationship and classification of tea germplasm resources. Tea In Fujian,(1):10-14. (in Chinese) |
| 黎星辉, 施兆鹏, 唐和平, 李觅路. 2002. 茶树种质资源亲缘关系与分类的研究概况. 福建茶叶,(1):10-14. | |
| [17] |
Ma J, Huang L, Ma C, Jin J, Li C, Wang R, Zheng H, Yao M, Chen L. 2015. Large-scale SNP discovery and genotyping for constructing a high-density genetic map of tea plant using specific-locus amplified fragment sequencing(SLAF-seq). PLoS ONE, 10 (6):e0128798.
doi: 10.1371/journal.pone.0128798 URL |
| [18] |
McKenna A, Hanna M, Banks E, Sivachenko A, Cibulskis K, Kernytsky A, Garimella K, Altshuler D, Gabriel S, Daly M, DePristo M A. 2010. The Genome Analysis Toolkit:a MapReduce framework for analyzing next-generation DNA sequencing data. Genome Research, 20:1297-1303.
doi: 10.1101/gr.107524.110 pmid: 20644199 |
| [19] |
Miller M R, Dunham J P, Amores A, Cresko W A, Johnson E A. 2007. Rapid and cost-effective polymorphism identification and genotyping using restriction site associated DNA(RAD)markers. Genome Research, 17:240-248.
doi: 10.1101/gr.5681207 URL |
| [20] | Min Tianlu. 1992. A revision of Camellia sect. Thea. Acta Botanica Yunnanica, 14 (2):115-132. (in Chinese) |
| 闵天禄. 1992. 山茶属茶组植物的订正. 云南植物研究, 14 (2):115-132. | |
| [21] | Min Tianlu. 2000. Monograph of the genus Camellia. Kunming: Yunnan Science and Technology Press. (in Chinese) |
| 闵天禄. 2000. 世界山茶属的研究. 昆明: 云南科技出版社. | |
| [22] | Ming T L, Bartholomew B. 2007. Theaceae. Flora of China. Beijing:Science Press & St. Louis: Missouri Botanical Garden Press. |
| [23] |
Niu S, Koiwa H, Song Q, Qiao D, Chen J, Zhao D, Chen Z, Wang Y, Zhang T. 2020. Development of core-collections for Guizhou tea genetic resources and GWAS of leaf size using SNP developed by genotyping-by-sequencing. PeerJ, 8:e8572.
doi: 10.7717/peerj.8572 URL |
| [24] |
Niu S, Song Q, Koiwa H, Qiao D, Zhao D, Chen Z, Liu X, Wen X. 2019. Genetic diversity,linkage disequilibrium,and population structure analysis of the tea plant(Camellia sinensis)from an origin center,Guizhou plateau,using genomewide SNPs developed by genotyping-byse quencing. BMC Plant Biology, 19:328.
doi: 10.1186/s12870-019-1917-5 URL |
| [25] |
Price A L, Patterson N J, Plenge R M, Weinblatt M E, Shadick N A, Reich D. 2006. Principal components analysis corrects for stratification in genome-wide association studies. Nature Genetics, 38:904-909.
doi: 10.1038/ng1847 pmid: 16862161 |
| [26] | Sealy J R. 1958. A revision of the genus Camellia. London:The Royal Horticultural Society. |
| [27] | Sun Meihua. 2020. Gene analysis of fasciated locus regulating the locule number of tomato[Ph. D. Dissertation]. Shenyang: Shenyang Agricultural University. (in Chinese) |
| 孙美华. 2020. 调控番茄心室数量的fasciated位点基因解析[博士论文]. 沈阳: 沈阳农业大学. | |
| [28] |
Sun X, Liu D, Zhang X, Li W, Liu H, Hong W, Jiang C, Guan N, Ma C, Zeng H, Xu C, Song J, Huang L, Wang C, Shi J, Wang R, Zheng X, Lu C, Wang X, Zheng H. 2013. SLAF-seq:an efficient method of large-scale de novo SNP discovery and genotyping using high-throughput sequencing. PLoS ONE, 8:e58700.
doi: 10.1371/journal.pone.0058700 URL |
| [29] | Suzuki T, Miyoshi N, Hayakawa S, Imai S, Isemura M, Nakamura Y. 2016. Health benefits of tea consumption. Beverage Impacts on Health and Nutrition. Germany: Springer International Publishing. |
| [30] |
Wang R J, Gao X F, Yang J, Kong X R. 2019. Genome-wide association study to identify favorable SNP allelic variations and candidate genes that control the timing of spring bud flush of tea(Camellia sinensis)using SLAF-seq. Journal of Agricultural and Food Chemistry, 67 (37):10380-10391.
doi: 10.1021/acs.jafc.9b03330 URL |
| [31] |
Wang X, Feng H, Chang Y, Ma C, Wang L, Hao X, Li A l, Cheng H, Wang L, Cui P, Jin J, Wang X, Wei K, Ai C, Zhao S, Wu Z, Li Y, Liu B, Wang G-D, Chen L, Ruan J, Yang Y. 2020. Population sequencing enhances understanding of tea plant evolution. Nature Communications, 11:4447.
doi: 10.1038/s41467-020-18228-8 pmid: 32895382 |
| [32] |
Xia E, Tong W, Hou Y, An Y, Chen L, Wu Q, Liu Y, Yu J, Li F, Li R, Li P, Zhao H, Ge R, Huang J, Mallano A I, Zhang Y, Liu S, Deng W, Song C, Zhang Z, Zhao J, Wei S, Zhang Z, Xia T, Wei C, Wan X. 2020. The reference genome of tea plant and resequencing of 81 diverse accessions provide insights into its genome evolution and adaptation. Molecular Plant, 13:1013-1026.
doi: S1674-2052(20)30134-9 pmid: 32353625 |
| [33] |
Xia E, Zhang H, Sheng J, Li K, Zhang Q J, Kim C, Zhang Y, Liu Y, Zhu T, Li W, Huang H, Tong Y, Nan H, Shi C, Shi C, Jiang J, Mao S, Jiao J, Zhang D, Zhao Y, Zhao Y, Zhang L, Liu Y, Liu B, Yu Y, Shao S, Ni D, Eichler E, Gao L. 2017. The tea tree genome provides insights into tea flavor and independent evolution of caffeine biosynthesis. Molecular Plant, 10:866-877.
doi: 10.1016/j.molp.2017.04.002 URL |
| [34] | Xiong Jihua. 2005. Flora of Chongqing Jinyun Mountain. Chongqing: Southwest Normal University Press. (in Chinese) |
| 熊济华. 2005. 重庆缙云山植物志. 重庆: 西南师范大学出版社. | |
| [35] |
Yamashita H, Uchida T, Tanaka Y, Katai H, Nagano A J, Morita A, Ikka T. 2020. Genomic predictions and genome-wide association studies based on RAD-seq of quality-related metabolites for the genomics-assisted breeding of tea plants. Scientific Reports, 10:17480.
doi: 10.1038/s41598-020-74623-7 pmid: 33060786 |
| [36] |
Yang H, Wei C, Liu H, Wu J, Li Z, Zhang L, Jian J, Li Y, Tai Y, Zhang J, Zhang Z, Jiang C, Xia T, Wan X. 2016. Genetic divergence between Camellia sinensis and its wild relatives revealed via genome-wide SNPs from RAD sequencing. PLoS ONE, 11 (3):e0151424.
doi: 10.1371/journal.pone.0151424 URL |
| [37] | Yang Shixiong, Thinking on the taxonomy of Camellia sect. Thea. Journal of Tea Science, 41 (4):439-453. (in Chinese) |
| 杨世雄. 2021. 茶组植物的分类历史与思考. 茶叶科学, 41 (4):439-453. | |
| [38] | Zhang Hongda. 1981a. Thea-a section of beveragial tea-trees of the genus Camellia. Acta Scientiarum Naturalium Universitatis Sunyatseni,(1):87-99. (in Chinese) |
| 张宏达. 1981a. 茶树的系统分类. 中山大学学报(自然科学版),(1):87-99. | |
| [39] | Zhang Hongda. 1981b. A taxonomy of the genus Camellia. Guangzhou: Sun Yat-sen University Press. (in Chinese) |
| 张宏达. 1981b. 山茶属植物的系统研究. 广州: 中山大学出版社. | |
| [40] | Zhang Hongda. 1990. New species of Chinese Theaceae. Acta Scientiarum Naturalium Universitatis Sunyatseni, 29 (2):85-93. (in Chinese) |
| 张宏达. 1990. 中国山茶科植物新种(英文). 中山大学学报(自然科学版), 29 (2):85-93. | |
| [41] | Zhang Hongda. 1998. Flora Reipublicae Popularis Sinicae. Beijing:Science Press. (in Chinese) |
| 张宏达. 1998. 中国植物志. 北京: 科学出版社. | |
| [42] |
Zhang W, Zhang Y, Qiu H, Guo Y, Wan H, Zhang X, Scossa F, Alseekh S, Zhang Q, Wang P, Xu L, Schmidt M H-W, Jia X, Li D, Zhu A, Guo F, Chen W, Ni D, Usadel B, Fernie A R, Wen W. 2020. Genome assembly of wild tea tree DASZ reveals pedigree and selection history of tea varieties. Nature Communications, 11:3719.
doi: 10.1038/s41467-020-17498-6 pmid: 32709943 |
| [43] | Zhou Meng, Li Youyong, Sun Xuemei, Wang Jiajin, Xie Jin, Cheng Hao, Wang Yungang, Liu Benying. 2013. Genetic diversity analysis of wild tea plants in Yunnan based on EST-SSR markers. Jiangsu Agricultural Sciences, 41 (12):22-27. (in Chinese) |
| 周萌, 李友勇, 孙雪梅, 王家金, 谢瑾, 成浩, 汪云刚, 刘本英. 2013. 基于EST-SSR标记的云南野生茶树遗传多样性分析. 江苏农业科学, 41 (12):22-27. | |
| [44] |
Zúñiga-Mayo V M, Gómez-Felipe A, Herrera-Ubaldo H, Folter S d. 2019. Gynoecium development:networks in Arabidopsis and beyond. Journal of Experimental Botany, 70:1447-1460.
doi: 10.1093/jxb/erz026 pmid: 30715461 |
| [1] | ZHANG Meng, SHAN Yuying, YANG Yebo, ZHAI Feifei, WANG Zhaoshan, JU Guansheng, SUN Zhenyuan, LI Zhenjian. AFLP Analysis of Genetic Resources of Dendrobium from China [J]. Acta Horticulturae Sinica, 2022, 49(6): 1339-1350. |
| [2] | WANG Fusheng, LIU Xiaona, XU Yuanyuan, LIU Xiaofeng, ZHU Shiping, ZHAO Xiaochun. Cloning and Functional Analysis of Squalene Synthase Gene in Citrus [J]. Acta Horticulturae Sinica, 2021, 48(9): 1641-1652. |
| [3] | WANG Xin, LI Mingyang, TIAN Lin, LIU Dongyun. ISSR and rDNA-ITS Sequence Analysis of the Genetic Relationship of Clematis in Hebei Province [J]. Acta Horticulturae Sinica, 2021, 48(9): 1755-1767. |
| [4] | TANG Haixia, GAO Rui, WANG Zhongtang, ZHANG Qiong. High-density Genetic Linkage Map Reconstruction in Jujube Using SNP Markers [J]. Acta Horticulturae Sinica, 2021, 48(11): 2275-2285. |
| [5] | XIONG Yan1,2,ZHANG Jinzhu1,*,DONG Jie1,WANG Li1,GAO Peng1,JIANG Wan1,and CHE Daidi1,*. A Review of Reduced-representation Genome Sequencing Technique and Its Applications in Ornamental Plants [J]. ACTA HORTICULTURAE SINICA, 2020, 47(6): 1194-1202. |
| [6] | GAO Yuan, WANG Dajiang, WANG Kun, CONG Peihua, LI Lianwen, and PIAO Jicheng. Analysis of Genetic Diversity of Apple Germplasms of Malus Using SLAF-seq Technology [J]. Acta Horticulturae Sinica, 2020, 47(10): 1869-1882. |
| [7] | LI Beibei1,*,ZHANG Heng1,2,*,JIANG Jianfu1,ZHANG Ying1,FAN Xiucai1,FANG Jinggui2,**,and LIU Chonghuai1,**. Analysis of Genetic Diversity of Grape Germplasms Using SLAF-seq Technology [J]. ACTA HORTICULTURAE SINICA, 2019, 46(11): 2109-2118. |
| [8] | ZHANG Xiaoli,SHAN Xiaozheng,JIANG Hanmin,WEN Zhenghua,LIU Lili,YAO Xingwei,NIU Guobao,and SUN Deling*. Research Progress and Prospect of Wild Cabbage [J]. ACTA HORTICULTURAE SINICA, 2018, 45(9): 1715-1726. |
| [9] | FANG Qian,ZHANG Yuanyuan,YANG Yuting,HUANG Miaomiao,FU Qiaoli,ZHOU Huisha,CHEN Wenrong,ZONG Yu*,and GUO Weidong*. SSR Mining and Polymorphism Analysis in Leaf Transcriptome of Blueberry [J]. ACTA HORTICULTURAE SINICA, 2018, 45(7): 1359-1370. |
| [10] | LIU Lei,DENG Xuebin,FENG Jingjing,WANG Jing,SUN Xiaorong,SHU Jinshuai,and LI Junming*. Exploring Variant Sites Associated with Ve-1,I-2,Mi-1 and Pto Genes in Processing Tomato by Genome-wide Association Analysis [J]. ACTA HORTICULTURAE SINICA, 2018, 45(6): 1089-1100. |
| [11] | . Development of Retrotransposon-based Insertion Polymorphism Molecular Marker and Cultivar Identification of Blueberry [J]. ACTA HORTICULTURAE SINICA, 2018, 45(4): 753-763. |
| [12] | WANG Liyuan,SUN Peng,ZHANG Jiajia,FU Jianmin*,DIAO Songfeng,HAN Weijuan,SUO Yujing,and ZHANG Yue. Survey of Wild Male Germplasm Resources of Diospyros kaki and Their Genetic Diversity Analysis [J]. ACTA HORTICULTURAE SINICA, 2018, 45(2): 261-278. |
| [13] | QI Dan1,*,CHANG Yaojun1,*,CAO Yufen1,**,HU Hongju2,TIAN Luming1,DONG Xingguang1,ZHANG Ying1,HUO Hongliang1,XU Jiayu1,ZHANG Xiaoshuang1,and LIU Chao1. Genetic Diversity and Phylogenetics of Pear(Pyrus L.)Germplasm Resources from South China Revealed by Chloroplast DNA [J]. ACTA HORTICULTURAE SINICA, 2018, 45(12): 2308-2320. |
| [14] | WANG Qingbiao,ZHANG Li*,WEN Changlong,and YANG Jingjing. Development and Application of High-throughput SNP Markers for Ogura-CMS Fertility Restorer Gene in Radish [J]. ACTA HORTICULTURAE SINICA, 2017, 44(7): 1309-1318. |
| [15] | LIU Yuanxia1,LAN Jinhao2,BAI Suhua3,SUN Xiaohong3,LIU Chunxiao1,ZHANG Yugang1,and DAI Hongyi1,*. Screening of SNP and InDel Markers to Glomerella Leaf Spot Resistance Gene Locus in Apple Using HRM Technology [J]. ACTA HORTICULTURAE SINICA, 2017, 44(2): 215-222. |
| Viewed | ||||||
|
Full text |
|
|||||
|
Abstract |
|
|||||
Copyright © 2012 Acta Horticulturae Sinica 京ICP备10030308号-2 国际联网备案号 11010802023439
Tel: 010-82109523 E-Mail: yuanyixuebao@126.com
Support by: Beijing Magtech Co.Ltd